Abstract
The tightly regulated production of distinct erythrocyte protein 4.1R isoforms involves differential splicing of 3 mutually exclusive first exons (1A, 1B, 1C) to the alternative 3′ splice sites (ss) of exon 2′/2. Here, we demonstrate that exon 1 and 2′/2 splicing diversity is regulated by a transcription-coupled splicing mechanism. We also implicate distinctive regulatory elements that promote the splicing of exon 1A to the distal 3′ ss and exon 1B to the proximal 3′ ss in murine erythroleukemia cells. A hybrid minigene driven by cytomegalovirus promoter mimicked 1B-promoter–driven splicing patterns but differed from 1A-promoter–driven splicing patterns, suggesting that promoter identity affects exon 2′/2 splicing. Furthermore, splicing factor SF2/ASF ultraviolet (UV) cross-linked to the exon 2′/2 junction CAGAGAA, a sequence that overlaps the distal U2AF35-binding 3′ ss. Consequently, depletion of SF2/ASF allowed exon 1B to splice to the distal 3′ ss but had no effect on exon 1A splicing. These findings identify for the first time that an SF2/ASF binding site also can serve as a 3′ ss in a transcript-dependent manner. Taken together, our results suggest that 4.1R gene expression involves transcriptional regulation coupled with a complex splicing regulatory network.
Introduction
The regulated production of mRNA in higher eukaryotes requires several multicomponent cellular processes that include initiation of transcription, 5′ capping, splicing, polyadenylation, and export of mature mRNA to the cytoplasm. Although each process catalyzes distinct reactions, each also interfaces physically and functionally with the others.1-4 In this regard, coupling processes to others and their substrates could conceivably enhance the efficiency and accuracy of gene expression.4
A large number of eukaryotic genes contain multiple promoters5,6 ; the expression of alternative first exons is a common mechanism for achieving complex patterns of tissue-specific expression.7 Transcripts thus derived can either encode different amino termini with diverse functional properties8 or differ only in their 5′ noncoding regions while sharing the same open reading frames.5,6 However, in some cases, the first exons are noncoding, they differentially splice to alternative downstream splice sites, and they determine distinct amino-termini sequences.9,10 Alternative promoter usage also can modify alternative splicing of internal exons,11 thereby generating distinct isoforms with distinct physiologic functions, eg, the acetylcholinesterase (ACHE),12 connexin 43 (cx43),13 and bcl-X14 genes.
Two models have been proposed to explain promoter-specific alternative splicing. The first is the recruitment model, in which promoter-dependent recruitment of regulatory splicing factors to cognate enhancers or silencers modifies downstream splice site selection.15,16 The second is the kinetic coupling model, in which promoters control splice-site selection via the regulation of RNA polymerase II (pol II) elongation or processivity.17,18 In this case, the timing of presentation of the splicing regulatory elements contained in the nascent transcripts would determine splice site selection.11 Promoter-dependent regulation of alternative splicing contributes yet another layer of complexity to gene expression mechanisms.
Erythrocyte protein 4.1R (4.1R) is an example of how complex transcription and splicing mechanisms can produce diverse isoforms.10,19,20 Two major erythroid-differentiation stage-specific splicing events, an early event excluding exon 2′ and a late event including exon 16, lead to production of the erythrocyte spectrin-actin-binding 80-kDa isoform.21,22 The regulation of exon 16 expression is well studied,23-27 but the regulation of exon 2′ splicing in the context of erythroid cells remains poorly understood. Three mutually exclusive first exons, 1A, 1B, and 1C, are transcribed from their respective promoters and differentially splice to 2 alternative 3′ splice site (ss) at the 5′ end of exon 2′/2.10 Exons 1B and 1C splice to the proximal 3′ ss, include exon 2′, and encode high-molecular-weight isoforms (135 kDa) by allowing translation from an upstream start site (AUG-1) in exon 2′. Exon 1A splices to the distal 3′ ss, excludes exon 2′, and encodes low-molecular-weight “erythroid” (80-kDa) isoforms by translation from a start site in exon 4 (AUG-2). Splicing regulation at the 3′ end of the intron depends on the branchpoint followed by a polypyrimidine (Py)-rich sequence and a conserved AG dinucleotide to define the 3′ ss. The splicing factor U2AF65 binds to the Py tract,28 whereas U2AF35 contacts the AG dinucleotide29,30 for recruitment of U2 snRNP to pre-mRNAs. These processes are essential for splicing of AG-dependent substrates. How the exon 2′ and exon 2 3′ ss are differentially selected for 4.1R in erythroid cells has been an unresolved issue.
In this study, we investigated how the expression of exon 2′/2 in 4.1R might be regulated in murine erythroleukemia cells (MELCs), which have been used extensively as an in vitro model for erythroid differentiation.31,32 We demonstrated that exon 2′/2 selection is regulated by the coordinated actions of transcription and splicing. We further showed that the distal 3′ ss—in a transcription-dependent manner—also can provide a binding site for a splicing factor SF2/ASF. The differential usage of this site (ie, 3′ splice site vs SF2/ASF binding site) appears to depend on the promoter used; this governs the differential binding of splicing factors SF2/ASF and U2AF35 at the distal 3′ ss. These results provide the first direct evidence of a link between transcription and a complex splicing regulatory network in 4.1R gene expression.
Methods
Genomic DNA constructs
Genomic DNAs were isolated from MELCs by use of the standard protocol. All DNA constructs were made with standard cloning procedures and confirmed by sequencing.
4.1R minigenes controlled by a CMV promoter
Two-step polymerase chain reaction (PCR) was used to generate a series of murine-derived minigenes under cytomegalovirus (CMV) promoter control. These minigenes contained exon 1 and 200, 300, or 400 base pairs (bp) of its downstream intronic sequences joined with exon 2′/2 and equal lengths of its upstream intronic sequences. Two segments of mouse sequences, one for exon 1A and its downstream intron (Table 1, CMV minigenes M1A-S and M1A 200-As/M1A 300-As/M1A 400-As) and the other for exon 2′/2 and its upstream intron (Table 1, CMV minigenes M1 200-S/M1 300-S/M1 400-S and M2-As), were amplified by the use of indicated primers. These 2 halves with overlapping complementary sequences were annealed and amplified by a second-step PCR to obtain the entire M1A insert and cloned into pcDNA3 to generate the M1A minigene. M1B and M1C minigenes were constructed similarly by use of their respective primers.
Minigene under its respective promoter
To generate minigenes driven by the mouse 1A promoter, primers (Table 1, promoter constructs, pM1A-S, and CMV minigenes, M1A 400-As) were used to amplify the first segment spanning the DNA sequences 505 bp immediately preceding the most upstream exon 1A transcription start site to the 400 bp of intronic sequence downstream of exon 1A. The fragments were cloned into a pGL3-basic vector (Promega Inc) containing exon 2′/2 and 400 bp of its associated upstream intronic sequence. Mouse 1B and 1C promoter minigenes were similarly constructed.
UV cross-linking and SF2/ASF-shRNA constructs
Oligonucleotides (WT-2′UV-S or mutants Mut1-2′UV-S and Mut2-2′UV-S) were amplified with primers 2UV-S and 2′UV-As (Table 1, UV cross-linking oligos) and cloned into pCDNA3. RNAs were transcribed with T7 RNA polymerase (Novagen) in the presence of a Ribo m7G Cap Analog (Promega) and α-32P-UTP (Perkin Elmer). Competitor RNAs were synthesized without the addition of α-32P-UTP. The construction of SF2/ASF-specific shRNA expression vectors with target sequences sh-84, sh-584, and sh-726 (Table 1, SF2/ASF-shRNA constructs) was performed as described.33
Cell culture, transfection, and retroviral transduction
Purification and culture of human progenitor CD34+ cells to terminal erythroid differentiation was performed as described.34,35 MELC cells were cultured in Dulbecco modified Eagle medium with 10% fetal bovine serum. 5,6-Dichloro-1-b-D-ribofuranosylbenzimidazole (DRB) was added to cells at 50 μmol/L, 75 μmol/L, or 100 μmol/L for 24 hours before the RNA samples were collected. Cells were transfected with Lipofectamine 2000 (Invitrogen). A pool of stable transfectants was selected with 800 μg/mL G-418 or 2 μg/mL puromycin after cotransfection with pCDNA3- or pGL3-minigene–containing constructs and pBabe-puro (Addgene) into MELCs. Vector-alone stable transfectants served as controls. Four transfections were performed each experiment, and each experiment was repeated 3 times. Four pools were tested in each experiment.
Retroviral shRNA vectors were propagated in PirPlus DH10βF′DOT competent Escherichia coli and packed into retroviruses by the use of LinX viral packaging cell line (Open Biosystems). A total of 0.5 mL of shRNA retrovirus-containing medium was added to 1.5 mL of fresh medium with 0.75 × 106 MELC for infection. For each construct, 3 infections were performed per experiment, and each experiment was repeated 3 times.
5′ RACE and RT-PCR analyses
MELC mRNA was isolated with the Micro-FastTrack 2.0 Kit (Invitrogen), followed by DNase treatment. 5′ Rapid amplification of cDNA ends (5′ RACE) was performed by use of GeneRacer Kit (Invitrogen). For mapping of the transcription start sites upstream of exon 1A, M2-As was used for RT, and sense (GeneRace 5′ Primer and 5′ Nested Primer) and anti-sense primers (5′Race-MA1-As and 5′Race-MA2-As; Table 1, 5′ RACE) were used for nested PCR. The start sites for exons 1B and 1C were similarly mapped by use of their respective primer sets. Reverse transcription (RT)–PCR analysis of splicing products was performed with a limiting-cycle amplification protocol that obtains the PCR products within its linear range.27 RNAs were reverse-transcribed by SP6 for CMV-minigene, Luc RRV for native promoter-constructs, and mEx2-RT for mouse endogenous mRNAs (Table 1, Exon 2′/2 splicing analysis). PCRs were performed with M1A-S, M1B-S, or M1C-S and M2-As for pCDNA3-based minigenes (Table 1, CMV minigenes) and with the same sense primers and RRV2-As or M2-As for pGL3-based minigenes (Table 1, Exon 2′/2 splicing analysis). Similarly, CD34+ RNAs were reverse transcribed with HEx2-RT and PCR-amplified with primer sets H1A-S, H1B-S, or H1C-S and HEx2-As (Table 1, Exon 2′/2 splicing analysis). Spliced products were fractionated on 10% acrylamide gels and quantified by the use of analysis software from ChemiImager 5500 system (Alpha Innotech Co). Southern blot hybridization was performed with a digoxigenin-labeled Ex 2′ (Table 2, Southern probe) or M2-As.
Human exon 1A and 1C real-time PCR was performed on an Applied Biosystems 7900HT Real-Time PCR system by the use of Applied Biosystems SYBR Green Master Mix (Applied Biosystems). cDNA was generated with an oligo-dT primer. Primers HEx1A-S and Hex1A-As were used for exon 1A and HEx1C-S and Hex1C-As for exon 1C (Table 1, real-time PCR). The assays were performed in triplicate, and the obtained cycle threshold values were used to calculate the amount of amplified DNA with a standard curve that was calculated for each primer set by the use of serial dilutions (1:5). The ratio of Ex1A/Ex1C was then calculated with the approximated amounts of DNA.
Chromatin immunoprecipitation and immunoprecipitation of chromatin–RNA complexes
Chromatin immunoprecipitation (ChIP) was performed as described.36 Cells were cross-linked with formaldehyde, washed with phosphate-buffered saline, and resuspended in sodium dodecyl sulfate lysis buffer. Extracts were then sonicated to obtain approximately 500-nt chromatin fragments. A 50-μL sample was saved as input DNA. The remaining chromatin solution was diluted 1:10 in ChIP dilution buffer and incubated overnight with either 10 μg of mouse anti-SF2/ASF antibodies (#32-4500; Zymed) or nonimmune mouse IgG. Complexes were immunoprecipitated with protein G Sepharose beads and washed.36 The immune complexes were eluted, cross-links were reversed, digested with proteinase K, and the DNA extracted by use of the Qiagen PCR purification kit. Retrieved DNA templates and the saved input were analyzed by PCR by the use of primer sets M1 200-S and M2-As for the region between intron 1 and exon 2 (In/Ex2), Ex2′ and M2-As for exon 2′ and 2 (Ex2′/Ex2), and MEx2-S and M2-As for the region within exon 2 (Ex2/Ex2). Primer sets up1/up2 and up3/up4 were used to amplify the regions upstream of 1A promoter (Table 2, ChIP/ChRIP analysis).
For immunoprecipitation of chromatin–RNA (ChRIP), cells were cross-linked, harvested, and resuspended in RIPA buffer. Extract preparation, immunoprecipitation with rabbit anti-AcH4 Ab (#06-598, anti–acetyl-Histone H4; Upstate), rabbit anti–H3K4me1 antibody (ab8895, Abcam Inc), or nonimmune rabbit IgG, and washing were performed as described for ChIP. Chromatin was treated with proteinase K, eluted in 1% (wt/vol) SDS in Tris-ethylene diamine tetraacetic acid buffer, and the cross links were reversed. Recovered RNA segments were treated with DNase I and analyzed by RT-PCR for the presence of either unspliced (Table 1, MEx2-RT for RT and M1 200-S and M2-As for PCR) or spliced (Table 2, ChIP/ChRIP analysis, mEx17-RT for RT and Ex13-S and Ex17-As for PCR) 4.1R transcripts. The nature of each band was verified by Southern hybridization or by sequencing.
UV cross-linking assay
The purification of nuclear extracts from HeLa cells was performed as described.37 A total of 20 ng of α-32P–labeled RNA and 7.5 μL of nuclear extracts or 0.5 μg of purified SF2/ASF recombinant protein (Protein One) were incubated in a 25-μL splicing reaction26 at 30°C for 20 minutes. For competition assays, 25-fold molar excesses of unlabeled competitor RNAs were added to the preincubation reaction for 10 minutes before the addition of α-32P–labeled RNA. The samples were UV cross-linked, fractionated in a 10% denaturing polyacrylamide gel electrophoresis gel, and visualized by autoradiography as described.26
Western blot
SF2/ASF proteins were detected with anti-SF2 antibody (Zymed Lab) by use of the ECL detection kit (Amersham Pharmacia). Anti–β-actin antibody (Sigma) served as a loading control.
Results
Differential expression of first exons in human and mouse tissues/cells
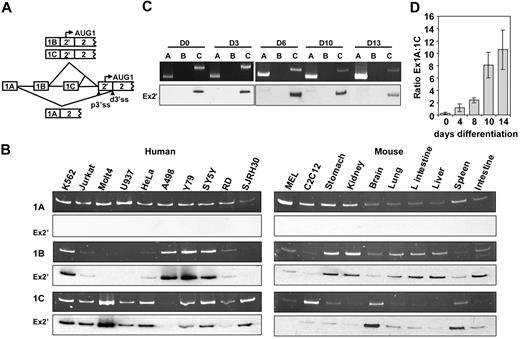
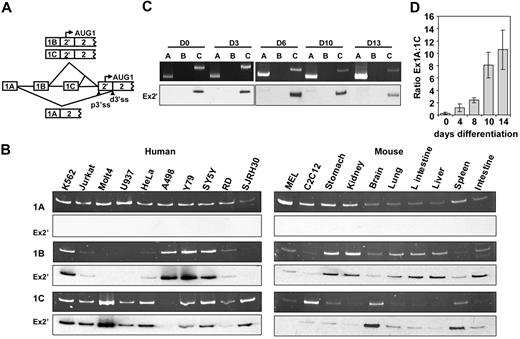
We first mapped the extreme 5′ untranslated region of mouse 4.1R mRNA. Although published data10 indicate that no other exons exist upstream of exons 1A, 1B, and 1C, it was not yet determined whether a true 5′ cap site was present in these exons. By the use of RNA ligase–mediated 5′ RACE, we found that exon 1A, 1B, and 1C were the most terminal 5′ exons. The 1B transcription start site is located 412nt upstream of the 3′ end of exon 1B, whereas both 1A and 1C have several transcription initiation sites (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Comparison of these start sites with the GenBank EST database suggests that these sites are likely to be representative of the major sites in MELCs. However, there may be other sites present in other tissues or cell types.
We then determined the relative expression levels of first exons in human and mouse cells (supplemental Table 1). In humans, exon 1A was expressed in all cell lines examined (Figure 1B). The expression levels of exons 1B and 1C were reciprocal with the exception of K562 and neuronal cells (Y79 and SY5Y), which exhibited equal 1B and 1C expression (Figure 1B left panel). In mouse, exon 1A also was detected in all cell lines and tissues examined, whereas exons 1B and 1C were expressed in complete reciprocal fashion (Figure 1B). Southern hybridization and sequencing verified that exon 1A spliced to the distal 3′ ss and excluded exon 2′, whereas both exon 1B and 1C spliced to the proximal 3′ ss and included exon 2′ (Figure 1B, Ex2′). Thus, exon 2′/2 expression is modulated at the transcriptional level, depending on which promoter—1A, 1B, or 1C—is selected.
Analyses of first exon expression. (A) Schematic representation of the splicing patterns from exon 1 to its downstream 3′ ss. p3′ss indicates proximal 3′ss; d3′ss, distal 3′ss. AUG-1 located within exon 2′. (B) First exon expression in human and mouse cell lines and tissues. A common anti-sense primer annealing to exon 2 and variable forward primers specific to each of the first exons were used for PCR on cDNA synthesized by use of a downstream exon 2 primer. The top panels are PCR; the bottom panels are Southern hybridization with exon 2′ as a probe (Ex2′). (C) First exon expression during cultured CD34+ erythroid differentiation by use of the same RT-PCR strategies. The top panels are PCR products; the bottom panels are Southern-blot hybridizations that use exon 2′ as a probe (Ex2′). (D) Real-time PCR analysis of ratios of 1A:1C expression during CD34+ differentiation. 0, 4, 8, 10, and 14 indicate days of differentiation.
Analyses of first exon expression. (A) Schematic representation of the splicing patterns from exon 1 to its downstream 3′ ss. p3′ss indicates proximal 3′ss; d3′ss, distal 3′ss. AUG-1 located within exon 2′. (B) First exon expression in human and mouse cell lines and tissues. A common anti-sense primer annealing to exon 2 and variable forward primers specific to each of the first exons were used for PCR on cDNA synthesized by use of a downstream exon 2 primer. The top panels are PCR; the bottom panels are Southern hybridization with exon 2′ as a probe (Ex2′). (C) First exon expression during cultured CD34+ erythroid differentiation by use of the same RT-PCR strategies. The top panels are PCR products; the bottom panels are Southern-blot hybridizations that use exon 2′ as a probe (Ex2′). (D) Real-time PCR analysis of ratios of 1A:1C expression during CD34+ differentiation. 0, 4, 8, 10, and 14 indicate days of differentiation.
We also measured the relative expression levels of first exons during human CD34+ erythroid-cell differentiation. Importantly, despite incomplete synchronization, the majority of the cells were in the same differentiation stage (supplemental Figure 2). 4.1R RNAs from differentiating cells contained exons 1A and 1C but not 1B (Figure 1C). Furthermore, exon 1A-containing mRNAs skipped exon 2′, whereas exon 1C mRNAs retained 2′ (Figure 1C, Ex 2′). A reciprocal expression of exon 1A and 1C was observed during CD34+ erythroid differentiation. The ratio of 1A/1C increased progressively as cells differentiated when analyzed by real-time PCR: 1A represents 23% of total 4.1R mRNA in day 0, 53% in day 4, 70% in day 8, 89% in day 10, and 91% in day 14 (Figure 1D). Exon 1C is largely used in early cells whereas exon 1A is mostly used in late erythroid cells. The erythroid differentiation stage-specific switch from the 135- to the 80-kDa form of human 4.1R is thus governed by a switch from use of the exon 1C promoter to use of the exon 1A promoter.
Cotranscriptional pre-mRNA splicing event in 4.1R gene expression
4.1R gene expression has been suggested to be regulated by a transcription-coupled splicing mechanism.10 We examined a cotranscriptional pre-mRNA splicing event via ChIP and ChRIP assays.
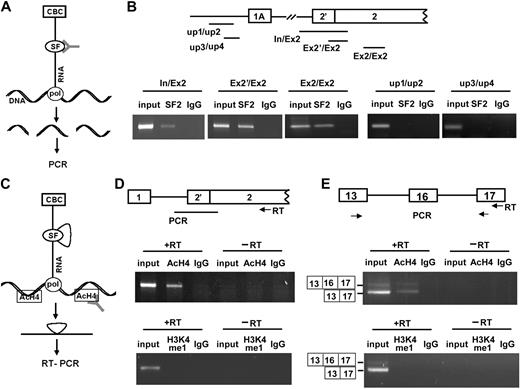
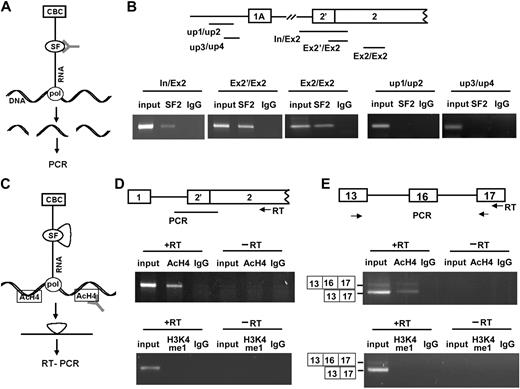
We performed ChIP to detect the recruitment of RNA-binding proteins to endogenous transcription units in MELCs, expecting ChIP—by use of an antibody against a potential splicing factor (Figure 2A, SF)—to pull down the chromatin regions to which the factor is attached through nascent transcripts (Figure 2A, RNA) and pol II (Figure 2A, pol). We26 showed that SF2/ASF binds to exon 16 of 4.1R. Analyses of 4.1R sequences by the use of ESEFinder (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process = home) also revealed more than 120 putative SF2/ASF binding sites situated on 4.1R. These findings implicate SF2/ASF as important in 4.1R mRNA splicing. Thus, we investigated cotranscriptional accumulation of SF2/ASF by use of an anti-SF2/ASF antibody. DNA templates retrieved by ChIP were analyzed by PCR with the primers that detect intron 1-exon 2, exon 2′-exon 2, or exon 2 (Figure 2B, In/Ex2, Ex2′/Ex2, Ex2/Ex2). 4.1R sequences were detected in both control input and anti-SF2/ASF precipitates (Figure 2B, In/Ex2, Ex2′/Ex2, Ex2/Ex2, lanes input and SF2) but not in control IgG precipitates (Figure 2B, lanes IgG). This finding strongly suggests that SF2/ASF associates with nascent 4.1R transcripts.
Cotranscriptional pre-mRNA splicing events detected by ChIP and ChRIP assays. (A) Schematic diagram of ChIP (adapted with permission from Listerman et al36 ); antibodies against splicing factors (SF) pull down chromatin regions attached to splicing factors through the nascent RNA and pol II (pol). CBC indicates cap-binding complex. (B) 4.1R DNA sequences detected in ChIP assays by the use of anti-SF2/ASF Ab. (Top panel) the 4.1R gene sequences amplified by indicated primer sets comprises the regions spanning intron upstream of exon 2′ and exon 2 (In/Ex2), exon 2′ and exon 2 (Ex2′/Ex2), and within exon 2 (Ex2/Ex2). DNA regions upstream of 1A promoter were amplified with primer sets up1/up2 and up3/up4. (Bottom panel) PCR. (C) Schematic diagram of ChRIP (adapted with permission from Listerman et al36 ); antibodies against AcH4 pull down the nascent RNA attached to the active chromatin through pol II. (D) Analyses of 4.1R pre-mRNA transcripts detected in anti-AcH4 (active chromatin) or anti-H3K4me1 (silenced chromatin) antibody precipitates. (Top panel) the region of primer annealing for RT-PCR is region. (Middle panel) 4.1R pre-mRNA spanning the region between the intron upstream of exon 2′ and exon 2 detected in input lysate, anti-AcH4, or IgG precipitates in the presence or absence of RT reaction. (Bottom panel) 4.1R pre-mRNA spanning the same region detected in input lysate, anti-H3K4me1, or IgG precipitates in the presence or absence of RT reaction. (E) 4.1R mRNA detected in anti-AcH4 or anti-H3K4me1 precipitates. (Top panel) the primer at exon 17 was used for RT and amplified by primer sets located at exons 13 and 17. (Middle panel) amplified products with or without exon 16 detected in anti-AcH4 precipitates in the presence or absence of RT reaction. Input lysate and IgG precipitates served as controls. (Bottom panel) amplified products with or without exon 16 detected in anti-H3K4me1 precipitates in the presence or absence of RT reaction. Input lysate and IgG precipitates served as controls.
Cotranscriptional pre-mRNA splicing events detected by ChIP and ChRIP assays. (A) Schematic diagram of ChIP (adapted with permission from Listerman et al36 ); antibodies against splicing factors (SF) pull down chromatin regions attached to splicing factors through the nascent RNA and pol II (pol). CBC indicates cap-binding complex. (B) 4.1R DNA sequences detected in ChIP assays by the use of anti-SF2/ASF Ab. (Top panel) the 4.1R gene sequences amplified by indicated primer sets comprises the regions spanning intron upstream of exon 2′ and exon 2 (In/Ex2), exon 2′ and exon 2 (Ex2′/Ex2), and within exon 2 (Ex2/Ex2). DNA regions upstream of 1A promoter were amplified with primer sets up1/up2 and up3/up4. (Bottom panel) PCR. (C) Schematic diagram of ChRIP (adapted with permission from Listerman et al36 ); antibodies against AcH4 pull down the nascent RNA attached to the active chromatin through pol II. (D) Analyses of 4.1R pre-mRNA transcripts detected in anti-AcH4 (active chromatin) or anti-H3K4me1 (silenced chromatin) antibody precipitates. (Top panel) the region of primer annealing for RT-PCR is region. (Middle panel) 4.1R pre-mRNA spanning the region between the intron upstream of exon 2′ and exon 2 detected in input lysate, anti-AcH4, or IgG precipitates in the presence or absence of RT reaction. (Bottom panel) 4.1R pre-mRNA spanning the same region detected in input lysate, anti-H3K4me1, or IgG precipitates in the presence or absence of RT reaction. (E) 4.1R mRNA detected in anti-AcH4 or anti-H3K4me1 precipitates. (Top panel) the primer at exon 17 was used for RT and amplified by primer sets located at exons 13 and 17. (Middle panel) amplified products with or without exon 16 detected in anti-AcH4 precipitates in the presence or absence of RT reaction. Input lysate and IgG precipitates served as controls. (Bottom panel) amplified products with or without exon 16 detected in anti-H3K4me1 precipitates in the presence or absence of RT reaction. Input lysate and IgG precipitates served as controls.
To confirm that anti-SF2 Ab did not pull down any nonspecific DNA sequences, we amplified isolated DNA fragments by using primers up1/up2 and up3/up4, located upstream of the 1A promoter in the regions for which no corresponding mRNA sequences exist (Figure 2B, up1/up2 and up3/up4). Although both sets of primers amplified DNA fragments from the input lysates, the primers did not amplify any fragments from ChIP of anti-SF2 Abs (Figure 2B, up1/up2 and up3/up4, lanes SF2). These results validate the specificity of the ChIP assay.
The accumulation of SF2/ASF on transcriptional complexes suggests that splicing may also occur cotranscriptionally. We investigated this by using ChRIP, in which active chromatin is immunopurified with acetylated histone 4 (AcH4) and the attached nascent RNA amplified by RT-PCR (Figure 2C) by the use of primers that detect either pre-mRNA (Figure 2D) or mRNA (Figure 2E). 4.1R pre-mRNA transcripts spanning the region between intron upstream of exon 2′ and exon 2 were detected in anti-AcH4 precipitates (Figure 2D, middle panel, +RT, lane AcH4) but not in control IgG precipitates (Figure 2D, middle panel, +RT, lane IgG). No products were detected in the absence of RT (Figure 2D, middle panel, −RT). The primers located in exons 13 and 17 amplified 2 products that corresponded to the alternatively spliced products either excluding or including exon 16 (Figure 2E, middle panel, +RT, lane AcH4). The specificity of ChRIP is evidenced by the failure of IgG to precipitate 4.1R mRNA (Figure 2E, middle panel, +RT, lane IgG). Furthermore, no products were detected in the absence of RT (Figure 2E, middle panel, −RT). These results suggest that 4.1R RNA is associated with chromatin and that 4.1R pre-mRNA can undergo splicing cotranscriptionally.
Transcriptionally active genes are trimethylated, whereas inactive genes are monomethylated at K4 of histone H3.38 Antibodies that recognize monomethylated histone H3K4 in ChRIP precipitate only silenced chromatin. We verified the specificity of ChRIP by using an anti-H3K4me1 antibody. In contrast to active chromatin detected with anti-AcH4 Abs, in which both unspliced pre-mRNA and spliced 4.1R mRNA were detected (Figure 2D-E, middle panels), no 4.1R transcripts were detected in precipitates obtained with anti-H3K4me1 Abs (Figure 2D-2E, bottom panels). These results further reassure us that the ChRIP assay was specific.
Promoter identity influences the exon 2′ splicing decision
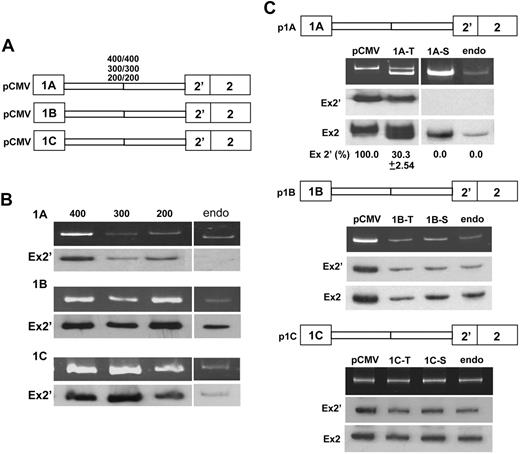
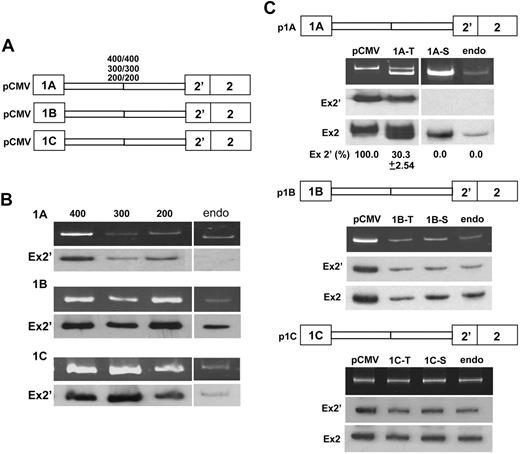
We constructed mouse exon 1 minigenes containing various intronic sequences downstream of each exon joined with equal lengths of intronic sequences upstream of exon 2′—all under the control of a CMV promoter (Figure 3A)—and measured exon 2′ expression in transfected MELCs. CMV-driven exon 1B and 1C minigenes replicated the endogenous splicing pattern with 100% exon 2′ inclusion (Figure 3B, 1B-1C, lanes 400, 300, 200, endo). CMV-driven exon 1A minigenes stimulated 100% inclusion of exon 2′ (Figure 3B, 1A, lanes 400, 300, 200), overriding the endogenous pattern of exon 1A expression in which exon 2′ is excluded (Figure 3B, 1A, lane endo).
Expression of exon 2′/2 under the control of CMV or its native promoter. (A) Minigene constructs under the control of a CMV promoter, containing exon 1 and its respective 200, 300, and 400 bp of downstream intronic sequences joined with exon 2′/2 and an equal length of its upstream intronic sequences. (B) Exon 2′ splicing patterns in minigene-transiently transfected or nontransfected MELC. Splicing products were analyzed for exon 2′ inclusion by RT-PCR by the use of a vector-specific primer for minigene or exon 2-specific primer for endogenous 4.1R. (Top panels) RT-PCR products. (Bottom panels) Southern blot with exon 2′ probe (Ex2′). Endo indicates MELC endogenous exon 2′/2 splicing pattern. (C) Exon 2′ splicing patterns in MELC transfected with minigene constructs under the control of their native promoters. RNAs were analyzed from either transiently (T) or stably (S) transfected MELCs. MELCs stably transfected with the respective minigenes under the control of CMV promoters (pCMV) or endogenous (endo) RNAs served as controls. (Top panels) RT-PCR products. (Middle panel) Southern blot with exon 2′ probe (Ex2′). (Bottom panels) Southern blot with exon 2 probe (Ex2). For each construct, 4 transfections were performed per experiment. Each experiment was repeated at least 3 times. Standard deviations are omitted in results that consistently have 0% or 100% of the same product in all 4 reproducible experiments.
Expression of exon 2′/2 under the control of CMV or its native promoter. (A) Minigene constructs under the control of a CMV promoter, containing exon 1 and its respective 200, 300, and 400 bp of downstream intronic sequences joined with exon 2′/2 and an equal length of its upstream intronic sequences. (B) Exon 2′ splicing patterns in minigene-transiently transfected or nontransfected MELC. Splicing products were analyzed for exon 2′ inclusion by RT-PCR by the use of a vector-specific primer for minigene or exon 2-specific primer for endogenous 4.1R. (Top panels) RT-PCR products. (Bottom panels) Southern blot with exon 2′ probe (Ex2′). Endo indicates MELC endogenous exon 2′/2 splicing pattern. (C) Exon 2′ splicing patterns in MELC transfected with minigene constructs under the control of their native promoters. RNAs were analyzed from either transiently (T) or stably (S) transfected MELCs. MELCs stably transfected with the respective minigenes under the control of CMV promoters (pCMV) or endogenous (endo) RNAs served as controls. (Top panels) RT-PCR products. (Middle panel) Southern blot with exon 2′ probe (Ex2′). (Bottom panels) Southern blot with exon 2 probe (Ex2). For each construct, 4 transfections were performed per experiment. Each experiment was repeated at least 3 times. Standard deviations are omitted in results that consistently have 0% or 100% of the same product in all 4 reproducible experiments.
We then examined whether native promoters would influence exon 2′/2 splicing. Parra et al10 reported that 1.5-kb regions of genomic DNA upstream of exons 1A, 1B, and 1C have robust promoter activity. Analyses of the 1.5-kb sequences upstream of mouse 4.1R exons 1A, 1B, and 1C in pGL3-basic luciferase reporter vectors yielded similar transcriptional promoter activity in MELCs (data not shown). Shortening the promoter region to ∼0.5 kb did not change promoter activity, leading us to use ∼500-bp promoters to test their effects on exon 2′/2 splicing decision. We positioned each native promoter upstream of its respective exon 1 minigene and measured exon 2′ expression in transfected MELCs. The amplified RT-PCR products were specifically derived from transcripts of the exogenously transfected minigene (supplemental Figure 3). We routinely analyzed 4 transfections, both transient and stable, from each experiment by RT-PCR and Southern blotting with exon 2′ and 2 specific probes (supplemental Figure 4). The 1A promoter caused approximately 70% of spliced products to exclude exon 2′ in transiently transfected MELCs (Figure 3C, p1A, lane 1A-T); the pattern shifted to complete exon 2′ exclusion in pooled stable transfectants (Figure 3C, p1A, lane 1A-S). In contrast, 1B and 1C promoter-driven minigenes resulted in complete exon 2′ inclusion in both transiently and stably transfected MELCs (Figure 3C, p1B and p1C). These results suggest that alternative splicing of exon 2′ is governed by promoter identity.
Pol II processivity does not determine exon 2′/2 splice-site selection
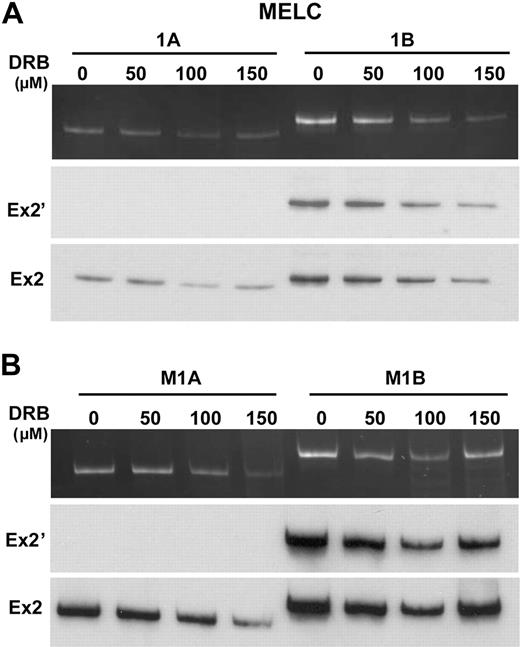
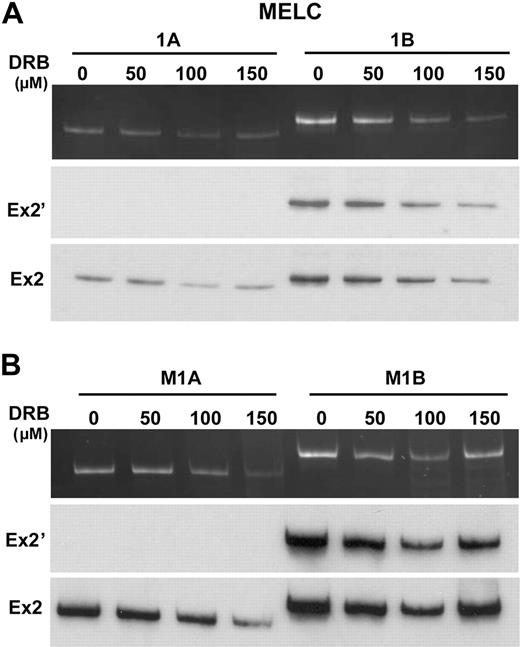
One existing model for the modulation of alternative splicing by promoters suggests that the promoter might control alternative splicing via the rate of pol II elongation (processivity).17,18 A rapidly elongating pol II would presumably favor the near simultaneous presentation of both 3′ ss to the splicing machinery, a situation in which the distal 3′ ss may out-compete the proximal 3′ ss, favoring exon 2′ exclusion. Conversely, slow pol II processivity would favor the selection of the proximal 3′ ss and inclusion of exon 2′. To test this hypothesis, we used an established protocol18 involving the treatment of cells with the potent transcription elongation inhibitor DRB. Upon inhibition of RNA pol II by DRB, SC35 relocalized from nuclear speckles to enlarged and rounder structures (supplemental Figure 5). This change in locale is a hallmark of impeded transcription elongation rate,39 and validates that DRB had the intended effect. However, when DRB-treated MELCs or minigene-transfected MELCs were analyzed for exon 2′/2 splicing, the splicing patterns of the endogenous (Figure 4A, MELC, 1A-B) or minigene RNAs (Figure 4B, M1A-B) were unaffected. Thus, the regulation of exon 2′ does not appear to be achieved through kinetic effects of pol II elongation.
Inhibition of transcription elongation did not alter 1A or 1B exon 2′/2 splicing patterns. (A) Analyses of exon 2′/2 expression in MELCs in the presence of 0, 50, 100, and 150 μmol/L of DRB. Cells were treated with DRB for 24 hours and RNAs analyzed for exon 2′ expression by the use of a common anti-sense primer located in exon 2 and sense primer located either at exon 1A or 1B. Ex2′ indicates Southern blot using exon 2′ as a probe; Ex2, Southern blot using exon 2 as a probe. (B) Analyses of exon 2′/2 expression in native promoter-driven 1A and 1B minigene stably transfected MELCs in the presence of 0, 50, 100, and 150 μmol/L of DRB. RNA collected from cells treated with DRB for 24 hours were analyzed for exon 2′ expression. Ex2′ indicates Southern blot using exon 2′ as a probe; Ex2, Southern blot using exon 2 as a probe. For each construct, 4 stable lines were performed per experiment. Each experiment was repeated at least 3 times.
Inhibition of transcription elongation did not alter 1A or 1B exon 2′/2 splicing patterns. (A) Analyses of exon 2′/2 expression in MELCs in the presence of 0, 50, 100, and 150 μmol/L of DRB. Cells were treated with DRB for 24 hours and RNAs analyzed for exon 2′ expression by the use of a common anti-sense primer located in exon 2 and sense primer located either at exon 1A or 1B. Ex2′ indicates Southern blot using exon 2′ as a probe; Ex2, Southern blot using exon 2 as a probe. (B) Analyses of exon 2′/2 expression in native promoter-driven 1A and 1B minigene stably transfected MELCs in the presence of 0, 50, 100, and 150 μmol/L of DRB. RNA collected from cells treated with DRB for 24 hours were analyzed for exon 2′ expression. Ex2′ indicates Southern blot using exon 2′ as a probe; Ex2, Southern blot using exon 2 as a probe. For each construct, 4 stable lines were performed per experiment. Each experiment was repeated at least 3 times.
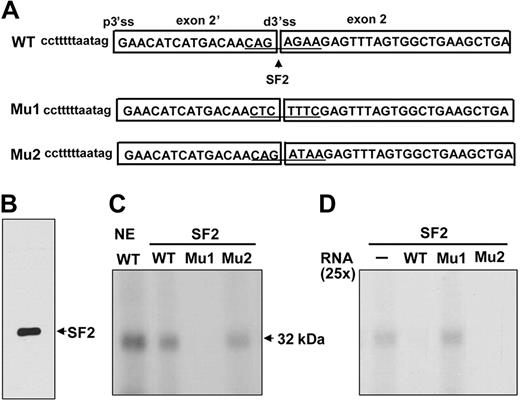
SF2/ASF binds to sequences at the junction of exon 2′ and 2
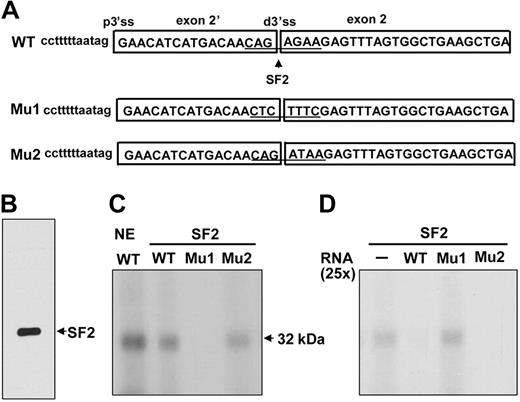
We searched for cis-elements that could potentially regulate exon 2′. The alternative 3′ ss at the exon 2′/2 junction contains a putative SF2/ASF binding site (Figure 5A, WT). Binding of SF2/ASF to an exonic splicing enhancer promotes exon definition.40 To test whether this site could bind to SF2/ASF even though it is a 3′ ss, we performed UV cross-linking assays. Wild-type RNA efficiently cross-linked with a ∼32 kDa protein from nuclear extracts and with purified SF2/ASF protein (Figure 5C, NE, lane WT; SF2, lane WT). Mutations in the SF2/ASF binding site theoretically abolishing both the 3′ ss and SF2/ASF binding (Figure 5A, Mu1) did not cross-link with SF2/ASF (Figure 5C, Mu1). The WT cross-linked band could be competed away by 25-fold molar excess of WT but not with Mu1 (Figure 5D, WT and Mu1), suggesting that SF2/ASF binds specifically to the sequences at the junction of exon 2′ and 2. This element could serve either as a distal 3′ ss or an SF2/ASF binding site. Binding of SF2/ASF to the site would block the usage of the distal 3′ ss by U2AF35, which must normally bind the conserved AG in a 3′ ss, thereby favoring use of the proximal 3′ ss.
SF2/ASF UV cross-linked to the sequences at the junction of exon 2′/2. (A) UV cross-linking templates consisting of the wild-type (WT) or mutated (Mu1, Mu2) junction of exon 2′/2 and its flanking sequences. (B) Purified SF2/ASF used in the cross-linking experiments. (C) WT or mutant transcripts cross-linked to HeLa nuclear extract or SF2/ASF. 32P-labeled transcripts were subjected to UV cross-linking by the use of indicated nuclear extracts or purified SF2/ASF in the presence of tRNA as a nonspecific competitor. NE indicates nuclear extracts. (D) For competition assay, 25-fold molar excess of unlabeled WT or Mu1 and Mu2 transcripts was added in binding reactions. − indicates that the probe was incubated in the absence of competitors.
SF2/ASF UV cross-linked to the sequences at the junction of exon 2′/2. (A) UV cross-linking templates consisting of the wild-type (WT) or mutated (Mu1, Mu2) junction of exon 2′/2 and its flanking sequences. (B) Purified SF2/ASF used in the cross-linking experiments. (C) WT or mutant transcripts cross-linked to HeLa nuclear extract or SF2/ASF. 32P-labeled transcripts were subjected to UV cross-linking by the use of indicated nuclear extracts or purified SF2/ASF in the presence of tRNA as a nonspecific competitor. NE indicates nuclear extracts. (D) For competition assay, 25-fold molar excess of unlabeled WT or Mu1 and Mu2 transcripts was added in binding reactions. − indicates that the probe was incubated in the absence of competitors.
To test whether reducing SF2/ASF binding activity while maintaining the strength of the distal 3′ ss would increase the utilization of the 3′ ss, a construct containing a mutant site that would impair SF2/ASF binding while still retaining the strength of the 3′ ss (Figure 5A, Mu2) was generated by the use of ESEFinder. Unfortunately, the mutated sequence still bound to SF2/ASF and competed efficiently with the wild-type sequence for SF2/ASF binding in vitro (Figure 5C-D, Mu2). Our attempt to address this issue was complicated by the unavailability of suitable mutations that could abolish SF2/ASF binding while retaining 3′ ss strength and not creating additional splicing enhancer or silencer binding sites.
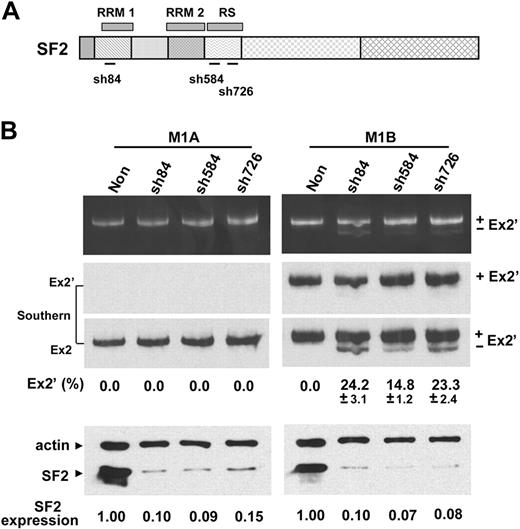
Depletion of SF2/ASF affects splicing of exon 1B to 3′ splice sites
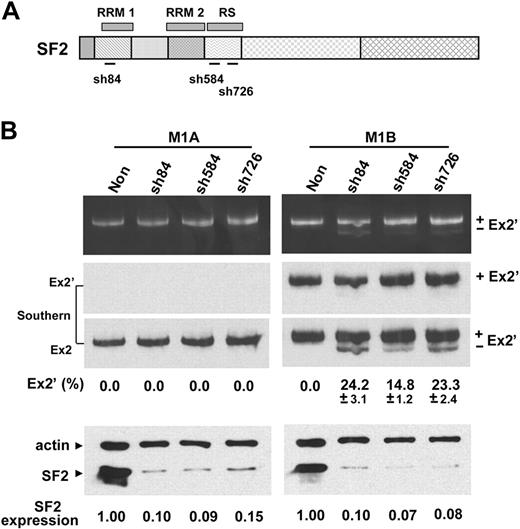
To test whether SF2/ASF could bind directly to the junction of exon 2′/2 and thereby block access to the distal 3′ ss, we examined whether reduction in SF2/ASF expression would allow preferential usage of the distal 3′ ss. Several SF2-shRNA constructs (Figure 6A) were evaluated for their reduction of SF2/ASF expression in MELCs (Figure 6B, bottom panel, SF2, lanes sh84, sh584, and sh726). A nonsilencing shRNA served as a control.
Depletion of SF2/ASF affects 3′ splice-site usages from exon 1B but not 1A. (A) Schematic of SF2/ASF structural organization and the regions targeted by SF2-shRNAs (sh84, sh584, and sh726). RRM indicates RNA recognition motif. RS, arginine/serine-rich domain. (B) The reduction of endogenous SF2/ASF expression by SF2-shRNAs and the effects on 3′ ss usage. (Top panel) RT-PCR analyses of exon 1A and 1B splicing patterns from RNA isolated from either control nonsilencing shRNA (Non) or SF2-shRNA–treated 1A or 1B minigene expressing MELC. (Middle panels) Southern blot hybridization with exon 2′ (Ex2′) or exon 2 (Ex2) probes. For each shRNA, 4 transductions were performed per experiment. Each experiment was repeated at least 3 times. Mean values ± SD of 3 independent experiments are shown. Standard deviations are omitted in results that consistently have 0% of exon 2′-inclusion product in all 3 reproducible experiments. (Bottom panel) equal amounts of cell lysates from control nonsilencing shRNA (Non)- and SF2-shRNA–treated cells were Western blotted for the presence of SF2/ASF. Expression, SF2/ASF expression levels in shRNA-treated cells relative to nonsilenced cells. β-actin served as loading control.
Depletion of SF2/ASF affects 3′ splice-site usages from exon 1B but not 1A. (A) Schematic of SF2/ASF structural organization and the regions targeted by SF2-shRNAs (sh84, sh584, and sh726). RRM indicates RNA recognition motif. RS, arginine/serine-rich domain. (B) The reduction of endogenous SF2/ASF expression by SF2-shRNAs and the effects on 3′ ss usage. (Top panel) RT-PCR analyses of exon 1A and 1B splicing patterns from RNA isolated from either control nonsilencing shRNA (Non) or SF2-shRNA–treated 1A or 1B minigene expressing MELC. (Middle panels) Southern blot hybridization with exon 2′ (Ex2′) or exon 2 (Ex2) probes. For each shRNA, 4 transductions were performed per experiment. Each experiment was repeated at least 3 times. Mean values ± SD of 3 independent experiments are shown. Standard deviations are omitted in results that consistently have 0% of exon 2′-inclusion product in all 3 reproducible experiments. (Bottom panel) equal amounts of cell lysates from control nonsilencing shRNA (Non)- and SF2-shRNA–treated cells were Western blotted for the presence of SF2/ASF. Expression, SF2/ASF expression levels in shRNA-treated cells relative to nonsilenced cells. β-actin served as loading control.
As predicted, reduction of SF2/ASF did not alter the splicing pattern of exon 1A to the distal 3′ ss in SF2-shRNA-treated cells (Figure 6B, M1A). In contrast, the reduction of SF2/ASF did affect the exon 1B splicing pattern in SF2-shRNA–treated cells (Figure 6B, M1B). Note that the nonsilencing control resulted in exclusive usage of the proximal 3′ ss and inclusion of exon 2′ (Figure 6B, M1B, “Non” lane) whereas the depletion of SF2/ASF resulted in clearly discernible usage of the distal site accounting for approximately 15% to 25% of the spliced products with exon 2′ exclusion (Figure 6B, M1B). The nature of the spliced products was confirmed by Southern blot analysis (Figure 6B, Southern). These results further suggest that promoter identity is involved in the determination of 3′ ss selection in 4.1R gene and that binding of SF2/ASF to the site, at least in part, mediates the usage of the proximal 3′ ss when transcribed from promoter 1B.
Discussion
The present study defines another of several pathways governing 4.1R exon 2′/2 expression: differential transcription by the use of alternative promoters, a coupled transcription-splicing mechanism, and differential binding of splicing factors to sequences located at the distal 3′ ss. Our findings suggest for the first time that a sequence element can serve as either an SF2/ASF binding site or the 3′ ss at which the splice occurs.
Consistent with increased 1A expression during Friend virus anemia-induced erythroblast differentiation,10 the expression of 1A drastically increased in differentiated human CD34+ cells. Thus, the differentiation-specific switch to “erythroid” 80-kDa 4.1R clearly depends on selective activation of promoter 1A. Constitutive expression of 1A encoding for 80-kDa isoforms in other cell types suggests its important role in cellular processes, as exemplified by its interaction with tubulin during mitosis41 and with nuclear actin in nuclear assembly.42 In most tissues, there is a reciprocal expression of 1B and 1C, both of which favor retention of exon 2′, permitting production of 135-kDa 4.1R isoforms. Although the exact exon composition of transcripts derived from these promoters remains to be determined, the different 5′ untranslated regions encoded by 1A, 1B, or 1C could provide a structural basis for differential regulation of mRNA turnover, translation, and/or subcellular localization.12-14
In our hands, discrepancy in exon 2′ exclusion was observed between transient and stable transfections of exon 1A-containing transcripts when transcribed from promoter 1A. Exon 2′ exclusion was much more prominent in stable lines than in transients. The number of copies of substrate per transfected cell may account for this difference; in transient lines, a minority of cells contains the transfected gene in high copy number and expresses the majority of transcripts. The essential splicing factors in transiently transfected cells may thus become limiting. The “naked” plasmid DNA sequences within transiently transfected cells also are known to be present in highly artificial configurations that can also profoundly influence transcription/splicing results.43 Incorporation of the DNA sequence into the genome in stably transfected cells, then, offers significant advantages in that it is a more “natural” chromatin environment and copy number. We avoided bias because of the accessibility of the sites into which the DNA was integrated by analyzing many pooled stable clones.
The alternative 3′ ss at the exon 2′/2 site is separated by 17 nucleotides, with a distal 3′ ss that is considerably weaker than the proximal 3′ ss. The distal 3′ ss with its poor Py tract could possibly suffer from weak interaction between U2AF35 and the 3′ ss AG dinucleotide, allowing easy competing-out by binding of SF2/ASF to the same site. The distal 3′ ss could thus function inherently better as an SF2/ASF-dependent enhancer than as a 3′ ss. However, transcripts derived from promoter 1A or 1B use different 3′ ss even though SF2/ASF is abundantly expressed. Thus, the functional effect of SF2/ASF on the 3′ ss is probably mediated by a promoter- or transcript-specific mechanism rather than by general cellular levels of SF2/ASF.
Normal levels of SF2/ASF completely suppress the use of the distal 3′ ss in transcripts derived from the 1B promoter. No exclusion of exon 2′ occurs unless SF2/ASF is depleted. Yet, a disproportionally modest use of the distal 3′ ss was found when SF2/ASF was depleted. Binding of SF2/ASF to the distal 3′ ss is thus unlikely to be the only regulatory mechanism needed to activate the weak distal 3′ ss. Parra et al44 suggest a 2-step intrasplicing mechanism by which alternative promoter choice can be coupled with subsequent splicing events. In the first step, the 5′ ss of exon 1B splices to the proximal 3′ ss. In the second step, exon 1A splices to the distal 3′ ss to form the mature product. This could well be the other downstream biochemical pathway by which the distal site is chosen when permitted by promoter choice.
Our results provide the first direct evidence that a link exists between transcription and splicing in protein 4.1R gene expression. Others have shown that the association of pol II with U1 snRNP and/or SR proteins can position key splicing factors close to the nascent pre-mRNA45 ; this coupling of transcription to splicing is thought to be mediated by the C-terminal domain (CTD) of the largest subunit of pol II.46 Differential phosphorylation of consensus repeats at CTD serines 2 and 5 modulates the association of transcription and splicing factors during the transcription cycle.47 Different promoters might then be bound by CTDs that are differentially phosphorylated.16,48
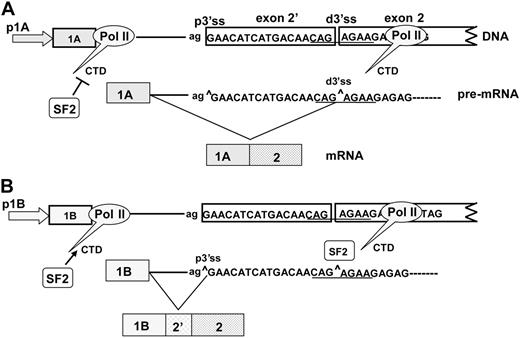
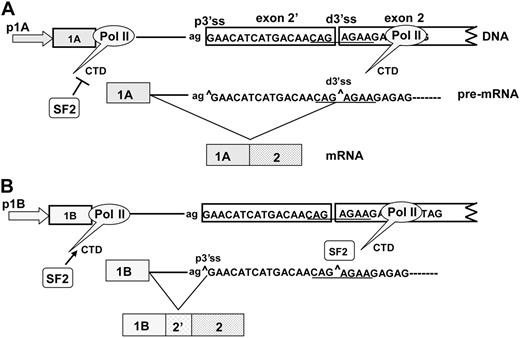
Our current working model for exon 2′ splicing is adapted from a recruitment model proposed by Cáceres and Kornblihtt49 (Figure 7). In this model, promoter 1A is less likely than 1B to recruit SF2/ASF to the CTD of pol II. Thus, SF2/ASF is absent from the CTD of pol II, and the resulting pre-mRNA exposes the distal 3′ ss and can be used to produce a mature mRNA lacking exon 2′. The 1B promoter recruits SF2/ASF, which “covers” the distal 3′ ss and favors use of the proximal 3′ ss and the inclusion of exon 2′. Inclusion or exclusion of exon 2′ is thus achieved through a coordinated action of transcription and splicing.
Recruitment model for cotranscriptional splicing of exon 2′/2. In this model, promoters 1A and 1B have differential abilities to bind SF2/ASF to the CTD of pol II. (A) In the 1A promoter, SF2/ASF is not recruited to the CTD; transcription of pre-mRNA exposes the distal 3′ss and allows for joining of exon 1A to produce a mature mRNA lacking exon 2′. (B) In the 1B promoter, SF2/ASF is recruited to the CTD of pol ll. The bound SF2/ASF blocks the distal 3′ss, favoring use of the proximal 3′ss, and results in the inclusion of exon 2′.
Recruitment model for cotranscriptional splicing of exon 2′/2. In this model, promoters 1A and 1B have differential abilities to bind SF2/ASF to the CTD of pol II. (A) In the 1A promoter, SF2/ASF is not recruited to the CTD; transcription of pre-mRNA exposes the distal 3′ss and allows for joining of exon 1A to produce a mature mRNA lacking exon 2′. (B) In the 1B promoter, SF2/ASF is recruited to the CTD of pol ll. The bound SF2/ASF blocks the distal 3′ss, favoring use of the proximal 3′ss, and results in the inclusion of exon 2′.
Alternative promoters and first exons are widespread among erythroid genes and may play a predominant role in tissue-specific gene expression regulation.6 Elucidation of 4.1R gene expression regulation is critical for a better understanding of the regulated expression of other key erythroid genes.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Shailja Pathania and David Livingston for assistance with the ChIP and ChRIP experiments, Karla Neugebauer for using modified Figures 2A and 2C illustrating the ChIP and ChRIP procedures, and Woan-Yuh Tarn for helpful discussion.
This study was supported by the National Institutes of Health grant HL24385 (to E.J.B.) and Claudia Barr Award (to S.C.H.).
National Institutes of Health
Authorship
Contributions: S.C.H., A.C., S.N., E.S.L., J.P., and E.J.B. designed research; A.C., S.N., E.S.L., J.P., G.Y., A.Y.Z., I.D.M., and A.C.O. performed research; A.W. and T.K.T. contributed reagents; S.C.H. wrote the paper; and A.C.O. and E.J.B. edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Shu-Ching Huang, Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA 02115; e-mail: shu-ching_huang@dfci.harvard.edu.
References
Author notes
*A.C., S.N., E.S.L., and J.P. contributed equally to this work.