Abstract
Megakaryoblastic leukemia 1 (MAL) is a transcriptional coactivator of serum response factor (SRF). In acute megakaryoblastic leukemia, the MAL gene is translocated and fused with the gene encoding one twenty-two (OTT). Herein, we show that MAL expression increases during the late differentiation steps of neonate and adult human megakaryopoiesis and localized into the nucleus after Rho GTPase activation by adhesion on collagen I or convulxin. MAL knockdown in megakaryocyte progenitors reduced the percentage of cells forming filopodia, lamellipodia, and stress fibers after adhesion on the same substrates, and reduced proplatelet formation. MAL repression led to dysmorphic megakaryocytes with disorganized demarcation membranes and α granules heterogeneously scattered in the cytoplasm. Gene expression profiling revealed a marked decrease in metalloproteinase 9 (MMP-9) and MYL9 expression after MAL inhibition. Luciferase assays in HEK293T cells and chromatin immunoprecipitation in primary megakaryocytes showed that the MAL/SRF complex directly regulates MYL9 and MMP9 in vitro. Megakaryocyte migration in response to stromal cell–derived factor 1, through Matrigel was considerably decreased after MAL knockdown, implicating MMP9 in migration. Finally, the use of a shRNA to decrease MYL9 expression showed that MYL9 was involved in proplatelet formation. MAL/SRF complex is thus involved in platelet formation and megakaryocyte migration by regulating MYL9 and MMP9.
Introduction
Serum response factor (SRF) is a widely expressed transcription factor required for the expression of immediate early, muscle-specific, and cytoskeletal genes.1-4 SRF contains a MADS domain that mediates homodimerization and DNA binding, and that allows recruitment of transcriptional cofactors. SRF binds to a CArG box present in promoter/enhancer regions of SRF-regulated genes.5 Depending on cell lines, different extracellular stimuli activate SRF through 2 main signaling pathways: the MAP-kinase pathway through members of the ternary complex factor (TCF)6,7 and the small GTPases pathway through the Rho family8 members regulating the myocardin-related transcription factors (MRTFs). The Rho-actin signaling pathway9-12 stimulates SRF by 2 ubiquitous MRTFs, megakaryoblastic leukemia 1 (MAL; MKL1, MRTF-A, BSAC) and MAL16 (MKL2, MRTF-B).
MAL was initially identified in acute megakaryoblastic leukemia (AMKL, M7) as a chromosome 22 encoded protein fused in 3′ with RNA-binding motif protein 15 (RBM15; OTT) located on chromosome 1.13-15 The translocation t(1,22)(p13;q13) leads to the in-frame fusion of the quasi-totality of OTT/RBM15 to the MAL gene. The OTT-MAL fusion protein is restricted to AMKL occurring de novo in infancy,16,17 in children older than 1 year or, occasionally, in Down syndrome patients.18,19
The subcellular localization of MAL is regulated through its association with globular actin by its RPEL motifs in the N-terminal region. The modification of the actin treadmilling by the Rho pathway results in the nuclear accumulation of MAL, and actin binding may also control MAL transcriptional activity.12,20 The 3 best-characterized Rho GTPase family members are Rho, Rac, and Cdc42. They regulate assembly and disassembly of actin cytoskeleton in response to a variety of extracellular stimuli. Rho regulates stress fibers and focal adhesion assembly, Cdc42 promotes filopodia, and Rac promotes lamellipodia formation. Different effectors of these 3 signaling pathways are involved in promoting actin polymerization. Except their direct role in actin cytoskeleton remodeling, Rho, Rac, and Cdc42 also control cell cycle, including G1 progression and cytokinesis, cell contraction, polarity and migration, vesicle transport (endocytosis, exocytosis), enzyme activation, and gene transcription via JNK, p38, nuclear factor-κB, and SRF.21,22 Hill et al reported that Rac1 and Cdc42 activate SRF in a RhoA-independent manner8 ; however, several studies showed a crosslink between these pathways in SRF regulation.23,24 In megakaryocytes (MKs), the platelet precursors, reorganization of actin cytoskeleton plays a crucial role not only in MK adhesion and migration25 but also at different stages during maturation, ie, during endomitosis,26 demarcation membrane formation, proplatelet formation, and platelet shedding.27-32 Recently, it has been shown that thrombopoietin (TPO) better stimulates Rho activity in immature than in mature MKs and that the Rho/ROCK pathway is a negative regulator of proplatelet formation. On the other hand, the use of constitutively active Cdc42 led to increased proplatelet formation33,34 (Y.C., unpublished data, February 2005). Interestingly, a recent study strongly supports the hypothesis that a local inhibition of the Rho/ROCK pathway is at the origin of MK ploidization by decreasing F-actin and myosin accumulation at midzone during cytokinesis.26
The involvement of MAL in the transformation of megakaryoblasts and recent experimental evidence35 strongly suggested a role in MK maturation. We therefore investigated its role in maturation, adhesion, migration, and proplatelet formation of MKs derived from neonate and adult CD34+ cells. The functional studies coupled to gene profiling after MAL depletion in MK revealed that the MAL/SRF complex regulates positively the known cytoskeletal genes and 2 new targets: metalloproteinase 9 (MMP-9) important for MK migration and MLC2 (MYL9) involved in platelet formation.
Methods
In vitro growth of MKs from CD34+ cells
Cord blood and cytapheresis samples were obtained after informed consent in accordance with the Declaration of Helsinki. Approval was obtained from the Assistance Publique des Hôpitaux de Paris. CD34+ cells were isolated using an immunomagnetic beads technique36 and grown in serum-free medium as previously reported.37 The culture medium was supplemented with recombinant human thrombopoietin (TPO; 10 ng/mL) or with a cytokine cocktail containing TPO (10 ng/mL), interleukin-3 (IL-3; 100 U/mL; Immunex), interleukin-6 (IL-6; 10 ng/mL; Tebu), stem cell factor (SCF; 25 ng/mL), and fetal liver tyrosine kinase 3 ligand (FLT3-L; 1 ng/mL; Amgen).
Lentiviral vector construction and production
For MAL, the published small interfering RNA (siRNA) sequence was used.9 For MYL9, the siRNA sequence was designed using siSearch software (http://www.dharmacon.com/DesignCenter/DesignCenterPage.aspx). Small hairpin sequences were created as previously described.38,39 Oligonucleotide MAL and MYL9 short hairpins (sh; supplemental Table 1A, available on the Blood website; see the Supplemental Materials link at the top of the online article) were synthesized (Eurogentec) and inserted into a pBlue Script vector containing the human H1 promoter. H1-shRNA specific for MAL (shMAL), H1-shMYL9, or H1-SCR (scramble control sequence) cassettes were inserted into a lentiviral vector (pRRLsin-PGK-eGFP-WPRE, Genethon). Lentiviral stocks were prepared and stored, and concentration was determined, as previously described.40
Cell transduction
CD34+ cells (105/mL) were prestimulated for 24 hours with TPO, IL-3, SCF, and FLT3-L. Lentiviral particles were added at a concentration corresponding to 125 ng viral p24/100 μL for 12 hours, followed by a second transduction. Cells were then cultured in the presence of TPO alone.
Cell sorting, flow cytometric, and cytologic analysis
Cells were incubated for 30 minutes at 4°C with an allophycocyanin-conjugated anti-CD41a monoclonal antibody and a phycoerythrin-conjugated anti-CD42a monoclonal antibody (BD Biosciences PharMingen). After washing, cells were incubated for 2 hours in 0.01 mM Hoechst 33342 (Sigma-Aldrich) at 37°C. To study endogenous MAL expression in the different differentiation classes, cells were sorted into CD34+41−; CD34−41+CD42−; CD34−41+CD42low; CD34−41+CD42high cell fractions for cord blood cultures, and into CD34+41−; CD34+41+CD42−; CD34+41+CD42 +; CD34−41+CD42 +cell fractions for cytapheresis cultures. To study the ploidy level, CD41+CD42+ cells stained with Hoechst were analyzed on a LSRII (BD Biosciences) with the CellQuest software package (BD Biosciences).
Real-time quantitative RT-PCR
Primers and internal probes for quantitative reverse-transcribed polymerase chain reaction (RT-PCR) are listed in supplemental Table 1B. PCR was carried out in the ABI Prism GeneAmp 7500 Sequence Detection System (Applied Biosystems) using TaqMan Universal PCR Master Mix (ABI) containing the specific primers (1.2 mM) and the specific probe (0.1 mM). Expression levels of all genes were expressed relatively to hypoxanthine phosphoribosyl transferase (HPRT).
Western blot analysis
Western blot analyses were performed as previously reported.41 The following antibodies were used: anti-MRTF-A (C-19), anti-HDAC-1 (H-51; Santa Cruz Biotechnology), and anti-HSC70 (Stressgen). In addition of anti-HDAC and anti-HSC70 staining, coloration with Ponceau S solution (Sigma-Aldrich) after transfer was used to control the quantity of proteins.
Proplatelet formation assay
CD41+CD42+GFP+ MK were sorted at day 8 of culture and plated in 96-well plates at a concentration of 2000 cells/well in serum-free medium containing TPO, and quantification of proplatelet MK was performed as previously described.42 Images were obtained using a Zeiss inverted microscope at a magnification of 40×, and the proplatelet network area was measured using the Axio Vision 4.6 software.
Luciferase assays
The 440-bp and 550-bp promoter regions upper transcription initiation of MMP9 and MYL9, respectively, were amplified (primer listed in supplemental Table 1C) and subcloned into the pLuc-MCS vector (Stratagene). MAL cDNA was cloned into the vector pMegix (gift of Dr Morgenstern from Millennium Pharmaceuticals). HEK293T cells were transfected with pLuc constructs (pLuc_MCS, pLuc_MMP9, pLuc_MYL9, pLuc_SRE, or pLuc_FLT3), a Renilla luciferase vector (for normalization) and MAL expression vector using Lipofectamine 2000 (Invitrogen). Cells were cultured in 12-well plates and transfected with 50 to 200 ng DNA. The total amount of DNA was kept constant by the inclusion of appropriate amounts of empty vectors. Cells were lysed with Passive Lysis Buffer 48 hours after transfection, and the luciferase activity was evaluated using the Dual Luciferase Reporter Assay System (Promega).
ChIP
Chromatin immunoprecipitation (ChIP) assays were performed with a ChIP assay kit (Upstate Biotechnology) using the anti-SRF antibody (clone H-65; Santa Cruz Biotechnology). Assays were performed using chromatin prepared from human MKs. Quantification of precipitated DNA fragments was carried out on an ABI PRISM 7500 sequence detection system using SYBR green (Applied Biosystems) in duplicate. Primers for MYL9, MMP9, and THSB1 are described in supplemental Table 1D. Fold enrichment is expressed as the ratio of target antibody signal to IgG signal calculated by extrapolation from a standard curve of DNA dilutions.
Migration assays
Cell migration was assessed with BD Bio-Coat Matrigel Invasion Chamber (BD Biosciences) according to the protocol provided by the manufacturer. Briefly, MKs were washed and prepared to a final concentration of 106 cells/mL. MK aliquots (100 μL) were seeded into each 8-μm pore transwell inserts, which were placed into the wells of a 24-well plate and incubated for 22 hours at 37°C. The lower compartment contained 600 μL serum-free media with or without 100 ng to stromal cell-derived factor 1α (SDF-1α; Abcys). The percentage of migrated CD41+CD42+GFP+ MK was determined by flow cytometry.
Cell adhesion and immunohistochemistry
Slides were coated overnight at 4°C with convulxin (kindly provided by M. Jandrot-Perrus, Hôpital Bichat, France) at 15 μg/mL in 1× PBS or with collagen type I (Nycomed) at 50 μg/mL. pRRL-SCR or pRRL-shMAL GFP+-sorted cells were seeded on coated slides for 2 hours (convulxin) or 1 hour (collagen type I) at 37°C. After a gentle wash, adherent cells were fixed in 2% paraformaldehyde for 10 minutes, permeabilized with 0.1% Triton X-100 for 5 minutes, and washed with 1× PBS for 5 minutes. Primary antibodies were subsequently applied to the slides at a concentration of 1 μg/mL and incubated for 1 hour at room temperature. Slides were then washed 3 times with 1× PBS for 5 minutes before and after application of the secondary antibody (20 μg/mL) for 30 minutes at room temperature. Slides were mounted using either Vectashield with 4,6-diamidino-2-phenylindole (Invitrogen) and observed under an epifluorescence microscope (Nikon Eclipse 600) equipped with a 60× objective (Nikon) or TOTO-3 iodide (Molecular Probes) and observed under a Zeiss LSM 510 laser scanning microscope (Carl Zeiss) with a 63×/1.4 numeric aperture oil objective. Images were processed using the Adobe Photoshop 6.0 and LSM 5 Image Browser software, respectively.
Primary antibodies were: anti-MRTF-A (clone C-19; Santa Cruz Biotechnology), anti-phalloidin–tetramethylrhodamine isothiocyanate (TRITC) (P-1951; Sigma-Aldrich). The secondary antibody was anti-goat TRITC (Jackson ImmunoResearch).
Electron microscopy
Samples were washed in 1× PBS, then fixed in 1.5% glutaraldehyde for 1 hour, and washed 3 times in 0.1 M phosphate buffer, pH 7.4. For morphologic examination, samples were postfixed in 1% osmic acid, dehydrated in ethanol, and embedded in Epon by standard methods. Samples were counterstained and were observed on a Philips Tecnai electron microscope.
Microarray analysis
We used Agilent long (60-bp) oligonucleotide microarrays and the dual-color analysis method in which probes from specimens and from the reference are differentially labeled with Cyanine 5 and Cyanine 3, as described previously.43 We performed a set of 4 dye swap experiments to compare RNAs obtained from MKs derived either from cytapheresis or cord blood CD34+ cells after transduction by shMAL. GFP+ cells were sorted at day 9 of culture. The purity of MKs derived from cytapheresis was approximately 97% (supplemental Figure 1). RNAs from scramble-lentivirus–transduced cells were used as the reference. From each of the 4 combined experiments, signatures (supplemental Table 2) were obtained by an analysis of variance test with P value of 10−3, and annotated with updated databases as Entrez-Gene. Data were analyzed using Resolver software, and gene selection was obtained by an analysis of variance test with P < 10−3 value.
Data availability
All data obtained by microarray analyses have been submitted to ArrayExpress44 with the accession number E-TABM-640.
Results
Expression of MAL and MAL16 in MKs
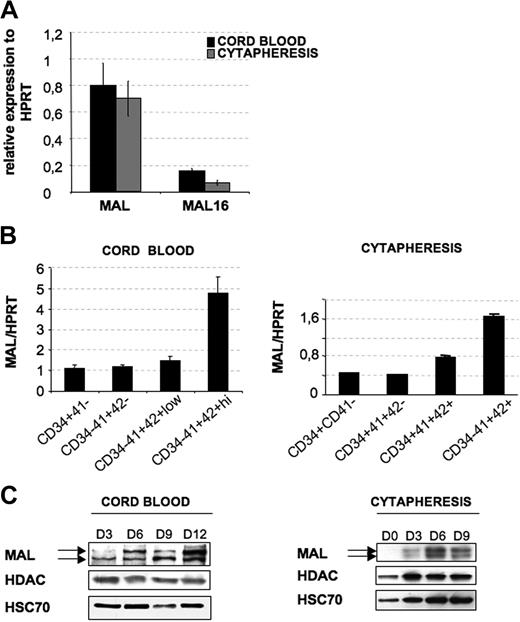
Different studies suggest a redundant role for 2 ubiquitous MRTFs, MAL and MAL16. Therefore, we first studied their relative expression in culture of MKs derived from CD34+ cells isolated from cord blood or cytapheresis and grown for 9 days in the presence of TPO. MAL mRNA level was approximately 7-fold higher than MAL16 mRNA in both types of MKs, taking HPRT as an internal control (Figure 1A). Because of the high expression of MAL and its direct involvement in megakaryoblastic transformation, we focused our study only on the role of MAL in megakaryopoiesis. To determine whether MAL expression varies through MK maturation during ontogenesis, CD34+ cells isolated from cord blood and cytapheresis were cultured in the presence of TPO and sorted at day 6 according to the expression of 3 differentiation markers (“Cell sorting, flow cytometric, and cytologic analysis”). No difference in MAL expression was found during ontogenesis. MAL transcripts remained stable during the first stages of differentiation in both neonate and adult-derived MKs but clearly increased in the most mature fraction (CD34−CD41+CD42+; Figure 1B). We next investigated the protein expression at different days of culture in the presence of TPO (until day 12 for cord blood and until day 9 for cytapheresis), as well as in platelets isolated either from cord blood or cytapheresis samples. Consistent with MAL mRNA level, the protein level increased continuously until day 12 of culture for cord blood-derived MKs and until day 9 of culture for cytapheresis-derived MKs (∼ 65% of CD41+CD42+ MKs; Figure 1C). In our culture conditions, proplatelet-forming MKs first appeared at day 12 for adult MKs and at day 15 for neonate MKs. Thereafter, MAL protein level decreased with the appearance of proplatelet-forming MKs and was undetectable in platelets (data not shown).
MAL expression in MKs. CD34+ cells from cord blood or cytapheresis were cultured with TPO. (A) At day 9 of culture, mRNA levels of MAL and MAL16 were evaluated by quantitative RT-PCR. Their expression was normalized with respect to HPRT mRNA. (B) To investigate MAL transcription level during MK differentiation, cells were sorted at day 6 of culture in 4 populations: for cord blood, CD34+CD41−, CD34−CD41+CD42−, CD34−CD41+CD42low, and CD34−CD41+CD42high; and for cytapheresis, CD34+CD41−, CD34+CD41+CD42−, CD34+CD41+CD42+, and CD34−CD41+CD42+. MAL mRNA level was evaluated as in panel A. Error bars in panels A and B represent the SD of the mean of 3 repeated experiments each performed in triplicate wells. (C) To investigate protein levels by Western blot analysis during MK differentiation, total cell populations were harvested on day 3 (D3), day 6 (D6), day 9 (D9), and day 12 (D12) for cord blood-derived MK and at day 0 (D0), day 3 (D3), day 6 (D6), and day 9 (D9) for cytapheresis-derived MKs. HDAC-1 and HSC70 were used as internal control. The figure illustrates representative data of 2 experiments with similar results.
MAL expression in MKs. CD34+ cells from cord blood or cytapheresis were cultured with TPO. (A) At day 9 of culture, mRNA levels of MAL and MAL16 were evaluated by quantitative RT-PCR. Their expression was normalized with respect to HPRT mRNA. (B) To investigate MAL transcription level during MK differentiation, cells were sorted at day 6 of culture in 4 populations: for cord blood, CD34+CD41−, CD34−CD41+CD42−, CD34−CD41+CD42low, and CD34−CD41+CD42high; and for cytapheresis, CD34+CD41−, CD34+CD41+CD42−, CD34+CD41+CD42+, and CD34−CD41+CD42+. MAL mRNA level was evaluated as in panel A. Error bars in panels A and B represent the SD of the mean of 3 repeated experiments each performed in triplicate wells. (C) To investigate protein levels by Western blot analysis during MK differentiation, total cell populations were harvested on day 3 (D3), day 6 (D6), day 9 (D9), and day 12 (D12) for cord blood-derived MK and at day 0 (D0), day 3 (D3), day 6 (D6), and day 9 (D9) for cytapheresis-derived MKs. HDAC-1 and HSC70 were used as internal control. The figure illustrates representative data of 2 experiments with similar results.
MAL localizes into the nucleus of MKs after Rho GTPase activation
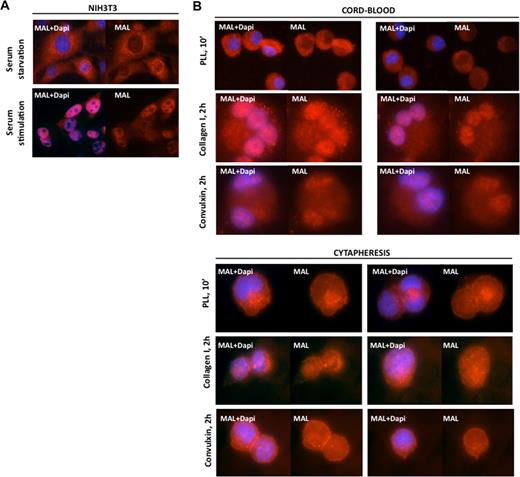
As previously described,8,12 we observed that MAL localized in the cytoplasm of serum-starved NIH3T3 cells and accumulated in the nucleus after serum stimulation (Figure 2A). This is mediated through small GTPase activation.8,12 To examine the effect of RhoA and Cdc42 activation on MAL localization in cord blood and cytapheresis-derived MKs, MKs were cultured for 9 days in serum-free medium supplemented by TPO and then allowed to adhere onto different extracellular matrix. After 10 minutes on polylysine, a substrate that does not allow Rho or Cdc42 activation,33 MAL remained cytoplasmic (Figure 2B). After 2 hours on collagen type I, a substrate that activates principally Rho/ROCK leading to fiber stress formation,33 MAL accumulated in the MK nucleus. After 2 hours onto convulxin, a substrate inducing mainly filopodia formation through activation of the Cdc42 pathway,33 MAL also accumulated in the MK nucleus (Figure 2B). The nuclear accumulation of MAL after Rho and Cdc42 pathway activation is observed in MKs derived either from cord blood or cytapheresis CD34+ cells.
MAL nuclear localization after Rho GTPase activation in MKs. (A) MAL cytoplasmic localization in serum-starved NIH3T3 cells and its nuclear localization after serum stimulation. (B) Cord blood-derived MKs (upper panels) and cytapheresis-derived MKs (lower panels). After adhesion to polylysine for 10 minutes (PLL, 10′), MAL is localized in the cytoplasm. After adhesion to collagen I or convulxin for 2 hours, MAL is localized in the nucleus. Both substrates induce Rho GTPase activation. MAL (red) and 4,6-diamidino-2-phenylindole (blue) staining was detected by immunofluorescence and visualized under a fluorescent light microscope at an original magnification ×60.
MAL nuclear localization after Rho GTPase activation in MKs. (A) MAL cytoplasmic localization in serum-starved NIH3T3 cells and its nuclear localization after serum stimulation. (B) Cord blood-derived MKs (upper panels) and cytapheresis-derived MKs (lower panels). After adhesion to polylysine for 10 minutes (PLL, 10′), MAL is localized in the cytoplasm. After adhesion to collagen I or convulxin for 2 hours, MAL is localized in the nucleus. Both substrates induce Rho GTPase activation. MAL (red) and 4,6-diamidino-2-phenylindole (blue) staining was detected by immunofluorescence and visualized under a fluorescent light microscope at an original magnification ×60.
MAL knockdown does not significantly impair the first stages of MK differentiation
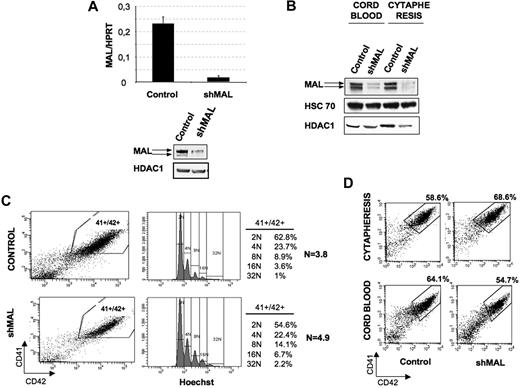
To investigate further the role of MAL in MKs, we developed a lentivirus encoding an shMAL under the H1 promoter. The vector efficiency was first tested in the MO7e leukemic cell line. Two days after transduction, MAL level was determined by RT-PCR and Western blot in cells sorted on the expression of GFP. The shMAL reduced MAL transcripts approximately 13-fold compared with the control shRNA (Figure 3A left panel), and the protein expression was similarly decreased (Figure 3A right panel). Then, CD34+ cells obtained from cord blood or cytapheresis were prestimulated in culture by IL-3, FLT3-L, SCF, and TPO and transduced at days 1 and 2 with the control shRNA or shMAL lentiviruses. Thereafter, cells were cultured in TPO alone. At day 9 of culture, GFP+ cells were sorted. The MK purity was approximately 90% in both cases (data not shown). As shown in Figure 3B, shMAL led to a major decrease of MAL protein level compared with the control shRNA.
Effect of MAL knockdown on ploidy level and MK differentiation. (A) MO7e cells were transduced with a control lentivirus or the lentivirus-encoding shRNA of MAL (shMAL). GFP+ cells were sorted and analyzed. MAL mRNA level (left panel) was measured as in Figure 1. MAL protein level (right panel) was analyzed at day 9 of culture by Western blot. HDAC-1 was used as internal control of a quantity. The figure illustrates representative data of 2 experiments with similar results. (B-D) Cord blood and cytapheresis-isolated CD34+ cells were transduced with the control lentivirus or the lentivirus-encoding shRNA of MAL (shMAL). (B) MAL protein level was analyzed at day 9 of culture by Western blot in GFP+ cells. HDAC-1 and HSC70 were used as internal controls. The figure illustrates representative data of 2 experiments with similar results. (C) The CD41+CD42+ cell population (left panel), corresponding to mature cytapheresis-derived MKs, was analyzed for ploidy level by Hoechst staining (right panel). The mean ploidy (N) was calculated from the number of cells at each ploidy level. The figure illustrates representative data of 3 experiments with similar results. (D) The percentage of mature MKs was evaluated as the percentage of cells coexpressing both CD41 and CD42 markers. Data illustrate analysis of 4 repeated experiments for cytapheresis and 3 for cord blood. Similar results were obtained in all experiments.
Effect of MAL knockdown on ploidy level and MK differentiation. (A) MO7e cells were transduced with a control lentivirus or the lentivirus-encoding shRNA of MAL (shMAL). GFP+ cells were sorted and analyzed. MAL mRNA level (left panel) was measured as in Figure 1. MAL protein level (right panel) was analyzed at day 9 of culture by Western blot. HDAC-1 was used as internal control of a quantity. The figure illustrates representative data of 2 experiments with similar results. (B-D) Cord blood and cytapheresis-isolated CD34+ cells were transduced with the control lentivirus or the lentivirus-encoding shRNA of MAL (shMAL). (B) MAL protein level was analyzed at day 9 of culture by Western blot in GFP+ cells. HDAC-1 and HSC70 were used as internal controls. The figure illustrates representative data of 2 experiments with similar results. (C) The CD41+CD42+ cell population (left panel), corresponding to mature cytapheresis-derived MKs, was analyzed for ploidy level by Hoechst staining (right panel). The mean ploidy (N) was calculated from the number of cells at each ploidy level. The figure illustrates representative data of 3 experiments with similar results. (D) The percentage of mature MKs was evaluated as the percentage of cells coexpressing both CD41 and CD42 markers. Data illustrate analysis of 4 repeated experiments for cytapheresis and 3 for cord blood. Similar results were obtained in all experiments.
Recently, it has been reported that inhibition of the Rho/Rock pathway leads to an increased MK ploidy level.26 Therefore, we examined whether the Rho effector MAL could be involved in the endomitotic process. To study the effect of MAL repression on MK ploidization, CD34+ cells isolated from cytapheresis were transduced at days 1 and 2 of culture and analyzed at day 9. The ploidy level of transduced (GFP+) cells was analyzed in the gate of mature CD41+CD42+ MKs. As illustrated in Figure 3C, MAL knockdown resulted in a moderate (21.3% ± 9%) increase in mean ploidy level that did not reach significance (n = 3, P ≤ .09), compared with the untransduced population.
Then, we analyzed the effect of MAL knockdown on maturation of MKs issued from cord blood and cytapheresis samples. Cells were transduced as at days 1 and 2, and GFP+ cells were analyzed at day 7 of culture for the expression of 2 MK markers, CD41 and CD42. As shown in Figure 3D, the percentage of CD41+CD42+ mature MKs slightly increased in cytapheresis samples (top panel, 26% ± 13.6%, n = 4, P ≤ .004) but decreased in cord blood samples (bottom panel, 16.8% ± 1.5%, n = 3, P ≤ .08) after MAL repression. The minor difference in the percentage of CD41+CD42+ MK in both cases after MAL repression and the slight increase in ploidy level suggest that MAL does not play an essential role in MK ploidization and during the early differentiation steps.
MAL plays an important role in adhesion, terminal maturation, and proplatelet formation of MKs
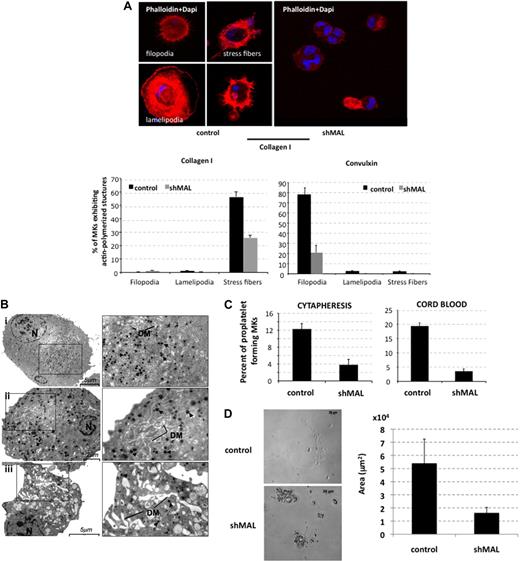
MAL and MAL16 are necessary for the formation of stress fibers and focal adhesion in NIH3T3 cells.4 Here, we investigated whether MAL had a similar role in MK. Control shRNA- or shMAL-transduced MKs were sorted at day 8 of culture on GFP+ and allowed to adhere for 1 hour on collagen type I or for 2 hours on convulxin. As illustrated in Figure 4A, the percentage of filopodia and stress fibers forming MKs was markedly decreased after MAL repression (n = 2, P ≤ .016; n = 2, P ≤ .05, respectively). These results suggested that, like in NIH3T3 cells, MAL acts as a mediator of actin-polymerized structures by regulating transcription of several cytoskeletal genes.
MAL knockdown alters actin polymerization, terminal maturation, and proplatelet formation of MKs. CD34+ cells isolated from cord blood or cytapheresis were transduced with the control lentivirus or the lentivirus encoding shRNA of MAL (shMAL). (A) GFP+ cells (day 8) were allowed to adhere on collagen I or convulxin for 2 hours. Stress fibers, filopodia, and lamellipodia were stained with TRITC-conjugated phalloidin (red) and nucleus with 4,6-diamidino-2-phenylindole (blue). The percentage of MK-forming stress fibers, filopodia, or lamellipodia was evaluated on a total of 500 cells using fluorescent light microscopy (original magnification ×40). (B) Ultrastructural aspect of control (i) and shMAL (ii-iii) transduced MKs. MKs were sorted at day 10 of culture on the expression of GFP and fixed. N indicates nucleus; DM, demarcation membranes. Arrowhead represents α granules. (i) Morphology of a typical normal MK. Bar represents 5 μm. (ii-iii) Morphology of shMAL-transduced MK. (ii) Bar represents 2 μm. (iii) Bar represents 5 μm. (C) GFP+ cells were sorted at day 9 on the coexpression of CD41 and CD42. Cells were seeded at 2 × 103 cells/well in a 96-well plate. The percentage of proplatelet-forming MKs was estimated by counting MKs exhibiting one or more cytoplasmic processes with areas of constriction. A total of 500 cells per well were counted at day 13 for cytapheresis-derived MKs and at day 15 for cord blood–derived MKs. Error bars in histograms represent the SD of one representative experiment performed in triplicate wells. Similar results were obtained in 3 repeated experiments with cytapheresis and 2 experiments with cord blood. (D) MKs derived from cytapheresis were plated as in panel C, and the area of proplatelet network estimated in microns squared was measured using Axio Vision 4.6 software. One representative control- and shMAL-transduced proplatelet-forming MKs is shown on left, and the area mean of 10 cells from each group is shown in the histogram on right. Error bars in histograms represent the SD obtained for 10 cells in one representative experiment (n = 2).
MAL knockdown alters actin polymerization, terminal maturation, and proplatelet formation of MKs. CD34+ cells isolated from cord blood or cytapheresis were transduced with the control lentivirus or the lentivirus encoding shRNA of MAL (shMAL). (A) GFP+ cells (day 8) were allowed to adhere on collagen I or convulxin for 2 hours. Stress fibers, filopodia, and lamellipodia were stained with TRITC-conjugated phalloidin (red) and nucleus with 4,6-diamidino-2-phenylindole (blue). The percentage of MK-forming stress fibers, filopodia, or lamellipodia was evaluated on a total of 500 cells using fluorescent light microscopy (original magnification ×40). (B) Ultrastructural aspect of control (i) and shMAL (ii-iii) transduced MKs. MKs were sorted at day 10 of culture on the expression of GFP and fixed. N indicates nucleus; DM, demarcation membranes. Arrowhead represents α granules. (i) Morphology of a typical normal MK. Bar represents 5 μm. (ii-iii) Morphology of shMAL-transduced MK. (ii) Bar represents 2 μm. (iii) Bar represents 5 μm. (C) GFP+ cells were sorted at day 9 on the coexpression of CD41 and CD42. Cells were seeded at 2 × 103 cells/well in a 96-well plate. The percentage of proplatelet-forming MKs was estimated by counting MKs exhibiting one or more cytoplasmic processes with areas of constriction. A total of 500 cells per well were counted at day 13 for cytapheresis-derived MKs and at day 15 for cord blood–derived MKs. Error bars in histograms represent the SD of one representative experiment performed in triplicate wells. Similar results were obtained in 3 repeated experiments with cytapheresis and 2 experiments with cord blood. (D) MKs derived from cytapheresis were plated as in panel C, and the area of proplatelet network estimated in microns squared was measured using Axio Vision 4.6 software. One representative control- and shMAL-transduced proplatelet-forming MKs is shown on left, and the area mean of 10 cells from each group is shown in the histogram on right. Error bars in histograms represent the SD obtained for 10 cells in one representative experiment (n = 2).
Electron microscopy studies revealed structural abnormalities of maturing MK after MAL knockdown (Figure 4B). Compared with the control shRNA (Figure 4Bi), the cytoplasm was disorganized with numerous vacuoles (Figure 4Bii-iii); the demarcation membrane system was heterogeneously dilated and abnormally distributed with its presence at the periphery of MKs (Figure 4Bii-iii); moreover, the α granules were scattered heterogeneously in the cytoplasm (Figure 4Bii).
To test whether MAL was involved in the process of proplatelet formation, control shRNA- and shMAL-transduced MKs were cultured in a liquid serum-free medium containing TPO. CD41+CD42+GFP+ MKs were sorted at day 9 and seeded in 96-well plates at 2 × 103 cells per well. At day 13 of culture, the percentage of proplatelet-forming MKs was 68% reduced in MAL knockdown MKs derived both from cytapheresis (n = 3, P ≤ .003) and cord blood (n = 2, P ≤ .1; Figure 4C). Moreover, we measured the total surface area of the proplatelet network in cells transduced with control or shMAL and found that it was decreased 3-fold after MAL inhibition (Figure 4D). Ten cells were analyzed in each group (P ≤ .008). These results suggest that the MAL/SRF complex plays a major role in terminal maturation, proplatelet formation, and adhesion of MK by transcriptionally regulating genes implicated in these processes.
Microarray analysis of genes deregulated after MAL knockdown in MKs
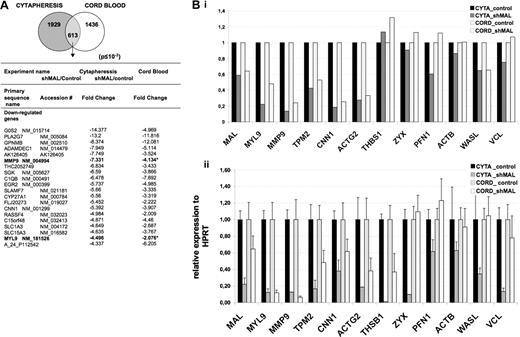
MAL is thought to regulate transcription through its interaction with SRF. To characterize MAL/SRF target genes in MKs derived from cord blood or cytapheresis samples, we compared control shRNA- and shMAL-transduced MKs by microarray analyses. CD34+ cells from cord blood or cytapheresis were transduced as described in the previous paragraph, cultured in the presence of TPO alone after transduction and GFP+ cells were sorted at day 9 of culture. The purity of MKs was approximately 97%, as illustrated for cytapheresis-derived MKs in supplemental Figure 1. Gene expression profiles were obtained using Agilent Human Whole Genome 44K oligonucleotide arrays allowing a direct comparison of 2 populations labeled by 2 different dyes (Cy3 and Cy5). Cell populations transduced by the control lentivirus were used as reference for transduction-induced changes. The signal from 1929 and 1436 probe sets (corresponding to 1037 and 773 genes, respectively) was significantly deregulated by the expression of the shMAL in MKs derived from cytapheresis and cord blood, respectively (Figure 5A). A total of 613 probe sets were common between cytapheresis- and cord blood–derived MKs (supplemental Table 2). The signal of 589 probe sets was found to be modified similarly in both cell types, and the expression of 24 probe set genes showed an opposite variation in cord blood and cytapheresis MKs (supplemental Table 3). Figure 5A shows the 20 most down-regulated genes found in MKs derived from cytapheresis and cord blood. They are listed according to the fold change observed in the experiment using cytapheresis MKs.
Gene profiling of MKs after MAL knockdown. CD34+ cells isolated from cord blood and cytapheresis were transduced with the control lentivirus or the lentivirus-expressing shMAL. At day 9 of culture, cells expressing GFP were sorted and mRNA was subjected to microarray analyses using Agilent Human Whole Genome 44K oligonucleotide arrays (A) and to quantitative RT-PCR (B). (A) A total of 1929 and 1436 probe sets were found significantly deregulated in MKs derived from cytapheresis and cord blood, respectively, with a 613 probe set common to both. The table shows primary sequence name, accession number, and fold change of the 20 most down-regulated genes after MAL knockdown in cytapheresis- and cord blood-derived MKs. (B) Validation of microarray data (i) by quantitative RT-PCR (ii) for 12 selected genes. The error bars represent the SD of the mean of 3 repeated experiments each performed in triplicate wells.
Gene profiling of MKs after MAL knockdown. CD34+ cells isolated from cord blood and cytapheresis were transduced with the control lentivirus or the lentivirus-expressing shMAL. At day 9 of culture, cells expressing GFP were sorted and mRNA was subjected to microarray analyses using Agilent Human Whole Genome 44K oligonucleotide arrays (A) and to quantitative RT-PCR (B). (A) A total of 1929 and 1436 probe sets were found significantly deregulated in MKs derived from cytapheresis and cord blood, respectively, with a 613 probe set common to both. The table shows primary sequence name, accession number, and fold change of the 20 most down-regulated genes after MAL knockdown in cytapheresis- and cord blood-derived MKs. (B) Validation of microarray data (i) by quantitative RT-PCR (ii) for 12 selected genes. The error bars represent the SD of the mean of 3 repeated experiments each performed in triplicate wells.
To confirm the microarray data, 12 genes were selected, most with a key role in cytoskeleton organization. Three of them (zyxin, profilin 1, and N-wasp) play a major role in actin polymerization and 3 others (β-actin, vinculin, and tropomyosin 2) are major structural proteins of the cytoskeleton and thus could be implicated in the defects in fiber stress and filopodia formation seen in MAL knockdown MKs. Gene expression was analyzed by quantitative RT-PCR (Figure 5B). For some genes (zyxin, thrombospondin, β-actin, and vinculin), a discrepancy was found between microarray (Figure 5Bi) and RT-PCR analyses (Figure 5Bii) that could be explained by methods sensitivity. Indeed, saturation of highly expressed genes may lead to their exclusion in microarray analyses.
We next focused our efforts on MMP9 and MYL9 (the myosin light chain associated with MYH9), 2 genes with a specific role in megakaryopoiesis.
MMP9 and MYL9 (MLC2) are direct targets of MAL/SRF complex in MKs
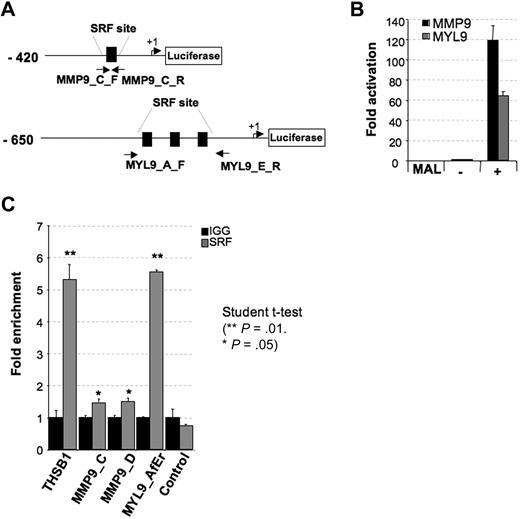
The MAL/SRF complex binds DNA at specific sites through the SRF DNA-binding domain. Using the ChipMapper software, we identified putative SRF-binding sites in both MMP9 and MYL9 promoters. To determine whether MMP9 and MYL9 are targets of the MAL/SRF complex, we cloned 420 bp of the MMP9 promoter (including one SRF-binding site) and 650 bp of the MYL9 promoter (including 3 SRF-binding sites) upstream of the firefly luciferase gene (Figure 6A) and tested the ability of MAL to modulate expression in transient reporter assays using HEK293T cells. Transcription from both MMP9 and MYL9 promoters was highly stimulated by MAL coexpression (Figure 6B). The baseline activity of MMP9 and MYL9 promoters compared with minimal promoter with or without SRF consensus-binding sites (pLUC_SRE and pLuc_MCS, respectively) or to a control promoter region without SRF consensus binding site cloned into the same luciferase vector (pLuc_FLT3) is shown in supplemental Figure 2.
MMP9 and MYL9 are directly regulated by MAL/SRF complex. (A) Schematic representation of MMP9 and MYL9 human promoter regions cloned into the pLuc-MCS reporter. Arrows represent the translation start site. (B) Luciferase assay performed by transient transfection of HEK293 cells with the 50 ng of Megix vector containing MAL. Luciferase levels are shown as fold change relative to cells transfected with the reporter construct alone. The total amount of transfected DNA was kept constant by addition of empty Megix vector. The histogram shows one representative experiment of 3, each in triplicate. Error bars represent the SD of triplicate. (C) ChIP assay performed in cytapheresis-derived MKs (day 10 in culture) with primer sets directed toward in silico predicted SRF-binding sites: MMP9_C_F and R, and MMP9_D_F and R for MMP9, and primer sets MYL9_A_F and MYL9_E_R for MYL9. Localization of primers MMP9_C_F and R for MMP9 and MYL9_A_F and MYL9_E_R for MYL9 are depicted in panel A. Primers MMP9_D_F and R are not localized in the cloned promoter region designed in panel A. Control primer sets allowing amplification of known SRF-binding sites (THSB1) or DNA region devoid of SRF sites were also used. Immunoprecipitation was performed with control rabbit IgG and anti-SRF antibodies. Histograms indicate relative occupancy of SRF-binding sites by SRF in the MMP9, MYL9, and THSB1 promoters. Error bars represent the SD of experiments performed in duplicate. The figure illustrates representative data of 2 independent experiments with similar results.
MMP9 and MYL9 are directly regulated by MAL/SRF complex. (A) Schematic representation of MMP9 and MYL9 human promoter regions cloned into the pLuc-MCS reporter. Arrows represent the translation start site. (B) Luciferase assay performed by transient transfection of HEK293 cells with the 50 ng of Megix vector containing MAL. Luciferase levels are shown as fold change relative to cells transfected with the reporter construct alone. The total amount of transfected DNA was kept constant by addition of empty Megix vector. The histogram shows one representative experiment of 3, each in triplicate. Error bars represent the SD of triplicate. (C) ChIP assay performed in cytapheresis-derived MKs (day 10 in culture) with primer sets directed toward in silico predicted SRF-binding sites: MMP9_C_F and R, and MMP9_D_F and R for MMP9, and primer sets MYL9_A_F and MYL9_E_R for MYL9. Localization of primers MMP9_C_F and R for MMP9 and MYL9_A_F and MYL9_E_R for MYL9 are depicted in panel A. Primers MMP9_D_F and R are not localized in the cloned promoter region designed in panel A. Control primer sets allowing amplification of known SRF-binding sites (THSB1) or DNA region devoid of SRF sites were also used. Immunoprecipitation was performed with control rabbit IgG and anti-SRF antibodies. Histograms indicate relative occupancy of SRF-binding sites by SRF in the MMP9, MYL9, and THSB1 promoters. Error bars represent the SD of experiments performed in duplicate. The figure illustrates representative data of 2 independent experiments with similar results.
To demonstrate that SRF binds to the MMP9 and MYL9 promoters in vivo, ChIP assays were performed using human MKs derived in culture from CD34+ cells. Thrombospondin, a previously described target of SRF,45 was used as a positive control for a ChIP experiment. Quantitative RT-PCR with primers amplifying the regions encompassing the predicted C (Figure 6A) and D (supplemental primer Table 1d) SRF-binding sites in MMP9 promoter and 3 SRF-binding sites in MYL9 promoter (Figure 6A) were performed. ChIP experiments revealed a high MYL9 promoter region enrichment and a slight but significant MMP9 promoter region enrichment using SRF antibody (Figure 6C). Our results confirmed that MMP9 and MYL9 are direct targets for MAL/SRF complex in MK.
MAL plays a key role in MK migration and platelet formation via MMP9 and MYL9 regulation
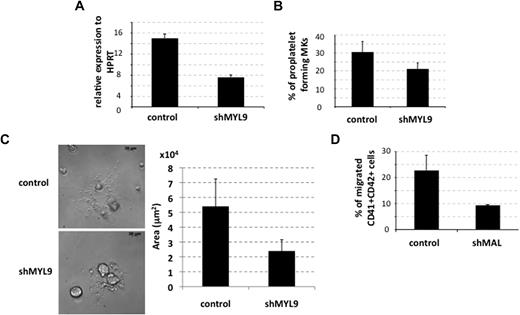
To confirm that the drop in MYL9 expression could be responsible for the defect in proplatelet formation observed on the knockdown of MAL, we constructed a lentivirus expressing an shRNA specific for MYL9 (shMYL9). The shMYL9 was expressed in primary MKs derived from cytapheresis, as described for MAL knockdown. Two independent experiments showed a 50% decrease in the MYL9 transcript level (n = 2, P ≤ .08; Figure 7A). At day 13 of culture, knockdown of MYL9 elicited an approximately 35% decrease in the percentage of proplatelet-forming MKs (n = 4, P ≤ .003; Figure 7B). Moreover, the total surface area of the proplatelet network in cells transduced with shMYL9 was decreased approximately 2-fold after shMYL9 transduction compared with control-transduced cells. Ten cells were analyzed in each group (P ≤ .009; Figure 7C).
MAL contribution to proplatelet formation and migration of MKs by targeting MYL9 and MMP9, respectively. (A-B) CD34+ cells isolated from cytapheresis were transduced at days 1 and 2 of culture with the lentivirus-encoding control shRNA (control) or MYL9 shRNA (shMYL9). (A) GFP+ cells were analyzed at day 9 of culture. MYL9 mRNAs were measured by quantitative RT-PCR (left panel). The histogram shows one of 2 representative experiments, each in triplicate. Error bars represent the SD of triplicate wells. (B) The percentage of proplatelet-forming MKs derived from cytapheresis samples was evaluated as described in Figure 4C at day 13 of culture. Error bars represent the SD of 1 representative experiment performed in triplicate wells. Similar results were obtained in 4 independent experiments. (C) MKs derived from cytapheresis CD34+ cells were plated as in panel C, and the area of proplatelet network estimated in microns squared was measured using Axio Vision 4.6 software. One representative control- and shMYL9-transduced proplatelet-forming MKs is shown on the left, and the mean area of 10 cells from each group is shown in the histogram on the right. Error bars in histograms represent the SD obtained for 10 cells in 1 representative experiment (n = 2). (D) MK migration through Matrigel-coated transwells in response to SDF-1. Cytapheresis isolated CD34+ cells were transduced at days 1 and 2 of culture with a control or shMAL-encoding lentivirus. The experiment was done on day 8 of culture. Data represent the percentage of migrated CD41+CD42+GFP+ cells compared with total CD41+CD42+GFP+ input. Error bars represent the SD of 1 representative experiment performed in triplicate. The figure illustrates representative data of 2 independent experiments with similar results.
MAL contribution to proplatelet formation and migration of MKs by targeting MYL9 and MMP9, respectively. (A-B) CD34+ cells isolated from cytapheresis were transduced at days 1 and 2 of culture with the lentivirus-encoding control shRNA (control) or MYL9 shRNA (shMYL9). (A) GFP+ cells were analyzed at day 9 of culture. MYL9 mRNAs were measured by quantitative RT-PCR (left panel). The histogram shows one of 2 representative experiments, each in triplicate. Error bars represent the SD of triplicate wells. (B) The percentage of proplatelet-forming MKs derived from cytapheresis samples was evaluated as described in Figure 4C at day 13 of culture. Error bars represent the SD of 1 representative experiment performed in triplicate wells. Similar results were obtained in 4 independent experiments. (C) MKs derived from cytapheresis CD34+ cells were plated as in panel C, and the area of proplatelet network estimated in microns squared was measured using Axio Vision 4.6 software. One representative control- and shMYL9-transduced proplatelet-forming MKs is shown on the left, and the mean area of 10 cells from each group is shown in the histogram on the right. Error bars in histograms represent the SD obtained for 10 cells in 1 representative experiment (n = 2). (D) MK migration through Matrigel-coated transwells in response to SDF-1. Cytapheresis isolated CD34+ cells were transduced at days 1 and 2 of culture with a control or shMAL-encoding lentivirus. The experiment was done on day 8 of culture. Data represent the percentage of migrated CD41+CD42+GFP+ cells compared with total CD41+CD42+GFP+ input. Error bars represent the SD of 1 representative experiment performed in triplicate. The figure illustrates representative data of 2 independent experiments with similar results.
MYL9 knockdown was less efficient in affecting the proplatelet formation than MAL knockdown. This is probably the result of the lower efficiency of the shMYL9 leading only to 50% reduction of MYL9 transcript, whereas shMAL resulted in a nearly complete abolition of MAL expression. Nevertheless, the data confirm that MAL/SRF complex is involved in proplatelet formation via a transcriptional regulation of MYL9.
MKs migrate through bone marrow endothelial cells to release platelets within the sinusoidal space in response to SDF-1.46,47 Because SRF was reported to be involved in cell migration,3 we investigated the effect of MAL knockdown on MK migration. MKs were transduced and grown as described in “Cell transduction,” and chemotactic assays in response to SDF-1 were performed on day 8 of culture. No difference was detected between control shRNA- or shMAL-transduced MKs derived from cytapheresis after migration through noncoated or collagen I–coated transwells (data not shown). However, when Matrigel-coated transwells were used, a 2-fold decrease in MK migration in response to SDF-1 was seen in shMAL-expressing cells (n = 2, P ≤ .08; Figure 7D). This defective MK migration through an extracellular matrix similar to the endothelial basement membrane suggests a deficiency in MMP activity.48 It could be explained by the marked decrease of MMP9 expression in shMAL-expressing cells (Figure 5A), which encodes for an important proteinase involved both in MK migration through bone marrow endothelial cells in response to SDF-1 and in the release of platelets into the blood flow.48
Discussion
MK differentiation is characterized by DNA endoreduplication, cytoplasmic maturation, and expansion. In addition, MKs must migrate in the bone marrow from the endosteal to the vascular niches, and then form long cytoplasmic extension that crosses the endothelial barrier to form platelets under the blood flow. During differentiation, MKs must progressively accumulate the main platelet proteins localized in the membrane or in secretory granules but also develop a complex cytoskeleton network to allow polyploidization, proplatelet formation, and adhesion and aggregation of future platelets. MK development is controlled by the concerted action of transcription factors, which form complexes to specifically activate or repress gene expression. The transcriptional regulation of platelet-specific genes is now well understood. It essentially depends on GATA-1/FOG-1 transcription complexes and on members of the Ets family, especially FLI-1.49 In contrast, the transcriptional regulation of cytoskeleton components as well as its relationship with the microenvironment-mediated signal transduction are still poorly understood. The role of this transcriptional regulation has been underscored by the knockout of p45NF-E2, which leads to a lethal thrombocytopenia with the absence of proplatelet formation. This profound defect is in large part the result of a defect in β1-tubulin, a direct target of p45NF-E2.49 These data have emphasized the major role of microtubules in the process of proplatelet formation.29 However, there is increasing evidence that the actomyosin cytoskeleton plays also an important function in efficient platelet production, not only by regulating proplatelet branching and amplification of platelet production but also by regulating MK polyploidization,26 demarcation membrane distribution, MK migration, and probably also platelet release.32,50
SRF is a major factor that transcriptionally regulates many cytoskeleton components. Transcriptional regulation of this set of genes by SRF depends on the myocardin family coactivators. We have used an shRNA strategy to knockdown MAL in human MKs derived from CD34+ cells and to show that MAL plays a major role in regulating target genes involved in proplatelet formation and MK migration.
In fibroblasts, MAL/SRF activation is dependent on the Rho pathway and controls many Rho-dependent cytoskeleton processes.51 Our data indicate that it might be the same in MK differentiation. We showed that, in suspension cultures, in the presence of TPO, a condition in which the Rho and Cdc42 pathways are only slightly activated, MAL is mainly distributed in the MK cytoplasm. In contrast, when the RhoA or Cdc42 pathways are activated after adherence on collagen 1 and convulxin, respectively, MAL accumulates in the nucleus. This result strongly suggests that MAL in MK, as in fibroblasts, is regulated by the actin treadmilling cycle through RhoA and Cdc42 effectors. Consequently, knockdown of MAL impairs adhesion, stress fiber, and filopodia formation in MK. This could be explained by down-regulation of β-actin, thrombospondin 1 (THSB1), and some proteins involved in a multicomponent focal adhesion complex (zyxin and vinculin). These cytoskeleton components are regulated by the MAL/SRF complex in other cell types, such as fibroblasts4 and vascular smooth muscle cells.52 THSB1 is an angiogenetic factor, which may negatively regulate MK progenitors,53 although THSB1 knockout mice have no MK/platelet defect.54 The expression of other focal adhesion proteins, such as talin, involved in GPIIb-IIIa–dependent platelet activation55 and focal adhesion kinase, a negative regulator of megakaryopoiesis,56 were not modified after MAL inhibition.
Recent reports performed in cell lines and CD34+ cells showed that MAL overexpression promotes MK differentiation and polyploidization.35 These results differ from ours in which knockdown of MAL has no significant effect on human MK differentiation, as judged by the expression of CD41 and CD42 and polyploidization. Furthermore, a tendency to increase ploidy was observed in MAL knockdown MKs, an expected result because inhibition of RhoA activity also leads to a slight increase in MK ploidy.26 It is possible that overexpression of MAL in MKs performed by Cheng et al35 modifies the SRF complexes and interferes with its association with TCFs. TCFs are important mediators of the MAP kinase pathway,6,7 which plays a major role in MK differentiation.57,58 Thus, a part of the effect of MAL overexpression might be related to an alteration in this pathway. However, Cheng et al reported also a decrease in ploidy level in bone marrow MKs of MKL1−/− mice.35 We cannot exclude that the regulation of MK polyploidization between mouse and human is different. However, it will be interesting to confirm these data in bone marrow MK from another MKL1−/− model developed by Li et al.59
In contrast, our study demonstrates a major effect of MAL knockdown on proplatelet formation and MK migration. This result is in agreement with the phenotype of MAL knockout mice,35 which have a decreased platelet count suggesting a defect in platelet biogenesis.
Gene expression profiling indicates that MAL knockdown affects the expression of 2.5% of the genes common between adult and neonate MK. To further work, we selected 2 of the 20 most down-regulated genes, MMP9 and MYL9. We have demonstrated by luciferase and ChIP experiments that the MAL/SRF complex directly regulates both MMP9 and MYL9. Very recently, Medjkane et al reported the decrease of MYL9 and MYH9 expression in breast carcinoma and melanoma cell lines as a result of siRNA-mediated MAL knockdown.60 Our study extends this report in another cell model and further supports the hypothesis that MAL/SRF regulates genes involved in actomyosin contractility.
MYL9 is a myosin light chain that is associated with MYH9, the only isoform of nonmuscle myosin II present in platelets.61 Phosphorylation of MYL9 activates MYH9 and negatively regulates proplatelet formation in normal MKs.62 In contrast, deficiency in myosin IIA leads to a defect in proplatelet formation in the MYH9 syndrome, and use of a shMYH9 induces a decrease in the percentage of proplatelet-forming MKs (Y.C. and N.D., unpublished results, February 2008). Again, a difference between mouse and human model should be noted because, although MYH9-deficient mice exhibited a marked thrombocytopenia, an increase in proplatelet-forming MKs was observed. The cytoplasm of murine MYH9 knockout MK is totally disorganized compared with WT MKs. The demarcation membrane system is abnormally dilated with a vacuole-like appearance.63 Interestingly, very similar ultrastructural abnormalities were observed in human MKs treated with the shMAL, strongly suggesting that basal regulation of MYL9 by MAL/SRF is an important determinant in the regulation of megakaryopoiesis by this transcriptional complex. This hypothesis was supported by MYL9 depletion in cultured MKs leading to inhibition of proplatelet formation.
In addition, we show that MAL is required for MK migration in response to SDF-1 through Matrigel-coated transwell but not through noncoated transwell (data not shown). This phenotype may be related to the MYL9 deficiency, which is implicated in cell motility, but even more by the marked decrease in MMP9 expression observed in MAL-deficient MKs. Indeed, MMP9 has been demonstrated to be necessary for in vitro MK migration through the basement membrane in response to SDF-1.48 In this study, the authors suggested that MMP9 may be also required for platelet release. MMP9 plays an important role in platelet functions, and its antiaggregant effect may be at the origin of hemorrhagic transformation of acute stroke. However, the large redundancy between various MMPs renders the study of their respective role in platelet functions difficult.64 Altogether, the defect in MYL9 and MMP9 induced by MAL knockout could explain the MK/platelet phenotype of MAL knockout mice recently described.35
The oncogenic OTT-MAL fusion protein has been shown to interfere with the activity of SRF in model cell lines.65 Acting as a dominant negative form of MAL, it could block the terminal stages of MK maturation affecting the expression of MMP9 and MYL9. Thus, it will be interesting to assess the role of these 2 new target genes of MAL in the phenotype of the t(1;22) acute megakaryoblastic leukemia, for example, in the context of recently developed knock-in OTT-MAL mouse model of AMKL.66
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank F. Wendling for helpful suggestions on the manuscript, Kirin Brewery (Tokyo, Japan) for human recombinant thrombopoietin, Dr Morgenstern for Megix vector (Millennium Pharmaceuticals, Inc, Cambridge, MA), M. Jandrot-Perrus (Hôpital Bichat, France) for the gift of convulxin, and T. Mercher for MAL cDNA (Hôpital Necker, France) and genomic platform of IGR for microarrays.
This work was supported by grants from Fondation de France and the Agence Nationale de la Recherche (ANR contrat blanc). L.G. was supported in part by the Association pour la Recherche sur le Cancer (ARC). D.B. was supported by a postdoctoral fellowship from ANR.
Authorship
Contribution: L.G. performed experiments, analyzed data, and wrote the paper; D.B. performed experiments and analyzed data; S.B., Y.Z., T.R., and N.D. performed experiments; Y.C. and O.A.B. discussed the results; P.D. analyzed data; W.V. designed the work and wrote the paper; and H.R. designed and supervised the experiments and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hana Raslova, Inserm, U790, 39 rue Camille Desmoulins, Villejuif, France; e-mail: hraslova@igr.fr.