Abstract
Myeloid cell leukemia-1 (MCL-1) is an essential survival factor for hematopoiesis. In humans, hematopoietic stem cells (HSCs) express MCL-1 at the highest level in response to FMS-like tyrosine kinase-3 (FLT3) signaling. We here show that this FLT3-dependent stem cell maintenance system also plays a critical role in survival of leukemic stem cells (LSCs) in acute myeloid leukemia (AML). The CD34+CD38− LSC fraction expresses high levels of FLT3 as well as MCL-1, even compared with normal HSCs. Treatment with FLT3 ligand induced further MCL-1 up-regulation in LSCs in all AML cases tested. Interestingly, the group of samples expressing the highest levels of MCL-1 constituted AML with FLT3–internal tandem duplications (ITD). In FLT3-ITD AML cell lines, cells expressed a high level of MCL-1, and an inhibition of MCL-1 induced their apoptotic cell death. A tyrosine kinase inhibitor suppressed MCL-1 expression, and induced apoptosis that was reversed by the enforced MCL-1 expression. Finally, transduction of FLT3-ITD into HSCs strongly activated MCL-1 expression through its signal transducer and activator of transcription 5 (STAT5)–docking domains. This effect was completely abrogated when STAT5 activation was blocked. Thus, the acquisition of FLT3-ITD ensures LSC survival by up-regulating MCL-1 via constitutive STAT5 activation that is independent of wild-type FLT3 signaling.
Introduction
Apoptosis is an evolutionarily conserved cell death pathway essential for homeostasis, development, and removal of damaged cells. Abnormality in genes affecting apoptosis has been found in the majority of cancers, suggesting that resistance to apoptosis, often achieved by overexpression of antiapoptotic proteins, is one of the critical factors for cancer development.1,2 B-cell leukemia 2 (BCL-2) family proteins regulate survival of many types of normal and malignant cells.3,4 In fact, in collaboration with other oncogenes, the enforced expression of BCL-2 can significantly enhance the development of acute myeloid leukemia (AML) in several murine models,5-7 suggesting that deregulation of cell survival plays a critical role in development of leukemias.
Recent studies have shown that myeloid cell leukemia-1 (MCL-1)8 is the most critical BCL-2 family molecule for hematopoietic stem cell (HSC) survival. We have reported that MCL-1 is expressed in HSCs at the highest level in murine hematopoiesis, and is essential for maintenance of cell survival in HSCs, based on the fact that conditional disruption of Mcl-1 in HSCs induced a complete ablation of hematopoiesis.9 Human HSCs also express MCL-1 at the highest level.10 In human lymphoid malignancies such as chronic lymphocytic leukemia and multiple myeloma, the high level of MCL-1 transcripts in malignant cells is a useful prognostic marker.11-14 Suppression of MCL-1 but not of BCL-2 or BCL-xL triggered rapid apoptosis in chronic lymphocytic leukemia and myeloma cells,13,15,16 suggesting that MCL-1 could be a therapeutic target in these lymphoid malignancies.
In early hematopoiesis, MCL-1 expression is regulated at least by cytokines. Human HSCs and early myelolymphoid progenitors express type III receptor tyrosine kinase family proteins including FMS-like tyrosine kinase-3 (FLT3) and steel factor receptor (c-KIT),10 and ligands for either receptor stimulate HSCs to express MCL-1, but not BCL-2 or BCL-xL.10 FLT3 ligation induces receptor dimerization, triggers 2 major signal transduction pathways including phosphatidylinositol 3-kinase (PI3K)/Akt, and RAS/mitogen-activated protein kinase (MAPK) pathways, and provides growth-stimulating and antiapoptotic signals.17
In AML, FLT3 is expressed in AML blasts at a high level in most cases.18 Of note, FLT3 mutation is one of the most frequent genetic alterations in AML19 and marks a fraction of patients with poor prognosis.20-22 FLT3 internal tandem duplication mutations (FLT3-ITD) induce constitutive activation of FLT3 signaling, and retroviral transduction of FLT3-ITD in murine bone marrow cells results in progression of myeloproliferative disorders,23-29 but these mutations themselves do not cause AML.24 In contrast to the wild-type FLT3 (FLT3-WT), FLT3-ITD is a potent activator of signal transducer and activator of transcription 5 (STAT5). Of note, the STAT5 Src homology 2 docking phosphorylation sites in the juxtamembrane domain of FLT3-ITD, tyrosines 589 and 591, which do not exist within the FLT3-WT, are necessary to recapitulate myeloproliferation in a FLT3-ITD knock-in mouse model.30
In this paper, we demonstrate that the leukemic stem cell (LSC) fraction of AML cells expresses a high level of MCL-1 even compared with normal HSCs, suggesting that AML LSCs use MCL-1 as a critical survival factor. AML LSCs expressed FLT3 and FLT3-ITD on their surface. MCL-1 expression in LSCs was further up-regulated by treatment with exogenous FLT3 ligands. Interestingly, LSCs with FLT3-ITD express extremely high levels of MCL-1, a process mediated by STAT5 signaling pathways that are not used in the FLT3-WT. Thus, both normal FLT3 and FLT3-ITD signaling maintain MCL-1 expression in LSCs presumably to prolong their survival, but FLT3-ITD exerts more potent effects on MCL-1 induction using a signaling pathway independent of FLT3-WT.
Methods
Patients
Bone marrow (BM) samples were analyzed from 30 adult patients with primary AML diagnosed according to French-American-British (FAB) criteria in Kyushu University Hospital. BM mononuclear cells were prepared by gradient centrifugation and cryopreserved until use for this study. Informed consent was obtained from all subjects in accordance with the Declaration of Helsinki. The Institutional Review Board of Kyushu University Hospital approved all research on human subjects.
Analysis of FLT3-ITD mutations
For detection of FLT3-ITD, polymerase chain reaction (PCR) amplification of genomic DNA was carried out using primers 11F and 12R located on FLT3 exons 14 and 15 as previously described.31 PCR products were separated by electrophoresis using 3% Nusieve GTG agarose gel (Cambrex Bio Science Rockland) and cut out from the gel, purified with a QIAquick Gel Extraction kit (QIAGEN), and cloned into the PCR II-TOPO vector (Invitrogen) according to the manufacturer's recommendations. Sequencing was performed using the ABI PRISM BigDye Terminator V 1.1 Cycle Sequencing Kit and 21M13 and T7 primer on an ABI 310 Prism sequencer (Applied Biosystems).
Antibodies, cell staining, and sorting
CD34+ cells were enriched from BM cells using the Indirect CD34 MicroBead Kit (Miltenyi Biotec) as described previously.32 For analyses and sorting, cell samples were stained with cyanin 5–phycoerythrin (PE)– or phycoerythrin-Cy5–conjugated lineage mixture, including cyanin 5–PE– or phycoerythrin-Cy5–conjugated CD3 (HIT3a), CD4 (RPA-T4), CD8 (RPA-T8), CD10 (HI10a), CD20 (2H7), CD11b (ICFR44), CD14 (RMO13), GPA (GA-R2), fluorescein isothiocyanate–conjugated anti-CD90 (Thy-1) (5E10) or anti-CD45RA (HI100), PE-conjugated anti-CD117 (c-Kit) (YB5.B8) or anti-FLT3 (CD135) (4G8), anti-CD123 (interleukin-3Rα [IL-3Rα]) (6H6), allophycocyanin–conjugated anti-CD34 (8G12), and biotin-conjugated anti-CD38 (HIT2) antibodies. Streptavidin-conjugated allophycocyanin was used for visualization of the biotinylated antibodies (BD Pharmingen). Nonviable cells were excluded by propidium iodide (PI) staining. HSCs (CD34+CD38−CD90+Lin− cells) and myeloid progenitors, including common myeloid progenitors (CD34+CD38+CD123+ CD45RAloLin−), granulocyte-macrophage progenitors (GMPs: CD34+CD38+CD123+ CD45RAloLin−), and megakaryocyte-erythroid progenitors (CD34+CD38+CD123−CD45RA−Lin−) were isolated from normal and AML BM cells by fluorescence-activated cell sorter (FACS, FACSAria; BD Biosciences).32,33 The sorted cells were subjected to an additional round of sorting using the same gate to eliminate contaminating cells and doublets. HSCs and GMPs in the murine bone marrow were purified according to the method described previously.34
Cell culture and apoptosis measurement
To test the effect of FLT3 stimulation on the leukemic cells, the sorted cells were prepared in the fetal calf serum (FCS)–free condition media with human (h) FLT3 ligand (FL, 20 ng/mL) or human stem cell factor (SCF, 20 ng/mL). After culture of 30 minutes, cells were subject to analysis of gene expression. Leukemic cell lines, including FLT3-ITD–positive cell lines, MV4-11, were cultured in RPMI-1640 medium containing 10% fetal calf serum (FCS) in the absence or presence of PKC-412 (LC Laboratories), Ly294002 (EMD Biosciences), AG490 (EMD Biosciences), or U0126 (Cell Signaling Technology). Cultured cell lines were picked up for analyses of changes in transcript and protein expression and apoptotic status. Apoptosis of cell lines treated with PKC-412 was evaluated using annexin V (BD Pharmingen) and PI staining. The living cells were defined as annexin V−/PI− among the live-gated cells.
Quantitative real-time PCR assay
Total RNA was extracted from 2000 purified cells using Isogen reagent (Nippon Gene) according to the manufacturer's protocols. RNA was reverse transcribed to cDNA using a TaKaRa RNA PCR Kit (Takara). The mRNA levels were quantified in triplicate using a real-time PCR (7500 Real-Time PCR system; Applied Biosystems). β2-Microglobulin mRNA was separately amplified in the same plate to be used for internal control. The primers and probes were designed by Primer Express software (Applied Biosystems) as follows: BCL-2 (the forward primer, 5-ctgggatgcctttgtggaa-3; the reverse primer, 5-cagccaggagaaatcaaacaga-3; and the probe, 5-FAM-tgtacggccccagcatgcgg-TAMRA-3), BCL-xL (the forward primer, 5-aatgaccacctagagccttgga-3; the reverse primer, 5-tgctgcattgttcccatagagt-3; and the probe, 5-FAM-cggcggctgggatacttttg-TAMRA-3), MCL-1 (the forward primer, 5-caaaacgggactggctagttaaa-3; the reverse primer, 5-ccttctaggtcctctacatggaaga-3; and the probe, 5-FAM-aaagaggctgggatgggtttgtggag-TAMRA-3), and β2-microglobulin (the forward primer, 5-tgactttgtcacagcccaagata-3; the reverse primer, 5-aatgcggcatcttcaaacct-3; and the probe, 5-FAM-ttaagtgggatcgagacatgtaagcagcatc-TAMRA-3). Collected data were analyzed with Sequence Detector software (Applied Biosystems). All the samples and PCR analyses were carried out in duplicate, and negative controls were included in each run. Data were expressed as fold changes in gene expression relative to those from normal BM or normal HSCs.
Intracellular flow cytometry
Cells were fixed and permeabilized, and labeled with the rabbit polyclonal antibody MCL-1 (S-19; Santa Cruz Biotechnology), followed by the second staining with fluorescein isothiocyanate– or PE-conjugated antirabbit antibodies for visualization.
Western blot analysis
Cells (109) were lysed as described.35 Lysates were then denatured in an equal volume of 2× sodium dodecyl sulfate (SDS) sample buffer, resolved by a 10% SDS–polyacrylamide gel electrophoresis gel, and electrotransferred to nitrocellulose membranes in non–SDS-containing transfer buffer (25mM tris(hydroxymethyl)aminomethane, 0.2M glycine, 20% methanol, pH 8.5). Western blotting was performed with rabbit anti–human MCL-1 sera (1:5000; Sigma-Aldrich), anti–human phospho-Akt (Cell Signaling Technology), anti–human phospho-mitogen-activated protein kinase kinase 1/2 (MEK1/2; Cell Signaling Technology), or mouse anti–β-actin monoclonal antibody (1:5000; Sigma-Aldrich), followed by 1:15 000 dilution of antirabbit or antimouse horseradish peroxidase–conjugated immunoglobulin G (Jackson ImmunoResearch Laboratories). Blots were developed with enhanced chemiluminescence plus kit (Amersham Biosciences). The chemiluminescence intensity was monitored by laser3000 (FujiFilm) instrument. We quantitated band intensity of the proteins using ImageGauge software (FujiFilm) and normalized their expression in reference to β-actin levels. Using these normalized data, relative expression (RE) is subsequently calculated as fold changes in protein expression compared with the controls.
Retroviral transduction of HSCs and progenitors
The constructs of FLT3-ITD N51and FLT3-ITD tyrosine to phenylalanine mutant at sites 589 and 591, which are occult STAT5 binding sites, were previously described.30 The retrovirus vector was transfected into 293T cells with gag and pol expression plasmids by CaPO4 coprecipitation. The supernatant from transfected cells was collected after 48 hours and immediately stored at −80°C until use. FACS-purified murine HSCs were prestimulated in RPMI-1640 medium containing 20% FCS in the presence of murine (m) SCF (20 ng/mL), murine leukemia inhibitory factor (10 ng/mL) and mFL (10 ng/mL) for 18 hours. Cells were subsequently plated onto recombinant fibronectin fragment-coated culture dishes (RetroNectin dish; Takara) with 1 mL of virus supernatant containing identical cytokine cocktail and cultured for 48 hours. For progenitor transduction, FACS-purified cells were cultured for 48 hours on RetroNectin dishes with 1 mL of virus supernatant containing mSCF and mIL-11 (10 ng/mL) for GMPs
The enforced expression and knockdown experiments for MCL-1
Human MCL-1 cDNA plasmid was purchased from Open Biosystems. The coding sequence of MCL-1 in this plasmid was amplified by PCR using primers containing additional EcoRI site (5′-cggaattcggcaatgtttggcctc-3′) and BamHI site (5′-gcggatcctcttattagatatgc-3′) and cloned into pEGFP-C3 vector (Clontech). To suppress the expression level of MCL-1, we generated a vector that possesses expression cassettes of short hairpin (sh) RNA against MCL-1 using the BLOCK-iT PolII miR RNAi Expression Kit (Invitrogen) according to the manufacturer's recommendation. Target sequences were selected using BLOCK-iT RNAi Designer (Invitrogen). Briefly, oligonucleotides were synthesized to form shRNA on transcription, in order of sense, loop (lower case), and antisense sequences as follows: MCL-1 top strand oligo, 5′-tgctgttgatgtccagtttccgaagcgttttggccactgactgacgcttcggactggacatcaa-3′; MCL-1 bottom strand oligo, 5′-cctgttgatgtccagtccgaagcgtcagtcagtggccaaaacgcttcggaaactggacatcaac-3′. These oligonucleotides were annealed with their complementary ones and cloned subsequently into pcDNA 6.2-GW/EmGFP-miR expression vector. After confirming their sequences, expression vectors were transduced into MV4-11 cell lines by Amaxa Nucleofector System (Lonza). Cells (2 × 106) were suspended in 100 μL of Nucleofector solution with 2 μg of negative control vector or subcloned plasmid. Pulses were delivered with Nucleofector device using program Q-001 (Lonza).
STAT5 knockdown experiment
Double-stranded STAT5 siRNA was purchased from Invitrogen. STAT5 siRNA was transduced into MV4-11 cell lines by Amaxa Nucleofector System (Lonza). Cells (2 × 106) were suspended in 100 μL of Nucleofector solution with 1.5 μg of siRNA. Pulses were delivered with Nucleofector device using program Q-001.
Statistical analysis
Levels of significance were measured using paired t test.
Results
The phenotype of AML LSCs is preserved irrespective of FLT3-ITD expression
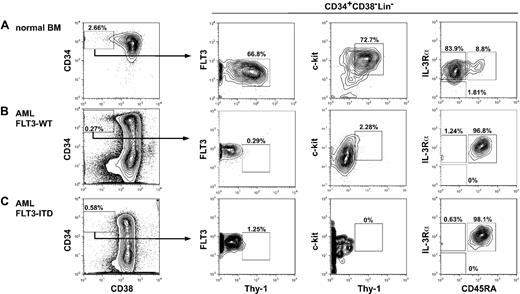
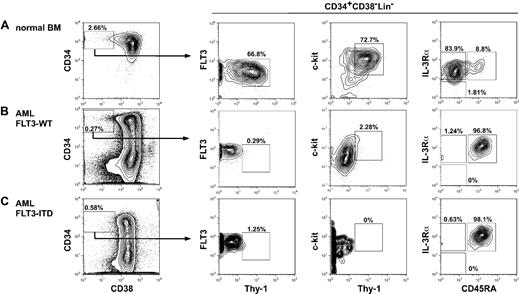
Among 30 AML samples enrolled in the study, FLT3-ITD was detected in 11 of 30 AML patients. The phenotypic characteristics are consistent in all AML cases analyzed irrespective of the presence of FLT3-ITD. Figure 1 shows the phenotypic analysis of normal and AML bone marrow cells. Patients' characteristics and their FACS data are summarized in Table 1. In normal BM, the HSC resides in the CD34+CD38−Lin− fraction that expresses Thy-1 (CD90) and c-KIT (CD117).36-38 These CD34+CD38− human HSCs uniformly express FLT3 (CD135), CD45RA−, and IL-3Rαlo (CD123)10 (Figure 1). Previous xenogeneic transplantation experiments showed that leukemia-initiating cells (leukemic stem cells [LSCs]) reside predominantly within the CD34+CD38−Lin− fraction of the AML bone marrow.39,40 Interestingly, the entire CD34+CD38− AML LSC fraction expressed FLT3 at a high level, whereas the majority of CD34+CD38− LSCs expressed diversified levels of c-KIT in all 30 AML cases studied. CD34+CD38− LSCs were negative for CD90 as reported.41,42 They expressed a high level of CD45RA and CD123, resembling the phenotype of human GMPs32 (Figure 1). Because anti-FLT3 antibodies react with the extracellular portion of FLT3-ITD as well as FLT3-WT, it is of note that FLT3-ITD protein is expressed in the CD34+CD38− AML LSC fraction, indicating that FLT3-ITD signaling should operate at the LSC stage.
Stem and progenitor analysis of normal and AML bone marrow cells. Representative phenotype of the bone marrow cells in a normal control (A), FLT3-WT AML (B), and FLT3-ITD AML (C). Human CD34+CD38− cells are enriched for normal HSCs with long-term reconstitution activity, and are Thy-1+FLT3loc-KIT+IL-3R+CD45RA−. In contrast, CD34+CD38− AML LSCs were Thy-1− and expressed a high level of FLT3 compared with normal HSCs. The LSCs expressed a variety of levels of c-KIT, and were IL-3R+CD45RA+, resembling GMPs' phenotype. No significant difference in expression level of these molecules was found between FLT3-ITD and FLT3-WT AML LSCs.
Stem and progenitor analysis of normal and AML bone marrow cells. Representative phenotype of the bone marrow cells in a normal control (A), FLT3-WT AML (B), and FLT3-ITD AML (C). Human CD34+CD38− cells are enriched for normal HSCs with long-term reconstitution activity, and are Thy-1+FLT3loc-KIT+IL-3R+CD45RA−. In contrast, CD34+CD38− AML LSCs were Thy-1− and expressed a high level of FLT3 compared with normal HSCs. The LSCs expressed a variety of levels of c-KIT, and were IL-3R+CD45RA+, resembling GMPs' phenotype. No significant difference in expression level of these molecules was found between FLT3-ITD and FLT3-WT AML LSCs.
The presence of FLT3-ITD marks a fraction of AML with highest levels of MCL-1 expression
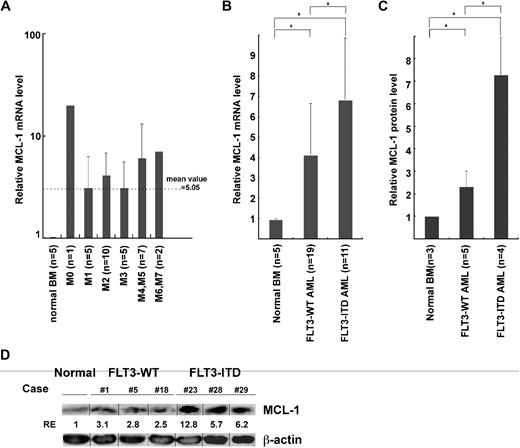
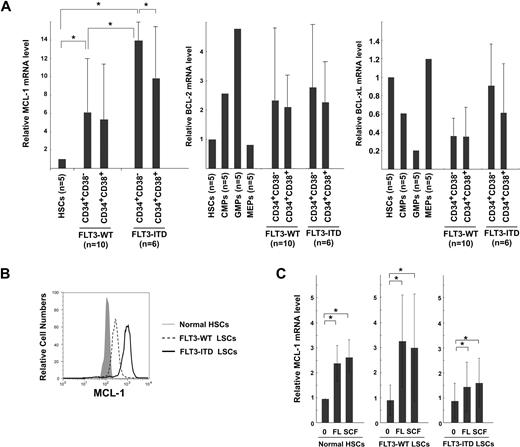
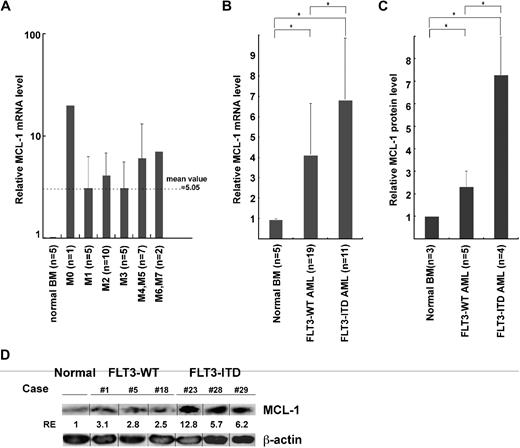
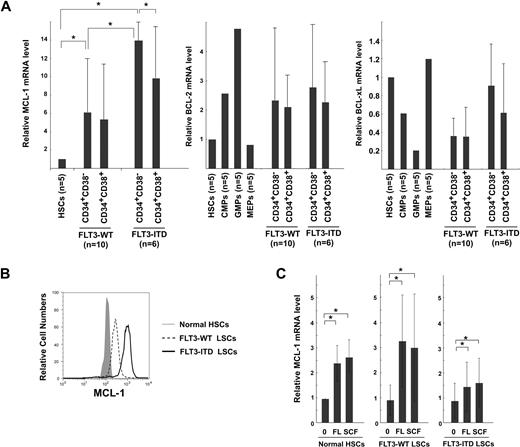
Figure 2 shows the real-time PCR analysis of MCL-1 mRNA levels in normal and leukemic BM mononuclear cells. AML BM cells expressed MCL-1 in all 30 cases at a level approximately 5-fold higher than that of normal BM cells, irrespective of their FAB types (Figure 2A). We have shown that, in normal human hematopoiesis, FLT3 signaling up-regulates MCL-1 but not BCL-2 and BCL-xL in HSCs and myeloid progenitors, promoting their cell survival.10 Therefore, we hypothesized that the constitutive expression of FLT3 signaling by FLT3-ITD mutation can induce up-regulation of MCL-1 in the AML LSC fraction. As shown in Figure 2B, the level of MCL-1 mRNA in FLT3-ITD AML BM cells was higher than that of FLT3-WT AML by approximately 2-fold, suggesting that the presence of FLT3-ITD can mark a fraction of AML with a high level of MCL-1. The high-level expression of MCL-1 protein in FLT3-ITD AML blasts was confirmed by Western blot analysis (Figure 2C-D). FLT3-ITD AML cells expressed approximately 7- and approximately 3-fold higher levels of MCL-1 protein compared with normal BM and FLT3-WT AML cells, respectively. In human hematopoiesis, HSCs express the highest level of MCL-1, and its expression gradually declines as they differentiate into mature myeloid or lymphoid cells.10 We therefore further subfractionated leukemic BM cells into CD34+CD38− LSCs and CD34+CD38+ leukemic progenitor cell fractions. We could obtain sufficient numbers of cells for such analysis in 16 samples (10 cases of FLT3-WT and 6 cases of FLT3-ITD AMLs). As shown in Figure 3A, the CD34+CD38− LSC fraction in FLT3-WT AMLs expressed approximately 6-fold higher, whereas those in FLT3-ITD AMLs had approximately 14-fold higher level of MCL-1, even compared with normal HSCs. In contrast, levels of BCL-2 and BCL-xL in the CD34+CD38− LSCs and normal HSCs were not significantly different (Figure 3A). Cytoplasmic staining of MCL-1 FLT3-ITD AML LSC fraction shows that this population uniformly expressed MCL-1 protein at a high level on FACS, indicating that all single cells possessed high levels of MCL-1 (Figure 3B). Collectively, the CD34+CD38− LSC fraction expressed the highest levels of MCL-1 in the leukemic BM.
The expression of MCL-1 in FLT3-ITD and FLT3-WT AML cells. The levels of Mcl-1 transcripts and proteins in AML bone marrow cells. (A) AML cells express ∼ 5-fold higher levels of MCL-1 compared with normal BM cells irrespective of their FAB types by real-time PCR assays. (B) FLT3-ITD AML cells expressed ∼ 2-fold higher levels of MCL-1 in average compared with those in FLT3-WT AML cells. (C) The level of MCL-1 protein on Western blot analysis in primary AML cells. Results are shown as mean ± SD (*P < .05). (D) Representative Western blot data of MCL-1 in FLT3-WT AML (cases 1, 5, and 8) and FLT3-ITD AML (cases 23, 28, and 29) samples. Vertical lines have been inserted to indicate a repositioned gel lane.
The expression of MCL-1 in FLT3-ITD and FLT3-WT AML cells. The levels of Mcl-1 transcripts and proteins in AML bone marrow cells. (A) AML cells express ∼ 5-fold higher levels of MCL-1 compared with normal BM cells irrespective of their FAB types by real-time PCR assays. (B) FLT3-ITD AML cells expressed ∼ 2-fold higher levels of MCL-1 in average compared with those in FLT3-WT AML cells. (C) The level of MCL-1 protein on Western blot analysis in primary AML cells. Results are shown as mean ± SD (*P < .05). (D) Representative Western blot data of MCL-1 in FLT3-WT AML (cases 1, 5, and 8) and FLT3-ITD AML (cases 23, 28, and 29) samples. Vertical lines have been inserted to indicate a repositioned gel lane.
MCL-1 expression and its regulation in AML LSCs. (A) Real-time PCR analysis of MCL-1, BCL-2, and BCL-xl expression within fractions of CD34+ cells. The LSC-enriched CD34+CD38− fraction in FLT3-WT AML (cases 1, 3-5, 8-10, 16, 18, and 19) expressed Mcl-1 at a level > 6-fold higher than that of normal HSCs, whereas LSCs in FLT3-ITD AML (cases 20, 22-24, 27, and 30) expressed even ∼ 2-fold higher levels of MCL-1 compared with FLT3-WT LSCs. In contrast, BCL-2 or BCL-xL expression was not significantly changed. (B) Cytoplasmic staining of MCL-1 in CD34+CD38− AML fractions with FLT3-ITD and FLT3-WT. A high amount of MCL-1 protein is detectable in FLT3-ITD LSCs. Representative FLT3-ITD and FLT3-WT LSCs are shown. (C) Changes in the expression level of MCL-1 in normal HSCs, LSCs of FLT3-WT AML, and of FLT3-ITD AML after incubation with FL or with SCF. Each bar represents an n-fold difference in the amount of MCL-1 expression prior to stimulation of normal HSCs. Results are mean ± SD (*P < .05).
MCL-1 expression and its regulation in AML LSCs. (A) Real-time PCR analysis of MCL-1, BCL-2, and BCL-xl expression within fractions of CD34+ cells. The LSC-enriched CD34+CD38− fraction in FLT3-WT AML (cases 1, 3-5, 8-10, 16, 18, and 19) expressed Mcl-1 at a level > 6-fold higher than that of normal HSCs, whereas LSCs in FLT3-ITD AML (cases 20, 22-24, 27, and 30) expressed even ∼ 2-fold higher levels of MCL-1 compared with FLT3-WT LSCs. In contrast, BCL-2 or BCL-xL expression was not significantly changed. (B) Cytoplasmic staining of MCL-1 in CD34+CD38− AML fractions with FLT3-ITD and FLT3-WT. A high amount of MCL-1 protein is detectable in FLT3-ITD LSCs. Representative FLT3-ITD and FLT3-WT LSCs are shown. (C) Changes in the expression level of MCL-1 in normal HSCs, LSCs of FLT3-WT AML, and of FLT3-ITD AML after incubation with FL or with SCF. Each bar represents an n-fold difference in the amount of MCL-1 expression prior to stimulation of normal HSCs. Results are mean ± SD (*P < .05).
FLT3 signaling stimulates MCL-1 expression in AML LSCs
Both FLT3 and c-KIT belong to the class III receptor tyrosine kinase family and share their major signaling cascade. Signals from either receptor play a critical role in maintenance of cell survival in normal HSCs and myeloid progenitors such as common myeloid progenitors and GMPs through MCL-1 up-regulation.10 In fact, both FLT3 and c-KIT were expressed in the CD34+CD38− LSC fraction, and their expression levels appear to be equal in FLT3-ITD and FLT3-WT AMLs (Figure 1). We thus tested whether ligation of FLT3 and c-KIT receptors can induce further up-regulation of MCL-1 in AML LSCs in 5 cases each of FLT3-WT and FLT3-ITD AMLs. As shown in Figure 3C, both FLT3 ligand and SCF, a ligand for c-KIT, significantly up-regulated MCL-1 in LSCs. This effect was observed in both FLT3-WT and FLT3-ITD AMLs. In contrast, BCL-2 or BCL-xL expression was not significantly changed (not shown). A combination of FLT3 ligand and SCF did not show further additive effects on MCL-1 levels on LSCs (not shown). These data strongly suggest that signals of tyrosine kinase receptor family such as FLT3 and c-KIT can stimulate MCL-1 transcription in AML LSCs, presumably to prevent their apoptotic cell death.
MCL-1 is required for FLT3-ITD leukemic cells to maintain cell viability
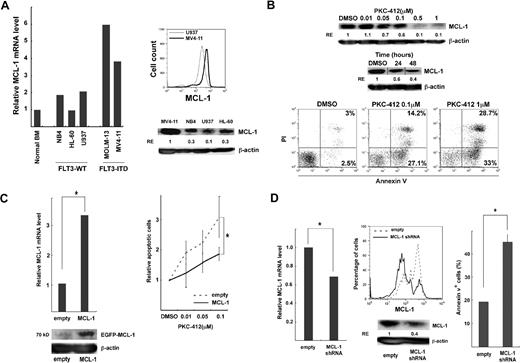
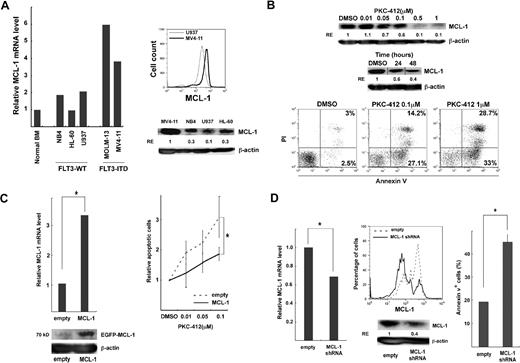
Leukemic cell lines with FLT3-ITD, such as MV4-11, were used to analyze antiapoptotic functions of FLT3-ITD. The level of MCL-1 transcripts and proteins in MV4-11 cells was higher than that in normal BM controls, or other cell lines without FLT3 mutations such as NB4, HL-60, and U937 (Figure 4A), suggesting that FLT3-ITD signals up-regulate MCL-1 expression. MV4-11 was then treated with the tyrosine kinase inhibitor, PKC-412. PKC-412 down-regulated the expression of MCL-1 protein in a dose-dependent fashion (Figure 4B top). At the concentration of 0.5μM, MCL-1 was progressively inhibited (Figure 4B top), and reached plateau at 48 hours (not shown). PKC-412 induced apoptotic cell death of MV4-11 cells 24 hours after initiation of culture in a dose-dependent manner (Figure 4B bottom).
Functional analysis of MCL-1 in FLT3-ITD AML. (A) MCL-1 expression in cell lines with FLT3-WT (NB4, HL-60, U937) and FLT3-ITD (MV4-11 and MOLM-13). High levels of MCL-1 mRNA and protein were seen in MV4-11 cells by real-time PCR analysis (left panel), the intracellular MCL-1 staining, and Western blot analysis (right panels). (B) The inhibition of MCL-1 protein (top) and cell survival (bottom) in MV4-11 after PKC-412 treatment. The inhibitory effect of PKC-412 reached plateau at a concentration of 0.5μM. Apoptotic cell death was evaluated by annexin/PI staining. Vertical lines have been inserted to indicate a repositioned gel lane. (C) The enforced expression of MCL-1 prevented FLT3-ITD cell lines from apoptosis in the presence of PKC-412. MCL-1 transcript and protein expression were significantly up-regulated 48 hours after transduction of MCL-1 into MV4-11 cells. PKC-412–induced apoptotic cell death is significantly inhibited in MV4-11 cells (right panel) (*P < .05). (D) Inhibition of MCL-1 by shRNA resulted in an apoptosis of FLT3-ITD cell lines. Forty-eight hours after transduction of MCL-1–shRNA–EGFP into the MV4-11 cells, MCL-1 transcript (left) as well as MCL-1 protein expression on FACS and Western blot analysis (middle) was significantly down-regulated. In parallel, the number of apoptotic cells increased by ∼ 2-fold compared with control (right). Results are shown as mean ± SD (*P < .05). RE indicates relative expression levels.
Functional analysis of MCL-1 in FLT3-ITD AML. (A) MCL-1 expression in cell lines with FLT3-WT (NB4, HL-60, U937) and FLT3-ITD (MV4-11 and MOLM-13). High levels of MCL-1 mRNA and protein were seen in MV4-11 cells by real-time PCR analysis (left panel), the intracellular MCL-1 staining, and Western blot analysis (right panels). (B) The inhibition of MCL-1 protein (top) and cell survival (bottom) in MV4-11 after PKC-412 treatment. The inhibitory effect of PKC-412 reached plateau at a concentration of 0.5μM. Apoptotic cell death was evaluated by annexin/PI staining. Vertical lines have been inserted to indicate a repositioned gel lane. (C) The enforced expression of MCL-1 prevented FLT3-ITD cell lines from apoptosis in the presence of PKC-412. MCL-1 transcript and protein expression were significantly up-regulated 48 hours after transduction of MCL-1 into MV4-11 cells. PKC-412–induced apoptotic cell death is significantly inhibited in MV4-11 cells (right panel) (*P < .05). (D) Inhibition of MCL-1 by shRNA resulted in an apoptosis of FLT3-ITD cell lines. Forty-eight hours after transduction of MCL-1–shRNA–EGFP into the MV4-11 cells, MCL-1 transcript (left) as well as MCL-1 protein expression on FACS and Western blot analysis (middle) was significantly down-regulated. In parallel, the number of apoptotic cells increased by ∼ 2-fold compared with control (right). Results are shown as mean ± SD (*P < .05). RE indicates relative expression levels.
We then tested whether reinforcement of MCL-1 can rescue FLT3-ITD cells from PKC-412–induced apoptosis. Expression vectors harboring MCL-1–enhanced green fluorescent protein and control EGFP were transduced into these FLT3-ITD AML cell lines. Forty-eight hours after transduction, an amount of MCL-1 mRNA in FLT3-ITD–EGFP+ cells was increased up to approximately 3-fold compared with those with a control vector (Figure 4C left). A significant increase in MCL-1 protein was also observed after transduction of MCL-1–EGFP vector (Figure 4C left). Purified FLT3-ITD–EGFP+ cells were incubated with graded concentrations of PKC-412 for 24 hours. PKC-412–induced apoptosis was significantly inhibited in cells transduced with MCL-1 transgene (Figure 4C right; supplemental Figure 1A, available on the Blood website; see the Supplemental Materials link at the top of the online article), suggesting that maintenance of MCL-1 level is critical for cell survival under this condition. We then inhibited MCL-1 expression in FLT3-ITD AML cell lines by transducing an shRNA expression vector. Vectors having MCL-1–shRNA–EGFP or control EGFP were transduced and the sorted live cells (EGFP+) were analyzed at 48 hours (Figure 4D). The amount of MCL-1 transcript reduced to approximately 60% of a control level. MCL-1 protein expression was also significantly reduced on FACS and Western blot analyses (Figure 4D middle). In parallel, numbers of apoptotic cells within MCL-1–shRNA–EGFP+ cells were increased up to approximately 2-fold compared with those in a control (Figure 4D right; supplemental Figure 1B). These results collectively suggest that FLT3-ITD supports leukemic cell survival by up-regulating MCL-1 production.
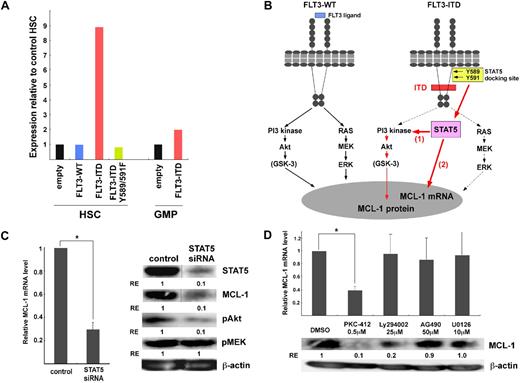
Enforced expression of FLT3-ITD stimulates MCL-1 expression in HSCs via the STAT5-dependent signaling pathway
To confirm that FLT3-ITD signals up-regulate MCL-1 expression, we transduced FLT3-ITD into purified murine HSCs and myeloid progenitors. Cells were purified from C57BL6/Ka-Thy1.1 mice and were infected with retrovirus vectors with FLT3-ITD-GFP or FLT3-WT-GFP constructs. Thirty-six hours after retroviral transduction, GFP+ cells were purified, and MCL-1 transcripts were quantified. In transducing retroviruses, we cultured cells with SCF, leukemia inhibitory factor, and FL, and these factors themselves have potential to stimulate MCL-1 expression.9 In fact, control HSCs infected with the empty vector up-regulated MCL-1 mRNA up to 4-fold after this treatment (not shown). HSCs transduced with FLT3-WT expressed an amount of MCL-1 transcript almost equal to HSCs with an empty vector, suggesting that HSCs with retroviral transduction have up-regulated MCL-1 expression presumably via normal FLT3 and c-KIT signaling during cultures. In contrast, HSCs transduced with FLT3-ITD further up-regulated MCL-1 expression at a level approximately 9-fold higher than those of HSCs infected with control or FLT3-WT retroviruses (Figure 5A). BCL-2 and BCL-xL mRNA levels were not significantly changed by this treatment (not shown). The up-regulation of MCL-1 transcripts by FLT3-ITD transduction was also seen in GMPs, but the effect was less significant compared with that in HSCs, suggesting that HSCs are the favorable target for FLT3-ITD signals to up-regulate MCL-1 transcripts. Importantly, because FLT3-ITD further stimulated MCL-1 expression in the presence of normal c-KIT and FLT3 signaling, it is suggested that a novel signaling cascade independent of the pathways induced by wild-type tyrosine kinase receptors should operate to up-regulate MCL-1 expression in HSCs transduced with FLT3-ITD (Figure 5A). Previous reports have shown that FLT3-ITD possessed 2 STAT5-docking domains, whereas FLT3-WT does not.27,43-45 We have shown that activation of STAT5 is necessary for FLT3-ITD to induce myeloproliferation, because transfection of the FLT3-ITD–Y589/591F mutant, in which tyrosine residues 589 and 591 located in each of these STAT5-docking sites (Figure 5B) were substituted by phenylalanine, could not induce myeloproliferative disorder in a mouse model.30 We therefore hypothesized that the FLT3-ITD–specific MCL-1 activation might occur via its STAT5-docking domains. As shown in Figure 5A, up-regulation of MCL-1 transcripts is completely abrogated by transducing the FLT3-ITD–Y589/591F mutant instead of FLT3-ITD.
FLT3-ITD activates MCL-1 expression via STAT5 signaling. (A) FLT3-WT, FLT3-ITD, and FLT3-ITD-Y589/591F mutants were transduced into purified murine HSCs and GMPs to test changes in the amount of MCL-1 transcripts. During the transduction, cells were cultured for 48 hours in the presence of FLT3 and SCF. In this condition, the amount of MCL-1 transcripts in HSCs transfected with FLT3-WT was almost equal to HSCs with an empty vector. Significant up-regulation of MCL-1 (∼ 9-fold higher) was seen when HSCs were transfected with FLT3-ITD. This effect was more pronounced in HSCs than GMPs. The up-regulation of MCL-1 was completely inhibited in HSCs transduced with FLT3-ITD-Y589/591F mutant, indicating that FLT3-ITD–specific STAT5-docking phosphorylation sites are necessary to stimulate MCL-1 transcription. (B) Schematic presentation of signaling pathways of FLT3-ITD to induce MCL-1 expression. In FLT3-WT, FLT3 ligation triggers both PI3K/Akt and RAS/MAPK pathways. In contrast, FLT3-ITD induces STAT5-dependent MCL-1 expression. STAT5 might induce MCL-1 expression by PI3K/Akt activation presumably through posttranscriptional mechanism (1), and by directly stimulating MCL-1 transcription (2). These 2 distinct FLT3-ITD signaling pathways for MCL-1 expression contribute to the development of AML. (C) STAT5 siRNA was transduced into MV4-11 cell lines to suppress STAT5 expression. The sorted live cells were analyzed at 48 hours after STAT5 siRNA transduction. STAT5 siRNA suppressed the expression of MCL-1 transcript and protein. STAT5 siRNA also suppressed phospho-Akt, but not phospho-MEK, pathway. Vertical lines have been inserted to indicate a repositioned gel lane. Results are shown as mean ± SD (*P < .05) (D) The expression of MCL-1 in MV4-11 24 hours after treatment with pharmacologic inhibitors of PKC (PKC-412), PI3K (Ly294002), JAK2 (AG490), or MEK (U0126). PKC-412 inhibited MCL-1 mRNA expression. PKC-412 and Ly294002 but not U0126 or AG490 inhibited the MCL-1 protein expression. RE indicates relative expression levels. Results are shown as mean ± SD (*P < .05).
FLT3-ITD activates MCL-1 expression via STAT5 signaling. (A) FLT3-WT, FLT3-ITD, and FLT3-ITD-Y589/591F mutants were transduced into purified murine HSCs and GMPs to test changes in the amount of MCL-1 transcripts. During the transduction, cells were cultured for 48 hours in the presence of FLT3 and SCF. In this condition, the amount of MCL-1 transcripts in HSCs transfected with FLT3-WT was almost equal to HSCs with an empty vector. Significant up-regulation of MCL-1 (∼ 9-fold higher) was seen when HSCs were transfected with FLT3-ITD. This effect was more pronounced in HSCs than GMPs. The up-regulation of MCL-1 was completely inhibited in HSCs transduced with FLT3-ITD-Y589/591F mutant, indicating that FLT3-ITD–specific STAT5-docking phosphorylation sites are necessary to stimulate MCL-1 transcription. (B) Schematic presentation of signaling pathways of FLT3-ITD to induce MCL-1 expression. In FLT3-WT, FLT3 ligation triggers both PI3K/Akt and RAS/MAPK pathways. In contrast, FLT3-ITD induces STAT5-dependent MCL-1 expression. STAT5 might induce MCL-1 expression by PI3K/Akt activation presumably through posttranscriptional mechanism (1), and by directly stimulating MCL-1 transcription (2). These 2 distinct FLT3-ITD signaling pathways for MCL-1 expression contribute to the development of AML. (C) STAT5 siRNA was transduced into MV4-11 cell lines to suppress STAT5 expression. The sorted live cells were analyzed at 48 hours after STAT5 siRNA transduction. STAT5 siRNA suppressed the expression of MCL-1 transcript and protein. STAT5 siRNA also suppressed phospho-Akt, but not phospho-MEK, pathway. Vertical lines have been inserted to indicate a repositioned gel lane. Results are shown as mean ± SD (*P < .05) (D) The expression of MCL-1 in MV4-11 24 hours after treatment with pharmacologic inhibitors of PKC (PKC-412), PI3K (Ly294002), JAK2 (AG490), or MEK (U0126). PKC-412 inhibited MCL-1 mRNA expression. PKC-412 and Ly294002 but not U0126 or AG490 inhibited the MCL-1 protein expression. RE indicates relative expression levels. Results are shown as mean ± SD (*P < .05).
To directly test whether STAT5 is necessary in FLT3-ITD–induced MCL-1 expression, we knocked down the expression of STAT5 by transducing STAT5 siRNA into FLT3-ITD MV4-11 cells (Figure 5C). Under these conditions, MCL-1 expression was significantly inhibited at both mRNA and protein levels, suggesting that STAT5 is critical for MCL-1 expression. Figure 5B schematizes the FLT3-ITD and FLT3 signaling pathways for MCL-1 expression. In FLT3-WT, FLT3 ligation triggers both PI3K/Akt and RAS/MEK/MAPK pathways.23 In FLT3-ITD cells, FLT3-ITD might directly activate STAT5 via its STAT5-docking sites. Treatment of MV4-11 cells with AG290, a Janus kinase 2 (JAK2) inhibitor, did not affect MCL-1 expression (Figure 5D). Thus, activated STAT5 triggers MCL-1 transcription without JAK2 activation. We then tried to test whether STAT5 induces MCL-1 expression by triggering activation of the PI3K/Akt pathway: Ly294002, a PI3K inhibitor, did not affect MCL-1 mRNA expression but significantly inhibited MCL-1 protein expression (Figure 5D), suggesting that the PI3K pathway regulates MCL-1 expression at the protein level. Of note, STAT5 inhibition significantly down-regulated the phosphorylation of Akt, a downstream molecule of PI3K, indicating that STAT5 is positioned upstream of the PI3K pathway (Figure 5C). On the other hand, STAT5 siRNA did not affect MEK phosphorylation (Figure 5C), and U0126, a MEK inhibitor, did not change the expression of either MCL-1 mRNA or protein (Figure 5D), suggesting that STAT5-induced MCL-1 expression is not mediated by the RAS/MEK/MAPK pathway. Collectively, in FLT3-ITD signaling pathway, STAT5 up-regulates MCL-1 by triggering MCL-1 transcription, and by activating PI3K to posttranscriptionally up-regulate MCL-1.
Discussion
FLT3 mutations are among the most common genetic alterations in AML. FLT3-ITD results in loss of function of a juxtamembrane autoinhibitory domain that maintains the kinase in an inactive state in the absence of ligand, resulting in ligand-independent receptor dimerization and constitutive FLT3 signaling. In addition to the normal FLT3 signaling, including activation of the RAS/MAPK and PI3K pathways, FLT3-ITD can activate STAT5 via FLT3-ITD–specific STAT5-docking domains. In a mouse line harboring FLT3-ITD knocked into the Flt3 locus, bone marrow cells displayed myeloproliferation resembling the phenotype of chronic myelomonocytic leukemia. Because this finding correlated at least with an increase in number and survival of multipotent progenitors,46 the reinforcement of cell survival might be one of the critical roles of FLT3-ITD.
The CD34+CD38− LSCs purified from FLT3-ITD AML patients can repopulate the disease in the immune-deficient mice after transplantation, confirming that FLT3-ITD mutations are acquired at the LSC level.47 We here show that the extracellular FLT3 is expressed at a high level in LSCs on FACS in all types of human AML irrespective of the presence of FLT3-ITD mutations. Because anti-FLT3 antibodies should react with FLT3-ITD, both FLT3 and FLT3-ITD are expressed in AML LSCs at a high level (Figure 1), suggesting that FLT3-ITD signaling should operate strongly at the LSC stage in AML. In normal human HSCs, FLT3 signaling is critical for maintenance of MCL-1 at a high level.10 Of note, freshly isolated AML LSCs express a higher level of FLT3 and MCL-1 compared with normal HSCs, which may lead to growth advantages of AML LSCs against normal HSCs in vivo. The high level of MCL-1 in purified AML LSCs (Figure 3A) may be derived from signals from a higher amount of FLT3 on LSCs' surface (Figure 1). The ligation of their FLT3 receptors induced even higher levels of MCL-1 in LSCs (Figure 3C). Freshly isolated FLT3-ITD LSCs already maintained a high level of MCL-1, but an addition of FLT3 ligand could cause further up-regulation of MCL-1 (Figure 3C). This additive effect could be due to ligation of normal FLT3 receptors generated from an intact FLT3 allele.
One of the major functions of FLT3-ITD signaling should prevent LSCs from apoptotic cell death via maintaining a high level of MCL-1, as directly shown in Figure 4. It has recently been demonstrated that the ratio of FLT3-ITD/WT mRNA expression shows a wide variation in the samples of AML blasts.21,48 Of note, higher FLT3-ITD/WT ratio manifests significantly shorter overall and disease-free survival of patients.21,48,49 We thus investigated the association between the ratio of FLT3-ITD/WT transcripts and MCL-1 expression in our AML samples. This ratio, however, did not correlate with the levels of MCL-1 expression in the samples analyzed so far (data not shown). Due to limited number of samples, further experiments are still needed to address this issue.
Our study also showed that the FLT3-ITD–specific STAT5-docking domains play a major role for FLT3-ITD, independent of JAK2 activation, to induce MCL-1 mRNA (Figure 5). This is of particular interest because these domains are necessary to cause myeloproliferative phenotype in a FLT3-ITD knock-in strain.46 STAT5 is well recognized as a common downstream pathway from cytokine receptors, and activates transcription of various target genes such as cyclin D1, p21, Pim-1, c-fos, and BCL-xL.46,50 Sustained activation of STAT5 in murine HSCs but not progenitors induced myeloproliferative disease, and knockdown of STAT5 impaired growth of normal and leukemic stem cells, indicating that STAT5 plays a crucial role in proliferation or survival of HSCs and LSCs.51,52 It has also been reported that AML cells with STAT5 phosphorylation display a lower percentage of spontaneous apoptosis, compared with AML cells without STAT5 phosphorylation, and constitutive STAT5 phosphorylation was observed with all AML cases with FLT3-ITD.44 Furthermore, AG1296, a tyrosine kinase inhibitor, strongly inhibited STAT5 phosphorylation and induced apoptosis in AML cells.44 In this study, we show that STAT5 is particularly critical for FLT3-ITD signaling to express MCL-1, because knockdown of STAT5 abrogates MCL-1 expression in FLT3-ITD cells at both mRNA and protein levels (Figure 5C). Of note, the STAT-binding motif is found in the promoter of the human MCL-1 gene,53 suggesting that STAT5 could directly activate the transcription of MCL-1 gene. Previous studies including our own have shown that MCL-1 expression is regulated also at the protein level by the PI3K pathway, because Akt, a downstream molecule of PI3K, up-regulates MCL-1 posttranscriptionally via regulating activation of glycogen synthase kinase 3.54,55 Interestingly, recent studies have shown that STAT5 interacts with and activates PI3K.56,57 We found that STAT5 knockdown inhibits phosphorylation of Akt and suppresses MCL-1 expression in FLT3-ITD cells (Figure 5C). It is thus likely that STAT5 could also activate the PI3K/Akt pathway to up-regulate MCL-1 at the protein level. Therefore, the STAT5/MCL-1 pathway downstream of FLT3-ITD could be a specific target to impair survival of FLT3-ITD LSCs, still sparing the effect on FLT3-dependent normal hematopoiesis.
The reinforcement of survival is one of the critical factors for leukemia development. We have reported that the myeloproliferative phenotype develops also in a mouse model expressing simply a Bcl-2 transgene at the myeloid progenitor stage using the myeloid-related protein-8 promoter.7 Furthermore, the myeloid-related protein-8–BCL-2 transgene accelerates the AML development in mice having leukemogenic mutations such as BCR-ABL7 and promyelocytic leukemia/retinoic acid receptor α (PML/RARα).6 Previous studies have shown that FLT3 mutations alone are not sufficient to cause AML, and that additional cooperating mutations are required for AML development.23,24 When FLT3-ITDs are combined with other genetic alterations such as PML-RARα,58 AML1-ETO,59 MLL-AF9,60 and NUP98-HOX,61 full transformation to AML can occur. Our data suggest that the primary role of FLT3-ITD in AML is at least to reinforce cell survival to promote AML development.
Thus, the CD34+CD38− AML fraction that concentrates LSCs possesses a high level of MCL-1 and might use this protein to ensure their survival. The acquisition of FLT3-ITD mutation provides LSCs with potent survival signals through further up-regulating MCL-1. This survival-promoting signal of FLT3-ITD is mediated by a STAT5-dependent pathway that is independent of normal FLT3 signaling. Targeting this pathway might be useful to trigger cell death process specifically in FLT3-ITD AML LSCs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We appreciate the medical and nursing staff working on the Fukuoka Blood and Marrow Transplantation Group.
This work was supported in part by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology in Japan (19659248 and 20390272 [T.M.], 17109010 and 17047029 [K.A.]) and the Takeda Science Foundation (T.M.).
Authorship
Contribution: G.Y., T.M., and K.A. designed and performed research, analyzed data, and wrote the paper; S.J.-T., T.I., Y.K., Y.M., T.S., H.N., and S.-i.M. performed research; H.I., K.T., and K.N. provided patients' samples and information; and J.L.R. and G.D.G. provided cDNA constructs and helped to write the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Koichi Akashi, Department of Medicine and Biosystemic Science, Graduate School of Medical Sciences, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan; e-mail: akashi@med.kyushu-u.ac.jp.