Abstract
Tonic B-cell receptor (BCR) signaling is a key survival pathway during normal B-cell ontogenesis and in a subset of diffuse large B-cell lymphomas (DLBCLs). We previously demonstrated that BCR-dependent DLBCL cell lines and primary tumors underwent apoptosis after treatment with an ATP-competitive inhibitor of the BCR-associated spleen tyrosine kinase (SYK). These “BCR-type” tumors also have more abundant expression of the transcriptional repressor, BCL6, and increased sensitivity to BCL6 inhibition. Herein, we evaluated potential connections between BCL6-mediated transcriptional repression and SYK-dependent BCR signaling. In transcriptionally profiled normal B-cell subsets (naive, germinal center, and memory B cells) and in primary DLBCLs, there were reciprocal patterns of expression of BCL6 and the SYK tyrosine phosphatase PTPROt. BCL6 repressed PTPROt transcription via a direct interaction with functional BCL6 binding sites in the PTPROt promoter. Enforced expression of BCL6 in normal naive B cells and RNAi-mediated depletion of BCL6 in germinal center B cells directly modulated PTPROt expression. In “BCR-type” DLBCLs, BCL6 depletion increased PTPROt expression and decreased phosphorylation of SYK and the downstream adaptor protein BLNK. Because BCL6 augments BCR signaling and BCL6 and SYK are both promising therapeutic targets in many DLBCLs, combined inhibition of these functionally related pathways warrants further study.
Introduction
Emerging data highlight the important role of B-cell receptor (BCR)–mediated survival signals in B-cell lymphomas. BCR engagement induces the phosphorylation of Igα and β immunoreceptor tyrosine-based activation motifs (ITAMS) by SRC family kinases and the subsequent recruitment and activation of the protein tyrosine kinase (PTK), SYK, and downstream pathways.1-3 Although BCR signaling is generally thought to be triggered by antigen binding, recent studies highlight the role of “tonic” BCR survival signals in the absence of receptor engagement. For example, in murine models, the inducible loss of the BCR or the selective excision of the Igα ITAM led to the death of peripheral B cells.4,5
The SYK PTK plays a central role in tonic BCR signaling, both transmitting downstream events and amplifying the original signal.2,3,6,7 SYK activity is tightly regulated by BCR-associated phosphorylation and protein tyrosine phosphatase (PTP)–mediated inhibition.8 We recently found that SYK is a major substrate of a tissue-specific and developmentally regulated PTP, PTPROt.6 PTPROt specifically inhibited BCR-triggered SYK tyrosyl phosphorylation, activation of associated adaptor proteins, such as BLNK, and downstream signaling events.6 In BCR-dependent lymphomas, PTPROt overexpression decreased cellular proliferation and induced apoptosis in the absence of BCR cross-linking, indicating that the phosphatase modulates SYK-dependent tonic BCR signaling.6
PTPROt is a member of the PTPRO family (also designated GLEPP, PTP-ϕ, PTP-OC, and PTPu2), a group of highly conserved receptor-type PTPs with a single catalytic domain and transmembrane region and a variably sized extracellular sequence.9,10 PTPRO includes an extended extracellular domain, whereas PTPROt contains a truncated extracellular region. The PTPROt 5′ untranslated region also functions as an intron that is spliced out of the larger PTPRO cDNA.11 These 2 isoforms have tissue-specific patterns of expression—PTPRO predominantly in epithelial cells and PTPROt primarily in B cells and macrophages.12 Initial studies suggest that PTPROt is developmentally regulated and decreased in abundance in normal germinal center (GC) B cells and a subset of B-cell lymphomas.12
We recently found that a subset of diffuse large B-cell lymphomas (DLBCLs) relies upon tonic BCR signaling as a survival mechanism and that PTPROt modulates SYK-dependent BCR signaling in these tumors.6,13 DLBCLs that were dependent upon BCR signaling had a notable transcriptional profile with increased expression of multiple components of the BCR signaling cascade, including SYK itself.13
These “BCR-type” DLBCLs also exhibit increased expression of the BTB/POZ domain transcriptional repressor, BCL6, and more frequent translocations of the BCL6 locus.14,15 BCL6 is required for normal GC development and is expressed at the highest levels in normal GC B cells.16,17 In previous studies, we found that “BCR-type” DLBCLs exhibit coordinate repression of the BCL6 target genes and increased sensitivity to BCL6 inhibitors.15 Because the same transcriptionally defined subset of DLBCLs relies upon SYK-dependent BCR signaling and exhibits coordinate BCL6-mediated transcriptional repression, we explored the relationship between these 2 processes. Herein, we report that BCL6 modulates PTPROt expression and associated SYK-dependent tonic BCR signaling.
Methods
Microarray analysis of BCL6 and PTPRO in normal B cells and in tumor samples
Two previously described datasets of transcriptionally profiled newly diagnosed DLBCLs with available comprehensive cluster and cell of origin designations14,18 and an additional series of profiled normal B-cell subsets (naive, centroblasts, and memory)18,19 were used to assess BCL6 and PTPROt transcript abundance. An additional series of highly purified normal B-cell subsets (naive, centroblasts, centrocytes, and memory) was isolated from normal human tonsils by magnetic cell separation (MidiMACS system; Miltenyi Biotec) and profiled using the Affymetrix U133Plus platform as previously described.20 Affymetrix probes 208121_s_at (PTPRO), 203140_at, 215990_s_at (BCL6), and the dChip 2007 program were used to assess PTPROt and BCL6 transcript abundance.
Computational analysis of PTPROt promoter, generation of PTPROt promoter constructs, and luciferase assays
Computational analysis of PTPROt promoter was performed with the publicly available MatInspector module of Genomatix suite (http://www.genomatix.de). The approximately 1.6-kb sequence spanning −1.1 kb upstream to +0.5 kb downstream from the previously identified PTPROt transcription start site (TSS) was interrogated, and 3 putative BCL6 binding sites were identified. To generate a PTPROt promoter reporter construct, a fragment spanning nucleotides −1108 to +381 was polymerase chain reaction (PCR)–amplified and cloned into the promoterless pGL3 luciferase vector (Promega). Deletions in BCL6 binding sites were generated using the GeneTailor Site-Directed Mutagenesis System (Invitrogen) as recommended by the manufacturer. BCL6 constructs encoding either the wild type protein (pMT2T-HA-BCL6) or a mutant protein, lacking either the N-terminal POZ domain or the C-terminal zinc fingers, (pMT2T-HA-BCL6-ZF and pMT2T-HA-BCL6-ΔZF, respectively) were used.21
For luciferase assays, HEK293T cells were maintained in Dulbecco-modified Eagle medium (Cellgro Mediatech) supplemented with 10% fetal calf serum (Cellgro Mediatech), 10mM HEPES buffer, 4 mM l-glutamine, 50 U/mL penicillin, and 50 U/mL streptomycin. HEK293T cells were seeded on 6 well-plates, grown to 60% to 80% confluency and cotransfected with 350 ng/well of the appropriate promoter pGL3 construct (wild-type or mutant PTPROt promoter construct), 150 ng/well of the control reporter plasmid, pRL-TK (Promega), and 5 to 100 ng of wild-type or 100 ng of mutant BCL6 construct using FuGENE 6 transfection reagent (Roche Applied Science) according to manufacturer's protocol. After 24 hours of incubation, cells were lysed, and luciferase activities were determined by chemoluminescence assay using the Dual Luciferase Assay kit (Promega) and Luminoskan Ascent luminometer (Thermo Lab Systems). Luciferase activities are presented as means from 3 experiments plus or minus SD.
Chromatin immunoprecipitation
DHL4, DHL6, or K422 cells were maintained in RPMI1640 medium supplemented with 10% fetal bovine serum (Cellgro Mediatech), 10 mM HEPES buffer, 4 mM l-glutamine, 50 U/mL penicillin, and 50 U/mL streptomycin. For chromatin immunoprecipitation (ChIP), 25 × 106 DHL4, DHL6, or K422 cells were fixed in 0.5% formaldehyde for 10 minutes at room temperature. Reactions were subsequently quenched in 0.2 M glycine for 5 minutes. Cells were then washed with 1× phosphate-buffered saline (PBS) and lysed in RIPA lysis buffer (150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris [pH 8.0], 2 mM EDTA) containing protease inhibitors (complete protease inhibitor cocktail; Roche Applied Science) and sonicated. Lysates were precleared and subsequently incubated with rabbit antisera directed against BCL6 (N3 antibody; Santa Cruz Biotechnology) or with normal rabbit IgG antibody (Santa Cruz Biotechnology). Immunocomplexes were captured with protein A/G Plus agarose preblocked with salmon sperm DNA (Abcam) and washed 3 times with RIPA buffer and once with final ChIP wash buffer (1% NP40, 0.1% SDS, 500 mM NaCl, 2 mM EDTA, pH 8.0, 20 mM Tris-HCl, pH 8.0, with protease inhibitors). Thereafter, immune complexes were eluted with 1% SDS in 100 mM NaHCO3 and cross-links were reversed by incubating samples for 4 hours at 65°C. Samples were then digested with proteinase K for 1 hour at 45°C. DNA fragments enriched by ChIP were recovered by standard phenol-chloroform extraction followed by ethanol precipitation and quantified by real-time PCR using PTPROt promoter and control region primers (F1: ACAGGCAGGGTTTTTTGGTCT; R1: TCGTAAAGGAAGCTGTGTTGTCTG; F2: CTCCACCTTTATTAAGTGCAGATTTTT; R2: ACTGAAGTTAGCATTGCCCTG ATA; F3: TGACTTTGCTGTGGGTGGAA, R3: TGGTTTTCTGACTGGAGCCAA; contF1: TGGCTTCACTCCCCTCTACC; contR1: CTGGGTTGTTGCTGGTGAAA; contF2: TGATGGAAAAGGCATCAGTCTG; contR2: AGACCCAGGAGAGATAATTCCAAA), a PowerSYBR green kit (Applied Biosystems), and an ABI 7700 thermal cycler (Applied Biosystems). Relative enrichment in putative BCL6 binding sites of PTPROt promoter and control regions in BCL6- over control IgG-immunoprecipitated samples was calculated using the 2−(ΔCT BCL6 − ΔCT IgG) method. Standard deviations were calculated from triplicate ΔΔ threshold cycle (CT) values.
Analysis of BCL6-mediated PTPROt repression in normal tonsillar naive B cells and centroblasts
Normal naive B cells and centroblasts were obtained from human tonsils by magnetic cell separation with the MidiMACS system (Miltenyi Biotec) as previously described.20 Expression of the BCL6 construct in naive B cells was achieved by infection of the cells with a FUGW lentivirus vector (Addgene) expressing BCL6 or an eGFP-only control as previously described.20 For BCL6 knockdown in centroblasts, a BCL6-specific or a negative control “scrambled” shRNA cloned into the pFIV-H1-copGFP vector was used.20 After 48 hours, total RNA was prepared with TRIzol (Invitrogen) and reverse-transcribed to cDNA with the Superscript III first-strand cDNA synthesis kit (Invitrogen). PTPROt transcript abundance was evaluated by quantitative 5′-nuclease assay PCR with the following primers and 5′-FAM labeled MGB probe: forward: ACTTTGTCTTTGCTCAGAACCAG; reverse: AGAAACAGCAA CTGGTTCCTGAAG; probe: CACTCTTCGCAGTGAAC. PCR was performed using an ABI 7700 thermal cycler (Applied Biosystems) and CT values were generated using the Sequence Detection Software Version 1.2 (Applied Biosystems). PTPROt transcript abundance was calculated relative to the housekeeping control cyclophilin A (Applied Biosystems) using the 2− (ΔCT PTPROt − ΔCT PPIA1) method. Standard deviations were calculated from triplicate ΔΔCT values. For absolute mRNA copy number quantification, PCR was performed with serial dilutions of plasmid DNA containing the PCR target sequence. CT values from the serial dilutions were plotted against log (no. of plasmid copies) to generate standard curves as described previously.22 Expression of the control BCL6 target gene, FCER2, was assessed relative to GAPDH using RT-PCR, a PowerSYBR Green kit, and the following primers: FCER2, F: ATGAATCCTCCAAGCAGGAG; FCER2, R: GACTTGAAGCTGCTCAGACTGCT; GAPDH, F: GATTCCACCCATGGCAAATTC; GAPDH, R: TGATTTTGGAGGGATCTCGCTC.
RNA-interference mediated BCL6 knockdown
BCL6-specific onTARGET plus siRNA and negative control scrambled (SCR) oligonucleotides (CCUCCAUAUCUCGCGCGUCUU) were obtained form Thermo Scientific. Oligos were resuspended in RNAse free water at 50 μM and stored at −70°C. For siRNA nucleofections, 4 × 106 of DHL4 or K422 cells were resuspended in 100 μL of AMAXA nucleofector solution V containing 75 pmoles of BCL6 or SCR oligonucleotides and treated with O-017 program in Nucleofector II device (AMAXA). Transduction efficiency was confirmed to be more than 90% by nucleofection of Cy3-labeled GAPDH oligonucleotides (Applied Biosystems/Ambion) and subsequent flow cytometry analysis. After nucleofections, cells were incubated for 72 hours and used for phosphor-specific flow cytometry or to prepare whole-cell extracts for immunoblotting or for immunoprecipitations.
Immunoprecipitation and immunoblotting
Previously described Tet-inducible Flag-tagged wild-type (WT) or mutant (CS) PTPROt clones were cultured with or without doxycycline, serum starved, and stimulated thereafter with goat anti–human IgG (10 μg/mL) for 10 minutes or left untreated.6 Cells were lysed in NP40 lysis buffer (1% NP-40, 50 mM Tris-HCl [pH7.4], 150 mM NaCl, and 2 mM Na3VO4) containing protease inhibitors. After centrifugation, supernatants were incubated with 2 μg/mL of α-SYK (4D10) or α-CD79a antibody (Santa Cruz Biotechnology) at 4°C for 1 hour with rotation. Thereafter, 50-μL protein G–Sepharose beads (50% slurry in lysis buffer) were added, and samples were rotated for 1 additional hour. Immunocomplexes were then recovered by centrifugation, washed with cold lysis buffer, resuspended in sample buffer, boiled at 95°C for 10 minutes, size-fractionated by PAGE, and transferred to polyvinylidene fluoride membranes (Millipore Corp). Blots were first incubated in blocking buffer (5% milk, 0.1% Tween in PBS) for 30 minutes and subsequently incubated overnight at 4°C with phospho-specific antibodies directed against SYK Y352 [BD Biosciences], Y525/526, Y323 [Cell Signaling], or with anti–pan-phospho-tyrosyl 4G10 antibody [Upstate]). After sequential washes with 0.1% Tween/PBS, blots were incubated with horseradish peroxidase–labeled secondary antibodies at room temperature for 1 hour, developed by enhanced chemiluminescence (Western Lighting Plus-ECL; PerkinElmer), and visualized with Kodak Biomax film (Carestream Health Inc). To reprobe with another antibody, the blots were stripped (0.063 M Tris-HCl [pH 6.8], 2% SDS, 0.026 M DTT) at 50°C for 30 minutes, washed, and analyzed with α-SYK and α-Flag antibodies. Densitometric analysis of developed films was performed with ChemiImager 4400 software (Alpha Innotech).
Whole-cell extracts from BCL6 siRNA-transduced cells were obtained by directly lysing cells in the RIPA lysis buffer. For immunoprecipitations, cells were first resuspended in 150 μL of PBS containing 1% FCS and left untreated or stimulated with goat anti–human IgG at 37°C for 1 minute. Cell suspensions were immediately lysed in 150 μL of 2× NP40 lysis buffer (2% NP-40, 100 mM Tris-HCl [pH 7.4], 300 mM NaCl, 4 mM Na3VO4 with protease inhibitors) and immunoprecipitations with α-SYK were performed. Lysates or immunoprecipitates were size-fractionated on NuPAGE Novex 4% to 12% Bis-Tris Gels (Invitrogen) and transferred to polyvinylidene fluoride membranes (Millipore). Blots were incubated with primary antibodies (αBCL6, αSYK, αBLNK, αCD79a [Santa Cruz Biotechnology], and α-βactin [Sigma-Aldrich]) in blocking buffer for 2 hours at room temperature. For phospho-specific SYK Y352, blots were incubated overnight at 4°C. Blots were washed, incubated with appropriate horseradish peroxidase–labeled secondary antibodies, developed, and visualized.
RNA-interference mediated PTPROt knockdown
PTPROt-specific siRNA was designed using the GeneScript RNAi selection algorithm23 (https://www.genscript.com/ssl-bin/app/rnai) and synthesized as single-stranded DNA oligonucleotides by Integrated DNA Technologies (IDT Inc) and annealed. PTPROt-specific oligonucleotide (GATCCGGATGACTTTGATGCCTATATTTCAAGAGAATATAGGCATCAAAGTCATCCTTTTTTG) or SCR oligonucleotide24 were ligated into the linearized pSIREN-RetroQ retroviral vector (Clontech). Generation of recombinant retrovirus and infection of DHL4 cells were performed as previously described.22 After infection, cells were subjected to puromycin selection (0.5 μg/mL) and subcloning by limiting dilution. Thereafter, RNA extracted from selected subclones was reverse transcribed and used to confirm PTPROt depletion by quantitative RT-PCR. The PTPROt-depleted subclone (90% diminution in PTPROt expression) and SCR control clone were subsequently used in phospho-specific flow cytometry.
Phospho-specific flow cytometry
Intracellular phospho-specific flow cytometry was performed as previously described13 according to the manufacturer's instructions. In brief, 2 × 106 cells were resuspended in 1 mL of cold PBS plus 1% FCS and left untreated or stimulated with goat anti–human IgG at 37°C for 1 minute. Thereafter, cells were fixed, permeabilized, and stained with phycoerythrin-conjugated α-pSYK (pY352), phycoerythrin-conjugated α-pBLNK (pY84; BD Biosciences), or isotype control antibodies. Flow cytometric analysis was performed using a FACS Canto II flow cytometer (BD Biosciences).
Results
Reciprocal patterns of PTPROt and BCL6 expression in normal B cells and primary DLBCLs
We previously found that PTPROt protein was less abundant in normal GC B cells than in naive or memory B cells.12 Because BCL6 expression is highest in normal GC B cells, we first compared the relative expression of PTPROt and BCL6 in 2 independent series of highly purified naive GC and memory B cells by transcriptional profiling (Figure 1A). PTPROt transcripts were significantly more abundant in normal naive and memory B cells than in GC B cells whereas BCL6 expression was highest in normal GC B cells (Figure 1A). In addition, there were reciprocal patterns of BCL6 and PTPROt expression in 2 large independent series of primary DLBCLs (Figure 1B and supplemental Figure 1). Of note, more than 78% of primary DLBCLs with high BCL6 and low PTPROt transcript levels were previously identified as “BCR-type” tumors (Figure 1B, χ2 test, P < .001).
Reciprocal patterns of PTPROt and BCL6 expression in normal B cells and primary DLBCLs. (A) Relative BCL6 and PTPROt transcript abundance in 2 independent series of highly purified normal B cells (naive, GC centroblasts [CB] and centrocytes [CC], memory). (B) Relative BCL6 and PTPROt transcript abundance in 176 newly diagnosed and previously profiled DLBCLs.14 Normal GC B cells that were profiled at the same14 time were included for comparison (left). Comprehensive cluster designations for the primary DLBCLs and normal GC cells are indicated above the heat map. The primary DLBCL series includes 77 BCR, 50 OxPhos and 49 HR tumors. Color scale at bottom indicates relative expression and SDs from the mean. Red connotes high-level expression; blue indicates low-level expression
Reciprocal patterns of PTPROt and BCL6 expression in normal B cells and primary DLBCLs. (A) Relative BCL6 and PTPROt transcript abundance in 2 independent series of highly purified normal B cells (naive, GC centroblasts [CB] and centrocytes [CC], memory). (B) Relative BCL6 and PTPROt transcript abundance in 176 newly diagnosed and previously profiled DLBCLs.14 Normal GC B cells that were profiled at the same14 time were included for comparison (left). Comprehensive cluster designations for the primary DLBCLs and normal GC cells are indicated above the heat map. The primary DLBCL series includes 77 BCR, 50 OxPhos and 49 HR tumors. Color scale at bottom indicates relative expression and SDs from the mean. Red connotes high-level expression; blue indicates low-level expression
PTPROt is a BCL6 target gene
The reciprocal patterns of BCL6 and PTPROt expression in normal B cells and primary DLBCLs and the identification of BCL6-high/PTPROt-low “BCR-type” DLBCLs raised the possibility that PTPROt was a target gene of BCL6. For these reasons, we examined the PTPROt regulatory region for BCL6 binding sites. The 1.5-kb PTPROt promoter region, which encompassed the previously identified TSS and TATA box,11,12 included 3 candidate BCL6 binding sites (−763 to −746; −124 to −107; and +300 to +317 nt from TSS). Of note, the 2 upstream candidate BCL6 binding sites (−763 to −746 and −124 to −107) are located within a region of the PTPROt promoter associated with repressed basal transcriptional activity.11
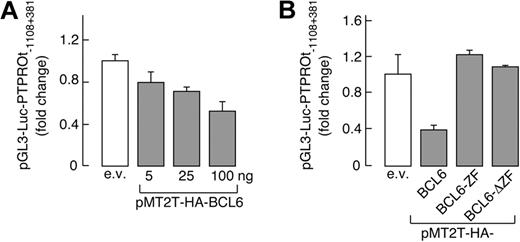
To assess the functional status of the candidate BCL6 binding sites, we generated a luciferase vector driven by the PTPROt promoter (pGL3-Luc-PTPROt-1108 + 381). This PTPROt-luciferase construct was cotransfected into HEK293T cells with a vector encoding either WT BCL6 or 1 of 2 inactive BCL6 mutants, BCL6-ZF or BCL6-ΔZF (lacking the amino-terminal transcriptional repressor domain or carboxy-terminal DNA-binding zinc-finger sequence, respectively). WT BCL6 repressed PTPROt promoter-driven luciferase activity in a dose-dependent manner (Figure 2A); in marked contrast, neither of the inactive BCL6 mutants decreased expression of the PTPROt reporter gene (Figure 2B).
PTPROt transcription is regulated by BCL6. (A) BCL6 represses PTPROt promoter-driven transcription in a dose-dependent manner; 350 ng of PTPROt promoter luciferase reporter construct (pGL3-Luc-PTPROt −1108 + 381) was cotransfected with empty vector or increasing doses (5-100 ng) of a BCL6 expression vector (pMT2T-HA-BCL6) into HEK293T cells. Luciferase activities were evaluated as described (see “Luciferase assays”). (B) WT-BCL6 but not BCL6 mutants repress PTPROt promoter-driven transcription. pGL3-Luc-PTPROt-1103 + 381 was cotransfected with 100 ng of vectors encoding either HA-BCL6 or 1 of 2 BCL6 mutants lacking either the amino-terminal transcriptional repressor domain (BCL6-ZF) or the carboxy-terminal DNA binding zinc-finger domain (BCL6-ΔZF) and luciferase activities were determined thereafter. In both panels A and B, representative luciferase activities from 3 independent experiments were normalized to Renilla luciferase activity and represented as fold change ± SD.
PTPROt transcription is regulated by BCL6. (A) BCL6 represses PTPROt promoter-driven transcription in a dose-dependent manner; 350 ng of PTPROt promoter luciferase reporter construct (pGL3-Luc-PTPROt −1108 + 381) was cotransfected with empty vector or increasing doses (5-100 ng) of a BCL6 expression vector (pMT2T-HA-BCL6) into HEK293T cells. Luciferase activities were evaluated as described (see “Luciferase assays”). (B) WT-BCL6 but not BCL6 mutants repress PTPROt promoter-driven transcription. pGL3-Luc-PTPROt-1103 + 381 was cotransfected with 100 ng of vectors encoding either HA-BCL6 or 1 of 2 BCL6 mutants lacking either the amino-terminal transcriptional repressor domain (BCL6-ZF) or the carboxy-terminal DNA binding zinc-finger domain (BCL6-ΔZF) and luciferase activities were determined thereafter. In both panels A and B, representative luciferase activities from 3 independent experiments were normalized to Renilla luciferase activity and represented as fold change ± SD.
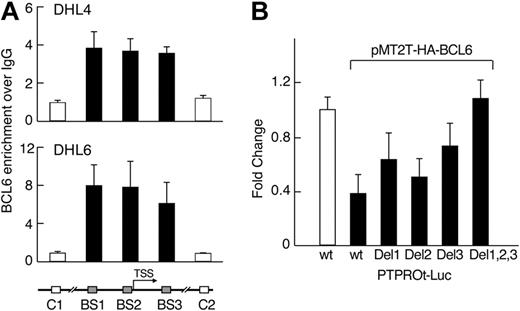
We next asked whether endogenous BCL6 binds the PTPROt promoter region in vivo using a chromatin immunoprecipitation assay. In 2 “BCR-type” DLBCL cell lines (DHL4 and DHL6), DNA fragments including the predicted BCL6 binding sites, but not 2 upstream/downstream control fragments, were significantly enriched in α-BCL6 chromatin immunoprecipitates (Figure 3A). These data suggest that each of the identified binding sites is occupied by BCL6 in vivo (Figure 3A). Consistent with these observations, individual mutations of each of the predicted BCL6 binding sites decreased BCL6-mediated repression of PTPROt, and combined mutations of all 3 BCL6 binding sites in the PTPROt promoter abolished its response to BCL6 (Figure 3B). Taken together, these results indicate that BCL6 represses the activity of the PTPROt promoter in the “BCR-type” DLBCL cell lines, DHL4 and DHL6, and that this function requires the identified BCL6 binding sites. In contrast, BCL6 does not bind the PTPROt promoter region in the non–BCR-type DLBCL cell line, K422, in chromatin immunoprecipitation assays (see supplemental Figure 2).
BCL6 represses PTPROt via direct interactions with the PTPROt promoter region. (A) BCL6 binds to the PTPROt promoter in vivo. Chromatin immunoprecipitation was performed in 2 DLBCL cell lines (DHL4 and DHL6) using BCL6 antibody or normal IgG as control. The target amplicons in the PTPROt promoter include the 3 predicted BCL6 binding sites (BS1-3, ■) and 2 distant upstream or downstream control regions (C1 and C2, □). The BCL6 versus IgG ratio was calculated for each region and normalized to control region 1. (B) BCL6-mediated repression of PTPROt promoter requires intact BCL6 binding sites. PTPROt-promoter-driven luciferase constructs with or without the individual or combined mutations in the predicted BCL6 binding sites were cotransfected with pMT2T-HA-BCL6 into HEK293T cells. Luciferase activities were determined thereafter.
BCL6 represses PTPROt via direct interactions with the PTPROt promoter region. (A) BCL6 binds to the PTPROt promoter in vivo. Chromatin immunoprecipitation was performed in 2 DLBCL cell lines (DHL4 and DHL6) using BCL6 antibody or normal IgG as control. The target amplicons in the PTPROt promoter include the 3 predicted BCL6 binding sites (BS1-3, ■) and 2 distant upstream or downstream control regions (C1 and C2, □). The BCL6 versus IgG ratio was calculated for each region and normalized to control region 1. (B) BCL6-mediated repression of PTPROt promoter requires intact BCL6 binding sites. PTPROt-promoter-driven luciferase constructs with or without the individual or combined mutations in the predicted BCL6 binding sites were cotransfected with pMT2T-HA-BCL6 into HEK293T cells. Luciferase activities were determined thereafter.
BCL6 regulates PTPROt expression in normal B cells and DLBCLs
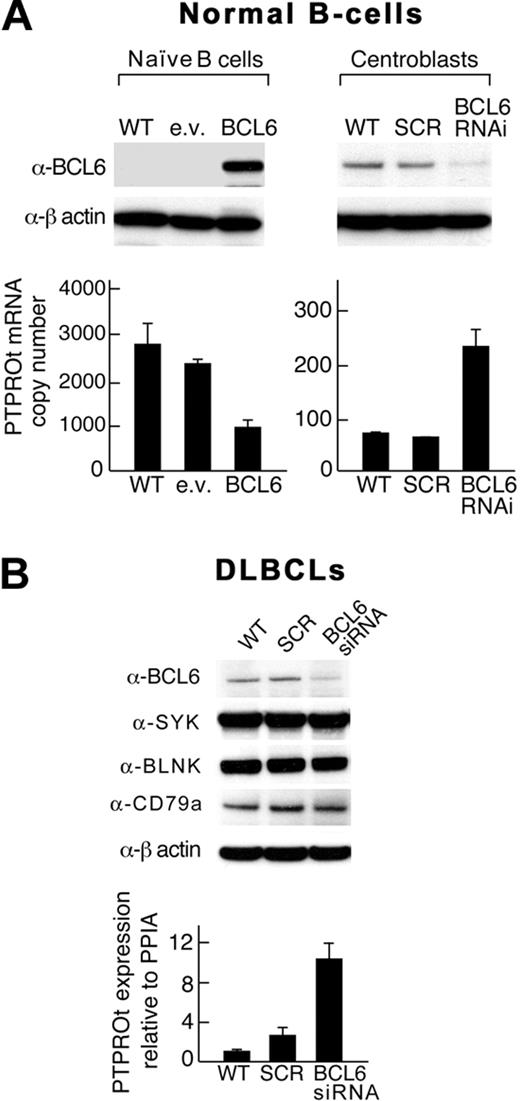
Given the reciprocal, developmentally regulated pattern of expression of BCL6 and PTPROt in normal B cells, we next asked whether PTPROt is a physiologic transcriptional target of BCL6 in highly purified naive B cells and GC centroblasts. After transducing naive B cells with a BCL6 lentiviral vector and centroblasts with a BCL6-shRNA construct, we assessed PTPROt expression by RT PCR (Figure 4A). In naive B cells, the forced overexpression of BCL6 markedly decreased PTPROt transcript abundance (Figure 4A left panel). Conversely, BCL6 depletion increased PTPROt expression in normal centroblasts (Figure 4A right panel).
BCL6 regulates PTPROt expression in normal naive B cells, GC centroblasts, and certain DLBCLs. (A) Normal B cells. Normal naive B cells (left) were transfected with a eGFP-BCL6 construct or empty vector and GC centroblasts (right) were transduced with a BCL6-shRNA lentiviral vector or scrambled control. (B) DLBCLs. A DLBCL cell line (DHL4) was transduced with BCL6 siRNA. In both the normal B cells (A) and the DLBCL cell line (B), BCL6 protein levels were assessed by Western blot (top panel), and PTPROt expression was evaluated by RT PCR (bottom panel). In DLBCLs, expression levels of total SYK, the upstream BCR pathway component, CD79a, and the downstream adapter protein, BLNK, were also evaluated. Error bars represent SDs from the mean for 3 independent RT-PCR assays for each condition and cell type in a representative experiment.
BCL6 regulates PTPROt expression in normal naive B cells, GC centroblasts, and certain DLBCLs. (A) Normal B cells. Normal naive B cells (left) were transfected with a eGFP-BCL6 construct or empty vector and GC centroblasts (right) were transduced with a BCL6-shRNA lentiviral vector or scrambled control. (B) DLBCLs. A DLBCL cell line (DHL4) was transduced with BCL6 siRNA. In both the normal B cells (A) and the DLBCL cell line (B), BCL6 protein levels were assessed by Western blot (top panel), and PTPROt expression was evaluated by RT PCR (bottom panel). In DLBCLs, expression levels of total SYK, the upstream BCR pathway component, CD79a, and the downstream adapter protein, BLNK, were also evaluated. Error bars represent SDs from the mean for 3 independent RT-PCR assays for each condition and cell type in a representative experiment.
After demonstrating that PTPROt is a physiologic target of BCL6 in normal B cells (Figure 4A), we asked whether BCL6 modulates PTPROt expression in a representative BCR-type DLBCL cell line (Figure 4B). Under conditions in which BCL6 siRNA increased the abundance of the known BCL6 target, FCER2 (data not shown), BCL6 depletion increased PTPROt transcript abundance 10-fold (Figure 4A). Of note, BCL6 depletion did not change the expression of total SYK, upstream BCR pathway components, such as CD79a, or downstream adapter proteins, such as BLNK. In a non–BCR-type cell line K422, BCL6 depletion did not up-regulate PTPROt expression (see supplemental Figure 2B). These data indicate that BCL6 modulates PTPROt expression in normal B cells and certain DLBCLs.
PTPROt dephosphorylates SYK Y352
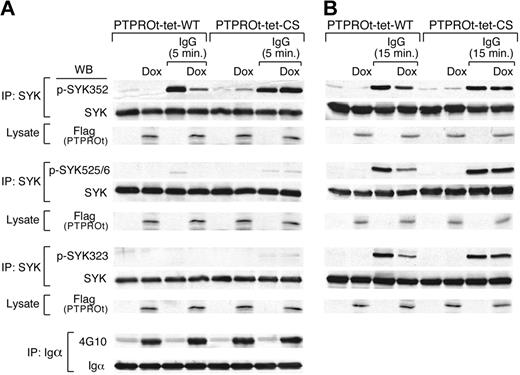
SYK activation requires LYN-mediated phosphorylation of SYK Y352 and Y348 in the linker region followed by autophosphorylation of SYK Y525/526 in the catalytic domain. Subsequent phosphorylation of SYK Y323 leads to Cbl-mediated ubiquitylation and degradation of SYK. To identify the PTPROt SYK tyrosine substrate, we overexpressed Flag-tagged WT or CS mutant PTPROt in DLBCL cells, cross-linked the BCR for 5 minutes, and compared BCR-induced phosphorylation at SYK Y352, Y525/526, and Y323 (Figure 5A). At this early time point, SYK Y352 phosphorylation was specifically inhibited by overexpression of WT-PTPROt but not by the inactive CS-PTPROt mutant (Figure 5A and supplemental Figure 3A). In the same experiments, WT-PTPROt overexpression did not alter the phosphorylation of upstream BCR pathway components such as CD79a (Igα; Figure 5A bottom panel).
PTPROt overexpression inhibits SYK Y352 tyrosyl phosphorylation. Tet-inducible Flag-tagged WT or mutant (CS) PTPROt clones were cultured with or without Dox and stimulated with goat anti–human IgG for 5 minutes (A), 15 minutes (B), or left untreated. Thereafter, cells were lysed and immunoprecipitated with anti-SYK or anti-CD79a antibody. Immunoprecipitates were size fractionated, blotted, and analyzed with the indicated phosphotyrosine antibodies (anti-pSYK352, -pSYK525/526, -pSYK323, and 4G10, respectively). The blots was subsequently stripped and blotted with an anti-pan SYK or anti-CD79a antibody. In each experiment, the corresponding whole cell lysates were simultaneously size-fractionated, blotted, and analyzed with Flag antibody to confirm Dox-induced overexpression of WT-PTPROt or CS-PTPROt.
PTPROt overexpression inhibits SYK Y352 tyrosyl phosphorylation. Tet-inducible Flag-tagged WT or mutant (CS) PTPROt clones were cultured with or without Dox and stimulated with goat anti–human IgG for 5 minutes (A), 15 minutes (B), or left untreated. Thereafter, cells were lysed and immunoprecipitated with anti-SYK or anti-CD79a antibody. Immunoprecipitates were size fractionated, blotted, and analyzed with the indicated phosphotyrosine antibodies (anti-pSYK352, -pSYK525/526, -pSYK323, and 4G10, respectively). The blots was subsequently stripped and blotted with an anti-pan SYK or anti-CD79a antibody. In each experiment, the corresponding whole cell lysates were simultaneously size-fractionated, blotted, and analyzed with Flag antibody to confirm Dox-induced overexpression of WT-PTPROt or CS-PTPROt.
To assess the effects of PTPROt on the subsequent phosphorylation of SYK Y525/526 and Y323, we performed the same experiments after 15 minutes of BCR cross-linking (Figure 5B and supplemental Figure 3B). At this later time point, when phosphorylation of all 3 SYK tyrosine residues is detectable, overexpression of WT-PTPROt, but not CS-PTPROt, inhibited phosphorylation of SYK Y352, Y525/526, and Y323 (Figure 5B and supplemental Figure 3B). Taken together, these results indicate that PTPROt dephosphorylates SYK Y352, limiting subsequent SYK Y525/526 autophosphorylation and activation. Consistent with these observations, shRNA-mediated down-regulation of PTPROt in DHL4 cells was associated with significantly higher SYK Y352 and BLNK Y84 phosphorylation than in mock-transfected cells (see supplemental Figure 4). Given the extremely low baseline expression of PTPROt, these results further highlight the role of this phosphatase in modulating tonic and Ig-cross-linking associated BCR signaling (see supplemental Figure 4).
BCL6-mediated repression of PTPROt increases SYK Y352 phosphorylation and promotes BCR signaling
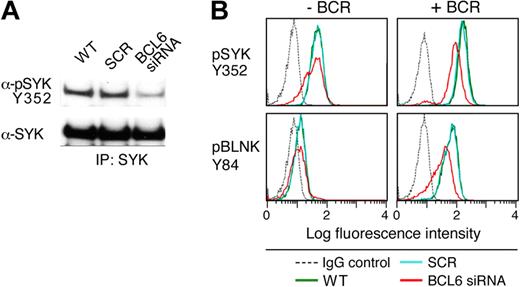
After demonstrating that PTPROt dephosphorylates SYK Y352, we assessed the consequences of BCL6 depletion on SYK phosphorylation and tonic BCR signaling in a representative BCR DLBCL cell line (DHL4). The DLBCL line was transduced with BCL6-siRNA or scrambled control; thereafter, cells were lysed, immunoprecipitated with pan SYK antibody, and immunoblotted with an anti-phospho SYK Y352 antibody. SYK phosphorylation was markedly lower in BCL6-depleted cells than in parental or mock-transduced cells (Figure 6A). We next used single-cell phospho-specific flow cytometry to specifically assess SYK Y352 phosphorylation and BCR signaling after BCL6 knockdown. Tonic and BCR crosslink-associated phosphorylation of SYK Y352 was much lower in BCL6-depleted cells than in control and parental cells (Figure 6B). Phosphorylation of the associated adaptor protein, BLNK, was similarly decreased in BCL6-depleted, but not in control or parental, cells (Figure 6B). Taken together, these data confirm that BCL6 regulates tonic and BCR-crosslink-induced signaling of the BCR pathway by repressing the SYK phosphatase, PTPROt (Figures 5–6).
BCL6-mediated repression of PTPROt increases tonic BCR signaling. The DLBCL cell line, DHL4, was transduced with BCL6-siRNA, SCR control oligonucleotides, or left untreated. Cells were subsequently incubated for 72 hours and stimulated with goat anti–human IgG (10 μg/mL) for 1 minute or left untreated. (A) Western analysis of SYK phosphorylation after BCL6 depletion. BCR-cross-linked cells were lysed and immunoprecipitated with anti-SYK. Immunoprecipitates were size-fractionated, blotted, and analyzed thereafter with α-pSYK Y352 antibody. The membrane was subsequently stripped and blotted with an anti-pan SYK antibody. (B) Phospho-specific flow cytometric analysis of tonic and α-Ig induced SYK Y352 and BLNK Y84 phosphorylation after BCL6 depletion. SYK Y352 and BLNK Y84 phosphorylation (top and bottom panels) was compared in cells transduced with BCL6-siRNA (red), SCR control oligonucleotides (blue), or left untreated (green) in the absence (left panel) or presence (right panel) of BCR cross-linking. Cells stained with an isotype-matched control Ig are also shown (gray dashed line). The x-axis denotes expression (log scale) and the y-axis indicates cell number.
BCL6-mediated repression of PTPROt increases tonic BCR signaling. The DLBCL cell line, DHL4, was transduced with BCL6-siRNA, SCR control oligonucleotides, or left untreated. Cells were subsequently incubated for 72 hours and stimulated with goat anti–human IgG (10 μg/mL) for 1 minute or left untreated. (A) Western analysis of SYK phosphorylation after BCL6 depletion. BCR-cross-linked cells were lysed and immunoprecipitated with anti-SYK. Immunoprecipitates were size-fractionated, blotted, and analyzed thereafter with α-pSYK Y352 antibody. The membrane was subsequently stripped and blotted with an anti-pan SYK antibody. (B) Phospho-specific flow cytometric analysis of tonic and α-Ig induced SYK Y352 and BLNK Y84 phosphorylation after BCL6 depletion. SYK Y352 and BLNK Y84 phosphorylation (top and bottom panels) was compared in cells transduced with BCL6-siRNA (red), SCR control oligonucleotides (blue), or left untreated (green) in the absence (left panel) or presence (right panel) of BCR cross-linking. Cells stained with an isotype-matched control Ig are also shown (gray dashed line). The x-axis denotes expression (log scale) and the y-axis indicates cell number.
Discussion
Herein, we demonstrate that BCL6 and the SYK phosphatase PTPROt exhibit a reciprocal pattern of expression in normal B cells and BCL6-dependent DLBCLs. In addition, we show that BCL6 binds to the PTPROt promoter and represses PTPROt transcription. Enforced expression of BCL6 decreases PTPROt expression in naive B cells, and BCL6 depletion increases PTPROt expression in normal centroblasts and certain DLBCLs. Consistent with these observations, BCL6-mediated repression of PTPROt leads to increased phosphorylation of SYK and enhanced tonic and ligand-dependent BCR signaling.
Tonic BCR signaling is thought to be initiated and regulated by stochastic interactions between PTKs and PTPs within cell membrane lipid rafts. In this homeostatic equilibrium model,2 positive regulators transiently and stochastically interact with the BCR complex and activate receptor-associated PTKs, such as SYK. This positive PTK-associated regulatory arm is counterbalanced by the recruitment of PTPs, such as PTPROt. Inhibition of the PTP-dependent negative-regulatory arm stabilizes and enhances tonic BCR signals. The role of PTPs in regulating tonic BCR signaling was first suggested by studies in which BCR-proximal PTKs were activated by treatment with the phosphatase inhibitor, pervanadate/H2O2, in the absence of BCR cross-linking.2,3,7,25 The current study identifies BCL6-mediated repression of PTPROt as one mechanism for enhanced SYK-dependent tonic BCR signaling.
PTPROt is a tissue-specific and developmentally regulated member of the PTPRO family, a group of highly conserved receptor-type PTPs that are thought to function as tumor suppressors. The epithelial isoform, PTPRO, has been implicated in lung and liver cancers.26,27 In model systems of these cancers, PTPRO inhibits anchorage-independent growth, delays cell cycle reentry, and increases susceptibility to apoptosis.26,27 The long PTPRO and short PTPROt isoforms are expressed from 2 distinct promoters.11 The PTPRO promoter, which is located approximately 220 kbp upstream of the PTPROt regulatory region, includes a CpG island. This feature is of note because the decreased expression of PTPRO in epithelial tumors is associated with tumor-specific hypermethylation of the PTPRO CpG island.26,27 Because the PTPROt promoter does not include a comparable CpG-rich region, other mechanisms are likely to control PTPROt expression.
In normal GC B cells, PTPROt is repressed by BCL6, a sequence-specific transcriptional repressor that functions as a master-regulator of the GC reaction.28 The finding that BCL6 suppresses PTPROt identifies a novel function for BCL6 in normal GC B cells. Within the GC, centroblasts proliferate rapidly and undergo somatic hypermutation of their immunoglobulin genes, which is the basis for affinity-maturation of antibodies.16,28 BCL6-mediated repression of the SYK phosphatase, PTPROt, likely lowers the threshold for tonic and ligand-induced signals from low-affinity BCRs in GC B cells, facilitating their survival. Once high-affinity BCRs are generated, enhanced ligand-induced BCR signaling promotes BCL6 down-regulation via MAPK-dependent phosphorylation of BCL6 PEST domains and associated proteasomal degradation, licensing exit from the GC.29
The tight spatio-temporal control of BCR signaling by BCL6 is likely altered in DLBCLs with deregulated BCL6 expression. In certain DLBCLs, the constitutive expression of BCL6 represses PTPROt and augments SYK-dependent BCR signaling. These observations identify a novel BCL6-dependent prosurvival pathway in B-cell lymphomagenesis. Because BCL6 and SYK are both promising rational therapeutic targets in the same group of DLBCLs, combined inhibition of these functionally related pathways warrants further study.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This paper was funded by National Institutes of Health grant 5 PO1 CA092625 and LLS SCOR grant 7391-07.
National Institutes of Health
Authorship
Contribution: P.J. designed and performed research, analyzed data, and wrote the paper; L.C. designed and performed research; E.O. performed research; J.M.P. designed and performed research and analyzed data; S.M.R. designed and performed research; R.D.F. contributed vital new reagents or analytical tools and analyzed data; A.M. designed research and analyzed data; and M.A.S. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Margaret A. Shipp, Dana-Farber Cancer Institute, 44 Binney St, Boston, MA 02115; e-mail: margaret_shipp@dfci.harvard.edu.

![Figure 1. Reciprocal patterns of PTPROt and BCL6 expression in normal B cells and primary DLBCLs. (A) Relative BCL6 and PTPROt transcript abundance in 2 independent series of highly purified normal B cells (naive, GC centroblasts [CB] and centrocytes [CC], memory). (B) Relative BCL6 and PTPROt transcript abundance in 176 newly diagnosed and previously profiled DLBCLs.14 Normal GC B cells that were profiled at the same14 time were included for comparison (left). Comprehensive cluster designations for the primary DLBCLs and normal GC cells are indicated above the heat map. The primary DLBCL series includes 77 BCR, 50 OxPhos and 49 HR tumors. Color scale at bottom indicates relative expression and SDs from the mean. Red connotes high-level expression; blue indicates low-level expression](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/26/10.1182_blood-2009-02-204362/4/m_zh89990945960001.jpeg?Expires=1767710935&Signature=gZfQ61ozlIupI8ZMwLfkP4LWUkkYtHvH01zevxo5EqLsD3s2EWp3~nuW1~H9hVLBOqCsDxutP8AMimCfRNGalCTqyZ3pTCDwUEpIvgK2NoLdLglx4p46GYxxjB8IS8feUOZgzj2Pc~vOBA-LkOH2XZVsSb3kZ3ieBEb~M7FGj5IaB-4VA35zy01ztoDH8DzflKSaH3urcyOLAFxUvJTAbvcfNDR3uU8PyGXBThGnzWPOtNgRPGm86vT0HOUVL4PZKz6TOeq1xeIOQlacd5aLSATAVnV2eKo72naCwENczTfQAIxdC0dXLqaDzOhCetH1gI2yqJ5GKcNDrGm0V6JCSQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)