Abstract
Despite continual improvement, morbidity and mortality after hematopoietic stem cell transplantation (HSCT) remain high. The importance of chemokines in HSCT lies in their regulation of immune responses that determine transplantation outcomes. We investigated the role of recipient and donor chemokine system gene polymorphisms by using a candidate gene approach on the incidence of graft-versus-host disease and posttransplantation outcomes in 1370 extensively human leukocyte antigen–matched, unrelated donor-recipient pairs by using multivariate Cox regression models. Our analysis identified that recipients homozygous for a common CCR5 haplotype (H1/H1) had better disease-free survival (DFS; P = .005) and overall survival (P = .021). When the same genotype of both the donor and recipient were considered in the models, a highly significant association with DFS and overall survival was noted (P < .001 and P = .007, respectively) with absolute differences in survival of up to 20% seen between the groups at 3 years after transplantation (50% DFS for pairs with recipient CCR5 H1/H1 vs 30% for pairs with donor CCR5 H1/H1). This finding suggests that donor and/or recipient CCR5 genotypes may be associated with HSCT outcome and suggests new diagnostic and therapeutic strategies for optimizing therapy.
Introduction
Hematopoietic stem cell transplantation (HSCT) from a human leukocyte antigen (HLA)–matched unrelated donor has become the definitive treatment for many patients with myelodysplastic syndrome and leukemia; however, despite marked improvements in pretransplantation donor-recipient matching and posttransplantation care, treatment-related morbidity (TRM) and mortality remain high.1 In particular, serious complications such as disease relapse, graft failure, life-threatening infection, and graft-versus-host disease (GVHD) are still difficult to completely prevent with current therapies. Genetic factors of the donor or recipient that might influence these outcomes, other than the need for precise HLA matching, are poorly understood.
Chemokines play critical roles in the maturation, maintenance, and activation of the immune system because leukocytes possess functional chemokine receptors that are the major mechanism directing their migration to damaged areas and normal lymphoid tissues.2-4 Inflammatory chemokines are small proteins secreted by almost all cells in response to injury or inflammatory stimuli, whereas homeostatic chemokines are expressed constitutively, especially in lymphoid tissues. Thus, the chemokine system is crucial to regulating the immune responses to a variety of traumatic, ischemic, and infectious injuries, as well as being intimately involved in the immune response to foreign antigens. Therefore, the chemokine system is a logical place to look for genetic factors that may affect the outcome of HSCT.
Hematopoietic malignancies express chemokine receptors, and these have effects on proliferation and response to chemotherapy.5,6 Induction chemotherapy and pretransplantation radiation therapy both induce tissue injury that cause chemokine release.5 CD34+ hematopoietic stem cells (HSCs) use chemokines to home to bone marrow and provide proliferative signals that allow them to reestablish the immune system. GVHD requires the recognition of the recipient as foreign and activation of the donor immune system, and infection and vaccination response are controlled by the chemokine system.7,8 Thus, the chemokine system may have a profound influence on many aspects of both successful and unsuccessful HSCT.
In this candidate gene study we focused on common, functional polymorphisms in the CX3CR1 and CCR5 chemokine receptors and CCL1 (a CCR8 ligand), CCL2 (a CCR2 ligand), and CCL5 (a CCR5 ligand) chemokine genes because these genes have been shown to be important in transplantation outcomes in previous animal and human studies.9-16 In particular, blockade or lack of the chemokine receptors CCR2 and CCR5 on donor leukocytes by administration of anti-CCR5 antibody or infusion of CCR2−/− T cells reduces the incidence of GVHD in animal models.10,11 Supporting the role of CCR5 in GVHD, a single nucleotide polymorphism (SNP) in chemokine ligand 5 (CCL5, also known as RANTES) has been significantly associated with a greater incidence of chronic graft-versus-host disease (cGVHD).12 The authors of another study13 demonstrated that recipients with the nonfunctional CCR5Δ32 allele were less likely to develop acute graft-versus-host disease (aGVHD), and other authors14-16 have found CCR5 promoter polymorphisms to be predictive of the response to chemotherapy in leukemia and in the likelihood of cytomegalovirus and Epstein-Barr virus reactivation after HSCT.
In this study we analyzed 1370 bone marrow transplantation recipients and their respective donors. To minimize any confounding effects of HLA mismatch, we only used those pairs that were high-resolution matched at the HLA A, B, C, and DRB1 and DQB1 loci (ie, 10/10 HLA match) by using samples from the US National Marrow Donor Program (NMDP). We found significant survival outcome associations with a genotype of the CCR5 gene that has been previously shown in multiple studies to be associated with an increased rate of HIV progression to AIDS. This association is believed to occur because persons homozygous for this haplotype have greater levels of expression of the chemokine receptor and HIV coreceptor, CCR5.17,18 This high-expressing CCR5 genotype, termed 59029 A/A,19 P1/P1,20 −2459A/A,21 or HHE/HHE,22 also has been associated with renal transplantation outcomes.23 Here we used 4 variants of the CCR5 gene to define the high-expressing haplotype, which in this study we term CCR5 H1. We found a strongly significant association of the homozygous CCR5 H1/H1 genotype with survival outcomes after HSCT when both the donor and recipient genotype was analyzed. Thus, this study provides additional evidence of the chemokine system's role in survival after transplantation and indicates that both the genotype of the donor and that of the recipient may have significant impact on HSCT outcomes.
Methods
Data source
The Center for International Blood and Marrow Transplant Research (CIBMTR) is a research affiliate of the International Bone Marrow Transplant Registry, Autologous Blood and Marrow Transplant Registry, and the NMDP, established in 2004. It comprises a voluntary working group of more than 450 transplantation centers worldwide that contribute detailed data on consecutive allogeneic and autologous HSCT to a statistical center at the Medical College of Wisconsin in Milwaukee and the NMDP coordinating center in Minneapolis. Participating centers are required to report all transplantations consecutively; compliance is monitored by on-site audits. Patients are followed longitudinally, with yearly follow-up. Computerized checks for discrepancies, physicians' review of submitted data, and on-site audits of participating centers ensure data quality. Observational studies conducted by the CIBMTR are performed in compliance with the Privacy Rule under the Health Insurance Portability and Accountability Act of 1996, as a Public Health Authority and in compliance with all applicable federal regulations pertaining to the protection of human research participants and the Declaration of Helsinki as determined by continuous review of the Institutional Review Boards of the National Marrow Donor Program and the Medical College of Wisconsin since 1985.
Patient population
Recipient/donor pairs from 1370 unrelated HLA-A, B, C, DRB1, and DQB1 allele-matched transplantations facilitated by the NMDP were included in the study. HLA typing was confirmed through the NMDP's ongoing retrospective high-resolution typing project as previously described.24 Transplantations were performed between 1990 and 2002. Eligible diagnoses included acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), chronic myeloid leukemia (CML), and myelodysplastic syndrome (MDS). Early-stage disease was defined as AML and ALL in first complete remission, CML in first chronic phase, and MDS subtype refractory anemia. Intermediate-stage disease was AML or ALL in second or subsequent complete remission or in first relapse, and CML in accelerated phase or second chronic phase. Advanced-phase disease was AML in second or greater relapse or primary induction failure, CML in blast phase, MDS subtypes refractory anemia with excess blasts or in transformation, or MDS not otherwise classified. The median follow-up of survivors was 96 months (range, 3-198 months). The median age was 37 years (range, 1-66 years). A total of 92% of patients received bone marrow as a graft source. A total of 45% had early disease, whereas 24% had advanced disease at the time of transplantation. Only 16% of the patients received T-cell depletion as part of the GVHD prophylaxis.
All surviving recipients included in this analysis were retrospectively contacted and provided informed consent for participation in the NMDP research program. Informed consent was waived by the NMDP Institutional Review Board for all deceased recipients. Approximately 4% of surviving patients would not provide consent for research. All observational studies were approved by the Institutional Review Boards of the National Marrow Donor Program and the Medical College of Wisconsin. To adjust for the potential bias introduced by exclusion of nonconsenting surviving patients, a modeling process randomly excluded approximately the same percentage of deceased patients by use of a biased coin randomization with exclusion probabilities on the basis of characteristics associated with not providing consent for use of the data in survivors.25
Definition of outcomes
The primary end points analyzed in the study were overall survival (OS), TRM, severe (grade III-IV), acute GVHD occurring within the first 100 days after transplantation (aGVHD), and chronic (cGVHD). Relapse was defined as clinical or hematologic relapse of primary disease with death without evidence of disease as a competing risk. For CML patients our definition of relapse included cytogenetic, molecular, and hematologic relapse as an event. TRM was defined as death during a continuous complete remission. For analysis of disease-free survival (DFS), failures were relapse or death from any cause with patients who were alive and in complete remission censored at time of last follow-up. Our analysis of OS treated death from any cause as the event, and surviving patients were censored at the date of last contact. Grades III to IV acute GVHD were defined by use of the Glucksberg scale.26 Extensive chronic GVHD was defined according to the Seattle criteria.27
Genotyping
A total of 10 individual genetic variants and the number of copies of a high-expressing CCR5 haplotype derived from the CCR5 gene variants were studied (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). These variants were chosen because they were thought to be common and functional variants of the chemokine system in relevant genes to HSCT and because power calculations revealed a high likelihood of finding an association. Investigators performing the genotyping procedure were blinded as to the identity of the specimens or their respective clinical outcomes. Genomic DNA was isolated from cryopreserved leukocytes from donors and recipients following the manufacturer's directions (Promega). The resultant DNA was then quantitated (Nanodrop; Thermo Scientific) and whole genome amplified with Phi29 DNA polymerase. Genotyping was then performed with SNPlex technology (Applied Biosystems) and automated genotype calling software. Samples that failed to yield a genotype for at least 6 of 10 SNPs presumably because of degraded or minimal genomic DNA yield (n = 122, 9% of total samples) were excluded from further analysis. In the remaining samples we were able to obtain a genotype call rate of 99%.
Statistical analysis
Univariate probabilities of DFS and OS were calculated by the use of the Kaplan-Meier method. The log-rank test was used for comparing survival curves. Probabilities of TRM, relapse, aGVHD, and cGVHD were calculated with cumulative incidence estimates. Apart from TRM, the cumulative incidence calculated for aGVHD and cGVHD treated death as a competing risk.28 Relapse was treated as a competing risk for TRM and vice versa.
The probabilities of OS, DFS, TRM, relapse, and aGVHD and cGVHD for all patients were estimated by use of the Cox proportional hazards regression model with a multivariate analysis performed to identify clinical variables that were associated with particular outcomes. Clinical variables considered in the models were disease, disease status, recipient age, donor age, and sex. For each outcome, all variables were examined for proportional hazards. Factors violating the proportional hazards assumption (P ≤ .05) were adjusted by stratification. The stepwise model building approach was then used in developing models for all the outcomes. OS and DFS were adjusted for disease, disease stage, cytomegalovirus match, patient age, and donor age and stratified by graft type and Karnofsky score. Relapse was adjusted for disease and donor sex and stratified by disease stage and Karnofsky score. TRM was adjusted for disease stage, patient age, and donor age and stratified by graft type and Karnofsky score. aGVHD was adjusted for disease, GVHD prophylaxis, and year of transplantation and stratified by disease stage. cGVHD was adjusted for sex match and patient age and stratified by disease.
Potential interactions between the genetic variants and clinical covariates were tested and were not present. Because of the possibility of type I error occurring from multiple comparisons, the genetic associations were referred to as statistically significant when a P value was .01 or less. SAS software version 9.1 (SAS Institute) was used in all the analysis. Our plan for analyzing the data included examining donors and recipients individually and in an interactive manner because our hypothesis was that donor chemokine/chemokine receptor variants could interact with recipient chemokine/chemokine receptor variants.
Results
Genotyping
We used automated technology to genotype 10 chemokine and chemokine receptor gene variants (9 SNPs and 1 mutation) and were able to determine the number of copies of a CCR5 gene haplotype (termed CCR5 H1) in 1248 recipients and their respective donors, whose characteristics are shown in Table 1. The median age at transplantation was 37 years in both groups, with a range of 1 to 56 years and 1 to 66 years, and the median follow-up of survivors was 96 months in both groups, with a range of 36 to 198 months and 3 to 196 months for the H1/H1 recipients versus all others, respectively. The characteristics (age at transplantation, Karnofsky score, sex, disease, stage, etc) of the H1/H1 recipients were compared with the χ2 against the remainder of the population and were not significantly different (Table 1). We found each of the 10 genetic variants to be in Hardy-Weinberg equilibrium in this cohort, indicating a lack of evolutionary selection and reliable genotyping (Table 2). In addition, the allele frequencies were comparable with those obtained in previous studies of white US subjects, and genotypes were consistent with previously reported genetic linkage.20,23,29 Taken together these indicate that our genotyping methodology was robust and reliable.
Analysis of CCR5 haplotypes
There has been an increasing emphasis on the importance of haplotypes in genetic association studies, associations that may not be observed from individual allele analysis either because multiple alleles have opposite effects or only have effects because of interactions. In this study we wanted to analyze the effect of a high-expressing CCR5 haplotype that is also in complete linkage disequilibrium with a nonfunctional CCR5 mutation, CCR5Δ32, and thus might have opposing effects. Therefore, although the analysis of individual genetic variants revealed no association with outcomes (supplemental Table 1), we did find a significant association with the CCR5 haplotype.
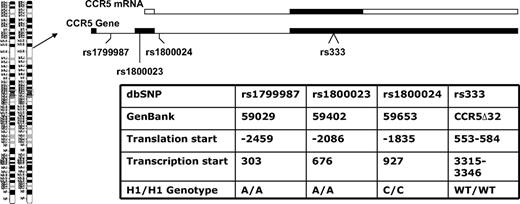
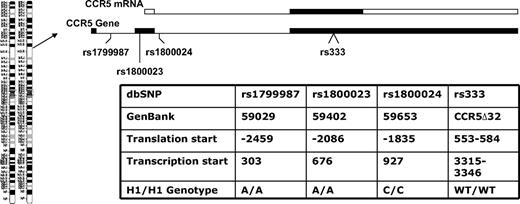
Donors and recipients who had the highest-expressing CCR5 haplotype were designated as CCR5 H1/H1 on the basis of having 2 copies of the high-expressing haplotype of the CCR5 promoter nearly identical to that designated HHE or P1 in previous epidemiologic studies in HIV patients. The CCR5 H1/H1 genotype was determined from the combination of genotypes by the use of 4 haplotype tagging SNPs (rs1799987, rs333, rs1800023, and rs1800024) that are in complete linkage disequilibrium because they are in close physical proximity on chromosome 3 (Figure 1). These persons have an A/A genotype at position 59029 (rs1799987), an A/A genotype at position 59402 (rs1800024), and a C/C genotype at position 59653 (rs1800023) in the CCR5 promoter and did not have the CCR5Δ32 mutation (Figure 1).
CCR5 gene organization and genotypes studied. The CCR5 gene is on chromosome 3p21 and is a 3-exon gene (filled boxes in the CCR5 gene diagram) with a proximal and distal promoter region (not shown). The relative positions of the CCR5 variants used to determine the CCR5 H1/H1 genotype are shown. The entire open reading frame of the gene is on exon 3 (filled box in the CCR5 mRNA diagram) and the majority of transcripts originate in exon 2 from the proximal promoter. Thus, the first 2 SNPs are promoter polymorphisms in the proximal promoter, the third SNP is in the intron, and fourth is the 32-basepair deletion in the open reading frame. Different nomenclatures have been used for the same position and genotype in different publications,19-22 so a table is provided with these designations and the nucleotide sequence that defines the CCR5 H1/H1 genotype in this study.
CCR5 gene organization and genotypes studied. The CCR5 gene is on chromosome 3p21 and is a 3-exon gene (filled boxes in the CCR5 gene diagram) with a proximal and distal promoter region (not shown). The relative positions of the CCR5 variants used to determine the CCR5 H1/H1 genotype are shown. The entire open reading frame of the gene is on exon 3 (filled box in the CCR5 mRNA diagram) and the majority of transcripts originate in exon 2 from the proximal promoter. Thus, the first 2 SNPs are promoter polymorphisms in the proximal promoter, the third SNP is in the intron, and fourth is the 32-basepair deletion in the open reading frame. Different nomenclatures have been used for the same position and genotype in different publications,19-22 so a table is provided with these designations and the nucleotide sequence that defines the CCR5 H1/H1 genotype in this study.
Donors and recipients homozygous for the CCR5 H1/H1 haplotype were compared with all others because persons with one copy (H1/−) were found to have very similar outcomes to those with none (−/−), which was the comparison group (defined as hazard ratio [HR], 1.0). For example, donors with one copy of CCR5 H1 (heterozygotes) had a HR of 0.94 (P = .666) for aGVHD and recipients with the same genotype had a HR of 0.92 (P = .283) for DFS. We thus focused additional analysis on a recessive genetic analysis of this genotype (CCR5 H1/H1 vs all others).
Donor CCR5 genotype analysis
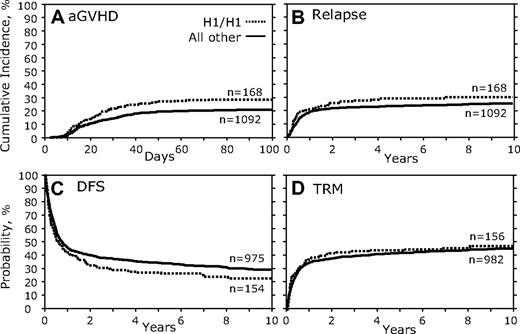
We first examined the association between donor genotypes and transplantation outcomes. There were no fully significant associations for the outcomes of HSCT in which persons with the CCR5 H1/H1 genotype were the donors in any of the measured outcomes (Table 3; Figure 2). However, a trend was noted for increased aGVHD (HR, 1.46; 95% confidence interval [95% CI], 1.070-1.982; P = .017; Table 3; Figure 2A) and decreased DFS (HR, 0.80; 95% CI, 0.651-0.973; P = .026; Figure 2C; Table 3). No significant association of donor CCR5 H1/H1 genotype was found with relapse (Figure 2B; Table 3), TRM (Figure 2D; Table 3), OS, or cGVHD (Table 3).
Donor CCR5 H1/H1 genotype versus all others. (A) Cumulative incidence curves of aGVHD grades III to IV for the first 100 days after transplantation. (B) Cumulative incidence of relapse 10 years after transplantation. (C) Kaplan-Meier curves for DFS. (D) Cumulative incidence of TRM.
Donor CCR5 H1/H1 genotype versus all others. (A) Cumulative incidence curves of aGVHD grades III to IV for the first 100 days after transplantation. (B) Cumulative incidence of relapse 10 years after transplantation. (C) Kaplan-Meier curves for DFS. (D) Cumulative incidence of TRM.
Recipient CCR5 genotype analysis
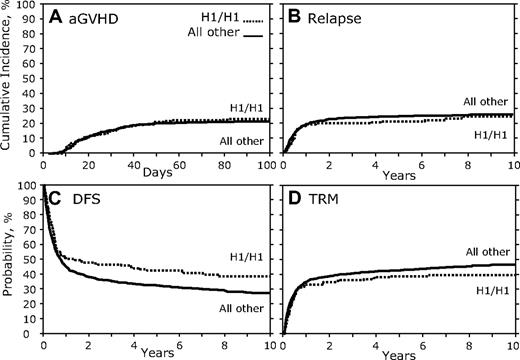
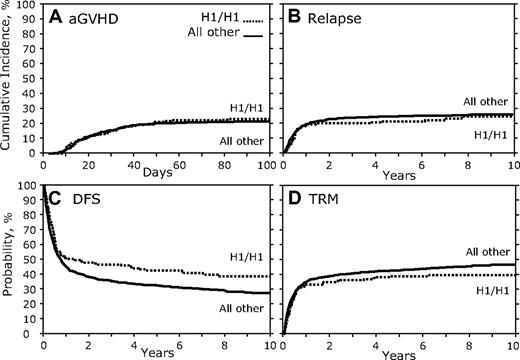
In contrast to the results of the donor genotype comparison, when recipients homozygous for the CCR5 H1/H1 genotype were compared with all others, we found a significant association (Table 3; Figure 3). There was no association with aGVHD, cGVHD, or relapse (Figure 3A-B; Table 3); however, there was a significant association with increased DFS (HR, 1.36; 95% CI, 1.098-1.684; P = .005; Figure 3C; Table 3). Recipients with the CCR5 H1/H1 genotype also showed a trend toward decreased TRM (HR, 0.76; 95% CI, 0.580-0.987; P = .039; Figure 3D; Table 3) and increased OS (HR, 1.28; 95% CI, 1.038-1.587; P = .021; Table 3). A secondary analysis of the reported causes of death of CCR5 H1/H1 recipients versus others failed to find a significant difference in the subset of persons for which these data were available (supplemental Table 2).
Recipient CCR5 H1/H1 genotype versus others. (A) Cumulative incidence curves of aGVHD grades III to IV for the first 100 days after transplantation. (B) Cumulative incidence of relapse 10 years after transplantation. (C) Kaplan-Meier curves for DFS. (D) Cumulative incidence of TRM.
Recipient CCR5 H1/H1 genotype versus others. (A) Cumulative incidence curves of aGVHD grades III to IV for the first 100 days after transplantation. (B) Cumulative incidence of relapse 10 years after transplantation. (C) Kaplan-Meier curves for DFS. (D) Cumulative incidence of TRM.
Donor and recipient combinatorial CCR5 genotype analysis
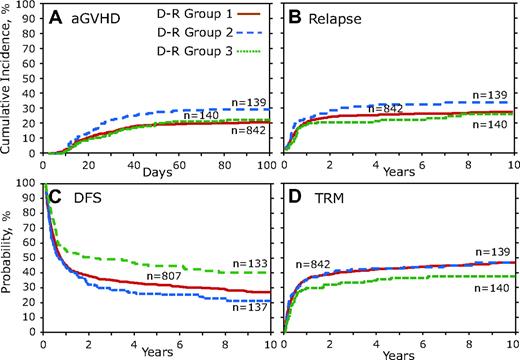
Because donor and recipient CCR5 genotypes could have important in vivo interactions, we examined the association of different donor and recipient genotype combinations. To accomplish this analysis, donors and recipients were combined into groups on the basis of CCR5 genotype (D-R groups). Donors and recipient pairs who both lacked the high-expressing CCR5 H1/H1 genotype were combined into D-R group 1, which was the reference group (HR, 1.0). The D-R group 2 was defined as those pairs in whom the donor had the CCR5 H1/H1 genotype but the recipient did not, and conversely D-R group 3 was those in whom the recipient had the CCR5 H1/H1 genotype but the donor did not. Finally, D-R group 4 was defined as when both the donors and recipients had the CCR5 H1/H1 genotype; however, the number of subjects (n = 17) in this category was of insufficient size to accurately determine risk estimates and was therefore removed from further consideration, although an analysis with this group included did not alter the conclusions reported here.
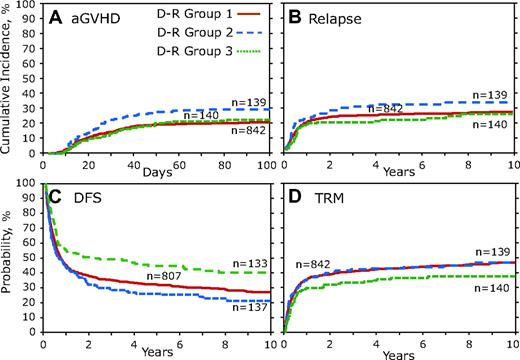
D-R group 1 was compared individually against D-R groups 2 and 3 by the use of multivariate analysis and the results presented as the overall 2 degree of freedom (2DF) test for a difference between all 3 groups and as HR, 95% CI, and P value for each pair-wise comparison group (ie, D-R group 1 vs 2 or D-R group 1 vs 3; Table 4). Although the overall 2DF test for aGVHD did not reach our significance (P = .041), D-R group 2 was significantly associated with a greater risk of aGVHD (HR, 1.52; 95% CI, 1.097-2.098; P = .012; Figure 4A; Table 4). There was not a significant difference in relapse (Figure 4B), but multivariate analysis of combinatorial D-R groups demonstrated that all 3 groups were significantly associated with DFS (P < .001). We observed significantly increased DFS in D-R group 3 (HR, 1.49; 95% CI, 1.176-1.894; P = .001; Table 4), and there was a large (>25%) difference in mortality at 3 years after transplantation between D-R group 3 and D-R group 2 (Figure 4C). Although the 2DF test was not significant (P = .033), in the pairwise comparisons TRM was significantly lower in D-R group 3 (HR, 0.69; 95% CI, 0.512-0.925; P = .013; Table 4). Multivariate analysis of the D-R groups showed a significant association with OS (P = .007). D-R group 3 had a significant increase in OS (HR, 1.41; 95% CI, 1.114-1.783; P = .004; Table 4). At 3 years after transplantation D-R group 3 had 52% survival compared with only 36% for D-R group 2.
Donor-recipient CCR5 genotype analysis. Donor-recipient pairs who both lacked the high-expressing CCR5 H1/H1 genotype were combined into D-R group 1. D-R group 2 was defined as those pairs in which the donor had the CCR5 H1/H1 genotype but the recipient did not; conversely, in D-R group 3 the recipient had the CCR5 H1/H1 genotype but the donor did not. D-R group 4 was not included because of small number of persons (n = 17). (A) Cumulative incidence curves of aGVHD grades III to IV for the first 100 days after transplantation. (B) Cumulative incidence of relapse 10 years after transplantation. (C) Kaplan-Meier curves for DFS. (D) Cumulative incidence of TRM.
Donor-recipient CCR5 genotype analysis. Donor-recipient pairs who both lacked the high-expressing CCR5 H1/H1 genotype were combined into D-R group 1. D-R group 2 was defined as those pairs in which the donor had the CCR5 H1/H1 genotype but the recipient did not; conversely, in D-R group 3 the recipient had the CCR5 H1/H1 genotype but the donor did not. D-R group 4 was not included because of small number of persons (n = 17). (A) Cumulative incidence curves of aGVHD grades III to IV for the first 100 days after transplantation. (B) Cumulative incidence of relapse 10 years after transplantation. (C) Kaplan-Meier curves for DFS. (D) Cumulative incidence of TRM.
Discussion
In this study, we examined the association between common functional chemokine system variants with 6 HSCT outcomes (aGVHD, cGVHD, relapse, TRM, DFS, and OS) in 1370 extensively HLA-matched (10 of 10 alleles) pairs of unrelated donors and recipients. All of the recipients were transplanted since 1990 and received state-of-the-art medical care. The majority of our recipients underwent myeloablative conditioning regimens (total body irradiation and antineoplastic drugs). Initially we developed Cox proportional hazard models by using clinical variables previously shown to have significant effects on HSCT outcomes. Using the genetic data and the models, we were then able to analyze each variant in a multivariate controlled manner and discover associations related to both the donor and recipient genotype and their interaction.
When the CCR5 donor and recipient genotype, defined by possessing 2 copies of the H1 haplotype, was put into the models, we found highly significant associations with both OS and DFS. In our pairwise comparison, a difference was observed in aGVHD between D-R group 2 (donors with the CCR5 H1/H1 genotype, recipients without it) compared with D-R group 1. Because there was no association between recipient genotype and aGVHD, donor genotype should chiefly contribute to this association. Chemokines and chemokine receptors have been shown to a play a critical role in regulating the trafficking of alloreactive donor T cells into GVHD target organs. Therefore, various strategies have been applied in mouse models to reduce the severity of GVHD through blockade of the chemokine system. Anti-CCR5 antibody reduced inflammatory cell trafficking and resulted in protection from hepatic GVHD.10
Our findings would be consistent with increased CCR5 expression in donor lymphocytes causing increased inflammatory responses in affected parenchymal tissues by more efficiently trafficking to them and the lymphoid tissue, such as intestinal Peyer patches where they are activated. An increase in the risk of severe aGVHD (grade III or IV) can directly affect survival or indirectly enhance mortality as a result of the additional treatment-related immunosuppression. Moreover, the greater probability of developing severe aGVHD observed in patients who receive donor CCR5 genotype H1/H1 bone marrow stem cells may contribute to the observed increase in the incidence of TRM in our pairwise comparison, which was especially apparent after the first 6 months after transplantation. The decreased DFS and OS seen in non-H1/H1 patients receiving CCR5 genotype H1/H1 donor bone marrow cells further support this explanation.
HSCT recipients who had CCR5 genotype H1/H1 demonstrated a significant association with increased DFS and a potential association with decreased TRM and increased OS. However, no significant association was found between the recipient CCR5 genotype H1/H1 and aGVHD or relapse as shown in Table 3. The lack of effect on aGVHD by recipient CCR5 is in accordance with the lack of host T cells after the pretransplantation conditioning regimen and the replacement with donor CCR5-expressing cells. Consequently, we would not expect the recipient genotype to correlate with the risk of aGVHD. A biologic mechanism of action of recipient CCR5 levels on survival outcomes remains to be elucidated, but one could speculate that it might involve better engraftment, more favorable host or tumor chemotherapy/radiotherapy response, or improved host defenses against posttransplantation infections.
The importance of genetic variability on individual response to disease pathogenesis is becoming increasingly recognized. Because there is great variation in the mortality of patients after transplantation despite available tools, there is a clear need to develop a comprehensive panel of predictive biologic markers for pretransplantation testing. Our data, with an emphasis on results observed in the combinatorial analysis, could identify high-risk patients not revealed even with the most extensive HLA matching. This finding raises the possibility that profiling donors and recipients for the CCR5 genotype investigated could improve the morbidity and mortality associated with HSCT therapy by providing individualized predictive biomarkers associated with outcomes. At this time, more studies are needed to better understand the biologic mechanisms explaining the differential outcomes observed in this study.
However, this study shares some of the common limitations of genetic association studies, and therefore the findings will need to be interpreted with some degree of caution until they can be replicated in another suitably sized cohort. This study represents the largest to date of extensively HLA-matched HSCT patients, and our design has allowed us to investigate the interaction of both donor and recipient genotype and adjust for other known clinical variables. Multiple comparisons creating false-positive associations have been considered, but with our number of comparisons based on testing a limited number of candidate genes, our biostatisticians determined that P .01 or less would be an appropriate cutoff for the level of significance. Like any similar study, we are unable to conclude that any associations found are not actually attributable to closely linked genetic variants that are the true causal variant. Despite all these limitations, if these data are proven to be significant in a second cohort, they could have substantial clinical impact on the morbidity and mortality seen in HSCT recipients.
Identification of individual predictive CCR5 or other immune system genetic variants could allow physicians, through a personalized medicine approach, to tailor preventative therapies and better anticipate the likelihood of complications for patients with a variety of diseases. Plerixafor and maraviroc, inhibitors of 2 chemokine receptors (CXCR4 and CCR5), have recently been approved by the Food and Drug Administration. These new drugs show that the chemokine system has become an attractive target to control undesirable immune reactions and identify that further understanding of the mechanism of these associations may allow pharmaceutical manipulation of this system to improve HSCT outcomes. In summary, if our findings are confirmed regarding the association of donor and recipient CCR5 genotypes with HSCT outcomes, this could contribute to the development of important predictive clinical markers for high-risk patients, reduce the highly variable outcomes observed in HSCT, and also provide a potential individualized therapeutic target for effective pharmaceutical intervention.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Intramural Division of the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and a grant to R.A. by the National Marrow Donor Program (NMDP).
The CIBMTR is supported by Public Health Service Grant/Cooperative Agreement U24-CA76518 from the National Cancer Institute (NCI), the National Heart, Lung, and Blood Institute (NHLBI), and the National Institute of Allergy and Infectious Diseases (NIAID); a Grant/Cooperative Agreement 5U01HL069294 from NHLBI and NCI; a contract HHSH234200637015C with Health Resources and Services Administration (HRSA/DHHS); grants N00014-06-1-0704 and N00014-08-1-0058 from the Office of Naval Research; and grants from AABB; Aetna; American Society for Blood and Marrow Transplantation; Amgen Inc; anonymous donation to the Medical College of Wisconsin; Astellas Pharma US Inc; Baxter International Inc; Bayer HealthCare Pharmaceuticals; Be the Match Foundation; Biogen IDEC; BioMarin Pharmaceutical Inc; Biovitrum AB; BloodCenter of Wisconsin; Blue Cross and Blue Shield Association; Bone Marrow Foundation; Canadian Blood and Marrow Transplant Group; CaridianBCT; Celgene Corporation; CellGenix, GmbH; Centers for Disease Control and Prevention; Children's Leukemia Research Association; ClinImmune Labs; CTI Clinical Trial and Consulting Services; Cubist Pharmaceuticals; Cylex Inc; CytoTherm; DOR BioPharma Inc; Dynal Biotech, an Invitrogen Company; Eisai Inc; Enzon Pharmaceuticals Inc; European Group for Blood and Marrow Transplantation; Gamida Cell Ltd; GE Healthcare; Genentech Inc; Genzyme Corporation; Histogenetics Inc; HKS Medical Information Systems; Hospira Inc; Infectious Diseases Society of America; Kiadis Pharma; Kirin Brewery Co Ltd; The Leukemia & Lymphoma Society; Merck & Company; The Medical College of Wisconsin; MGI Pharma Inc; Michigan Community Blood Centers; Millennium Pharmaceuticals Inc; Miller Pharmacal Group; Milliman USA Inc; Miltenyi Biotec Inc; National Marrow Donor Program; Nature Publishing Group; New York Blood Center; Novartis Oncology; Oncology Nursing Society; Osiris Therapeutics Inc; Otsuka America Pharmaceutical Inc; Pall Life Sciences; PDL BioPharma Inc; Pfizer Inc; Pharmion Corporation; Saladax Biomedical Inc; Schering Corporation; Society for Healthcare Epidemiology of America; StemCyte Inc; StemSoft Software Inc; Sysmex America Inc; Teva Pharmaceutical Industries; THERAKOS Inc; Thermogenesis Corporation; Vidacare Corporation; Vion Pharmaceuticals Inc; ViraCor Laboratories; ViroPharma Inc; and Wellpoint Inc.
S.E.C. is a MA candidate at Boston University, and this work is submitted in partial fulfillment of the requirement for the MA.
The views expressed in this article do not reflect the official policy or position of the National Institute of Health, the Department of the Navy, the Department of Defense, or any other agency of the US government.
National Institutes of Health
Authorship
Contribution: D.H.M. and R.A. proposed and were principally responsible for the design of the research; T.W., M.A.A., and S.S were the biostatisticians, H.T.B.T. and E.M. isolated the genomic DNA; S.M.R. and S.F.P. performed the genotyping; and all authors contributed to the writing of the manuscript and analysis of the data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Reza Abdi, 221 Longwood Ave, Transplantation Research Center, Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02115; e-mail: rabdi@rics.bwh.harvard.edu.