Abstract
The Ca2+ dependent transcription factor family known as nuclear factor of activated T cells (NFAT) has been shown to be important in T-cell immune responses. Because NFAT proteins have a weak DNA-binding capacity, they cooperate with other transcription factors at composite sites within the promoters of target genes. Recently, NFAT was shown to also be important for the induction of specific genetic programs that guide the differentiation and effector or regulatory activities of CD4+ T helper subsets via the transcriptional regulation of their lineage-specific transcription factors, specifically T-bet (Th1), Gata3 (Th2), RORγt (Th17), and Foxp3 (iTregs). In addition, the NFAT family governs the transcription of several signature cytokines, including their cytokine receptors. Subsequently, the integration of these complex intracellular signal transduction cascades is considered to critically determine the crosstalk between the T-cell receptor and receptors that are activated by both the adaptive and innate immune systems to determine pathways of T helper cell differentiation and function. Here, we carefully review the critical role of the established transcriptional partners and functional outcomes of these NFAT interactions in regard to the effector responses of these clinically relevant CD4+ T helper subsets.
Introduction
The Ca2+-dependent transcription factor family, known as nuclear factor of activated T cells (NFATs), originally identified by Shaw et al,1 regulates not only T lymphocytes but also a large number of growth factors, cytokines, and cell-to-cell interaction molecules essential for the morphogenesis, development, and function of many cell types and organs.2,3 In T lymphocytes, NFAT proteins govern gene expression that regulates T-cell development, activation, differentiation, as well as the induction and maintenance of T-cell tolerance.4-6
Briefly, the NFAT family consists of 5 family members that all share a highly conserved DNA-binding domain: NFAT1 (NFATc2; NFATp), NFAT2 (NFATc1; NFATc), NFAT3 (NFATc4); NFAT4 (NFATc3; NFATx), and NFAT5 (TonEBP; OREBP). In T lymphocytes, 2 or more splice forms of NFAT1, 2, and 4 are present, encoding the following domains4 : the NFAT homology region containing a transactivation (TAD-A) and regulatory domain (docking sites for calcineurin and the NFAT kinases), the Rel-homology region with the N-terminal DNA-binding domain that is also involved in protein-to-protein interactions, and the C terminal domain. The Rel-homology region contains the DNA-binding loop that confers base-specific recognition, and the C terminal domain, which makes contact only with the DNA phosphate backbone. Activation of the NFAT pathway occurs after receptor activation coupled to the calcium-signaling pathway.7 Calcium release from intracellular stores, together with cytosolic calcium, binds calmodulin, which in turn activates the calmodulin-dependent phosphatase calcineurin. NFAT proteins are dephosphorylated by activated calcineurin, leading to their nuclear translocation and the induction of NFAT-mediated gene transcription.8,9
In the nucleus, the NFATc subunits bind to the DNA with protein partners, initially termed NFATn, to induce the active NFAT transcriptional complexes, thereby combining the T-cell receptor (TCR) pathway with other receptor transactivation pathways. This binding to different protein partners enables the diversification of NFAT function by combinatorial association with different pathways.10 NFAT is also regulated by an autoregulatory loop at the transcriptional level.11,12 Although NFAT proteins are themselves induced in a relatively nonspecific manner, the differential access of NFATs to specific regulatory elements on the different cytokine loci results in differences in the chromatin dynamics and recruitment of lineage-specific transcription factors.7,13 The choice to differentiate into different T-cell subsets is determined by the antigenic stimulus, the type of antigen-presenting cell that delivers it, and the signals received from specific cytokines produced by either the surrounding tissue, already differentiated T cells, or the innate immune system.4,14 Epigenetic changes induced by TCR signaling and cytokine-mediated signaling can lead to the establishment of specific patterns of cytokine expression and the subsequent induction of different lineage commitments, which then selectively produce large amounts of specific effector cytokines on antigen encounter. After this initiation phase, powerful feedback mechanisms reinforce the subset lineage decision, together with changes in chromatin structure, DNA methylation status, cis-regulatory elements, and silencing mechanisms, including endogenous RNAi involvements.13,15,16
Nevertheless, NFAT transcription factors govern gene expression at several key gene loci that become open and available for immediate transcription. Subsequently, remodeling of the chromatin structure takes place to regulate access of NFAT and its transcriptional partners.15,17,18 Therefore, T helper (Th) differentiation can be divided into initiation, specific reinforcement/amplification, and maintenance.16 The cytokine milieu provided by the surrounding tissue, dendritic cells, and cells from the innate immune system, together with antigen specificity and the network of transcription factors induced downstream of the TCR and cytokine receptors, subsequently determines the Th lineage. The transcriptional partners of NFAT have been well reviewed,4 but the transcriptional partners and different target genes, such as Il2, Nfat, Cd25, Gitr, Il4, Tbx21, Twist, Il12Rβ2, Il5, Il13, Il10, c-Maf, Gata3, Rorγt, Il17, Il12, Il21, Il22, Il6, and Foxp3 (Table 1), have not been outlined in the context of the different CD4+ T helper subsets.
In addition, adding to the already well-established Th1 and Th2 subsets,52 the Th17 subset was identified in 2006,53,54 as well as the iTregs.6,55 From the medical viewpoint, the importance of the NFATs is highlighted by the fact that the most widely used method for immune suppression to avoid transplant rejection is the inhibition of the calcineurin-NFAT signaling cascade via cyclosporine (CsA) or tacrolimus (FK-506). In the context of autoimmunity, organ transplantation, infectious disease, or antitumor immunity, the ratio of effector versus regulatory CD4+ T cells is highly relevant for the outcome of the given immune response. Currently used immunosuppressive drugs, such as CsA and tacrolimus (both inhibiting the calcineurin/NFAT transactivation pathway) or rapamycin/sirolimus (inhibiting the mammalian target of rapamycin, pathway activities downstream of IL-2/phosphatidylinositol 3-kinase signaling) are efficient by reducing effector T-cell expansion and effector functions via different signaling cascades.56,57 Calcineurin inhibitors have been shown to inhibit Th0, Th1, Th2, and the Th17 subset in humans, albeit with slightly different efficiency.56-58 Sirolimus has been shown to be significantly more effective than CsA in inhibiting the Th1 subset responses, whereas the inhibition on the Th2 subset did not differ in kidney transplantation patients.57,59 The ideal immunosuppressive agent should not impair Tregs. CsA has been shown to inhibit Treg function.57 In contrast, it has also been suggested that CsA might even enhance the Treg subset in transplantation medicine.60,61 Rapamycin not only enhances Treg proliferation but, in addition and in contrast to CsA, also fosters the dominance of the highly suppressive CD27+ Treg subset.62 A further advantage over current immunosuppressive regimens might be achieved by sotrastaurin (AEB-071, NVP-AEB-071), an orally bioavailable compound that exerts its effects through the inhibition of protein kinase C, thereby inhibiting the antigen receptor signaling to nuclear factor-κB (NF-κB) and NFAT transactivation.63 The immunosuppressive effects of oral sotrastaurin could be an effective novel treatment regimen for psoriasis.64 Because it represents an entirely new mechanism of drug action, it has the potential of an effective alternative and/or adjunct to calcineurin inhibitors for abrogating selective T-cell effector responses in future therapies.65
T-cell effector functions are relevant in the pathogenesis of different diseases, including asthma, rheumatoid arthritis, and multiple sclerosis, and the role of NFAT has been well established in these diseases. Understanding subset-specific NFAT protein–protein interactions in the clinically relevant CD4+ T helper subsets, and in the long run pharmacologic intervention of specific CD4+ T-cell effector functions, may greatly facilitate medical treatment, either for immune suppression in autoimmune diseases or for immune augmentation in cancer.
Naive T cells: NFAT transcriptional partners and targets
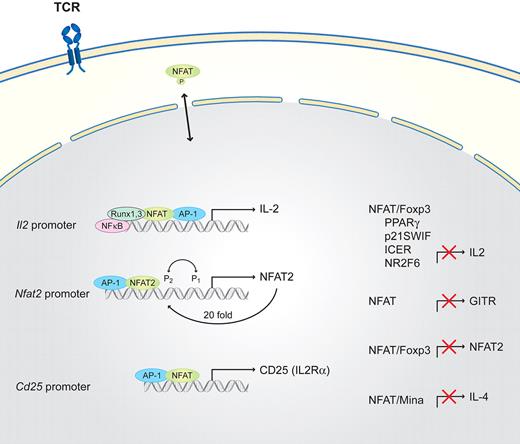
Naive T cells, which develop in the thymus, are the common precursors of helper T cells that have not yet encountered antigen. In naive cells, full T-cell activation (Figure 1) requires costimulatory receptors, including CD4+ or CD8+, together with LFA1, CD28, and ICOS. All of these receptors additionally activate one of the major signaling pathways converging at the activation of the NFAT, AP-1, or NF-κB pathways.10,19 NFATs, together with the originally described transcription factors NF-κB, AP-1, EGR, and Oct, induce interleukin-2 (IL-2). In addition, the transcription of Il2Rα (Cd25) is up-regulated in an NFAT-dependent manner.25 IL-2 itself, via the interleukin-2 receptor (IL-2R), activates signal transducer and activator of transcription 5 (STAT5) and, subsequently, cell-cycle entry. In naive T cells, only low concentrations of NFATs are present, but in combination with the other aforementioned transcription factors, together with an autoregulatory loop, lead to sufficient Il2 and Cd25 promoter induction.4,10,25 Different NFAT proteins having selective roles in T helper cell differentiation has been a controversial idea, but data suggest that NFAT1 and NFAT2 are functionally redundant.30 In contrast to the NFAT-AP-1 complex, the NFAT-Foxp3 complex suppresses Il2 expression as Foxp3 inhibits the activation of Nfat2 expression.22 Other additional factors that regulate NFAT-induced Il2 transcription are nuclear factor 90 (NF90), NF45, runt-related transcription factor 1 (Runx1), and Runx3.23,66
Schematic of NFAT interaction partners in naive T cells. T-cell activation requires TCR and costimulatory signals. NFAT is engaged via the TCR proximal signaling cascades and other signaling pathways, converges in the nucleus, and activates gene transcription. IL-2 via the IL-2R activates STAT5 and, subsequently, cell-cycle entry. Runx3 and Runx1 cooperate together with NF-κB with NFAT/AP-1 on the Il2 promoter to activate Il2, whereas the NFAT/Foxp3 complex suppresses Il2 transcription as well as the NFAT autoregulatory loop on the Nfat2 promoter. The expression of Il4 in naive cells is suppressed via the direct binding of Mina and NFAT to its promoter. In addition, NFAT/AP-1 activates the transcription of Cd25 (Il2Rα). NFAT functions as a negative regulator of GITR expression and is suppressed by PPARγ, p21SWIF, inducible cyclic adenosine monophosphate early repressor (ICER), and NR2F6.
Schematic of NFAT interaction partners in naive T cells. T-cell activation requires TCR and costimulatory signals. NFAT is engaged via the TCR proximal signaling cascades and other signaling pathways, converges in the nucleus, and activates gene transcription. IL-2 via the IL-2R activates STAT5 and, subsequently, cell-cycle entry. Runx3 and Runx1 cooperate together with NF-κB with NFAT/AP-1 on the Il2 promoter to activate Il2, whereas the NFAT/Foxp3 complex suppresses Il2 transcription as well as the NFAT autoregulatory loop on the Nfat2 promoter. The expression of Il4 in naive cells is suppressed via the direct binding of Mina and NFAT to its promoter. In addition, NFAT/AP-1 activates the transcription of Cd25 (Il2Rα). NFAT functions as a negative regulator of GITR expression and is suppressed by PPARγ, p21SWIF, inducible cyclic adenosine monophosphate early repressor (ICER), and NR2F6.
In naive T cells, NFAT functions as a negative regulator of glucocorticoid-induced TNF receptor family–related protein (GITR) expression, which is markedly up-regulated after TCR activation.26 Covalent modification of the NFAT cofactor protein NIP45 by arginine methylation is also an important mechanism involved in the regulation of NFAT-dependent cytokine gene expression.67 The inducible cyclic adenosine monophosphate early repressor interactions with NFAT/AP-1 composite DNA sites correlate with the ability to repress transcription.20 p21SNFT represses NFAT/AP-1 activity on the Il2 promoter by competing with Fos proteins for Jun dimerization.27 The nuclear orphan receptor peroxisome proliferator-activated receptor-gamma PPARγ (NR1C1) physically associates with NFAT, regulating the Il2 promoter and blocking NFAT DNA-binding and transcriptional activity.24
We identified another nuclear orphan receptor, NR2F6, which is a negative regulator of Il2 transcription, by interfering with NFAT binding to the Il2 promoter region. The inactivation of NR2F6 via phosphorylation of Ser 83 site results in the release of NR2F6 from DNA-binding sites. NR2F6 potently antagonizes the ability of Th0 CD4+ T cells to induce the expression of key cytokines, such as Il2; the loss of NR2F6 results in higher NFAT DNA-binding capabilities at the Il2 promoter and subsequently increases Il2 transcription.68 Another order of complexity was recently added in the regulation of the Il4 promoter region. Okamoto et al27 identified a previously unknown IL-4–repressive pathway in which Mina suppresses Il4 promoter activity in an NFAT-dependent interaction, thereby serving as a key molecule in regulating the amount of IL-4 produced by naive helper T cells and controlling the extent of the Th2 bias.
Th1 helper cells: NFAT transcriptional partners and targets
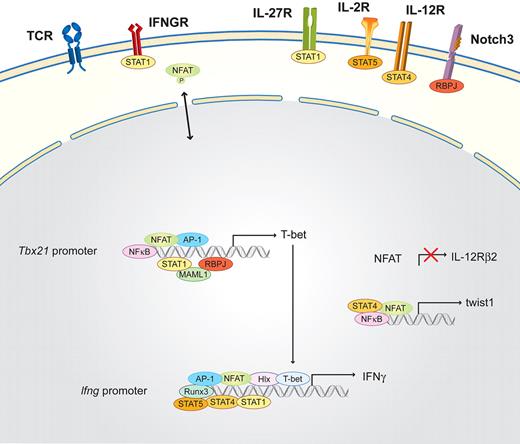
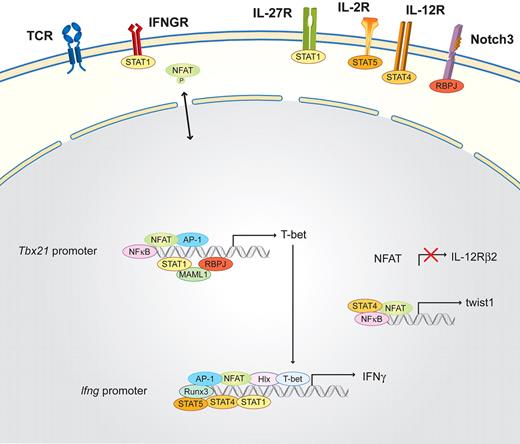
Th1 cells are essential for clearing intracellular bacteria and viruses; their signature cytokine is interferon-gamma (IFN-γ).69,70 Briefly, Th1 differentiation (Figure 2) is induced by signals from the antigen-presenting cells: IL-12, which is mainly produced by monocytes and dendritic cells; IFN-γ, which is secreted by already differentiated Th1 cells and by natural killer (NK) and NKT cells; and IL-27,71 which is produced by NK cells. IFN-γ activates signal transducer and activator of transcription 1 (STAT1) via the IFN-γ receptor. Together with the TCR-induced transcription factors (NFAT, NF-κB, and AP-1), these signals activate the master transcription factor of the Th1 lineage, T-bet (Tbx21), while repressing Il12Rβ2 subunit gene expression in an NFAT-dependent manner.28,29 Subsequently, T-bet induces the production of IFN-γ and the activation of the transcription factors H2.0-like homeobox (Hlx) and Runx3. IL-2 helps induce STAT5 via the IL-2R, which additionally triggers IFN-γ production. The termination of TCR signaling allows the up-regulation of Il12Rβ2. Therefore, IL-12 can additionally activate STAT4, which together with T-bet, Hlx, Runx3, AP-1, and NFAT activates the Ifng locus, positively enhancing STAT1 activation.72,73 The committed Th1 cells are then reactivated by the TCR receptor pathway plus the positive feedback loop of the IFN-γ-STAT1 signaling pathway.30 One regulator of this positive amplification loop was reported recently, the basic helix-loop-helix transcription factor twist1, which is induced by NFAT and NF-κB as a result of IL-12/STAT4 signaling in Th1 cells and transiently expressed in repeatedly activated Th1 cells.31 The Notch3 signaling pathway via the recombination signal binding protein for immunoglobulin kappa J region (RBPJ) and mastermind-like (MAML1) is also mandatory during Th1 subset differentiation, and its key role was reviewed recently.74
Schematic of the NFAT interaction partners in Th1 cells. Differentiation is initiated by the activation of the TCR and costimulatory signals, as well as the Th1-determining factors IFN-γ and/or IL-27, which induce via the IFNGR and the IL-27R the activation of STAT1. STAT1, in combination with NFAT, AP-1, NF-κB, and the Notch3/RBPJ/MAML1 signaling pathway, binds to the Tbx21 promoter and subsequently induces the transcription of the master transcription factor of Th1 cells, T-bet. T-bet induces the production of IFN-γ and the activation of the transcription factors Hlx and Runx3. IL-2 helps to induce STAT5 via the IL-2R, which additionally triggers IFN-γ production. The termination of TCR signaling allows the up-regulation of Il12Rβ2. Therefore, IL-12 can additionally activate STAT4, which together with STAT1, Hlx, Runx3, T-bet, AP-1, and NFAT, further induces IFN-γ transcription. In parallel, NFAT, together with STAT4 and NF-κB, binds to the promoter of the twist1 gene.
Schematic of the NFAT interaction partners in Th1 cells. Differentiation is initiated by the activation of the TCR and costimulatory signals, as well as the Th1-determining factors IFN-γ and/or IL-27, which induce via the IFNGR and the IL-27R the activation of STAT1. STAT1, in combination with NFAT, AP-1, NF-κB, and the Notch3/RBPJ/MAML1 signaling pathway, binds to the Tbx21 promoter and subsequently induces the transcription of the master transcription factor of Th1 cells, T-bet. T-bet induces the production of IFN-γ and the activation of the transcription factors Hlx and Runx3. IL-2 helps to induce STAT5 via the IL-2R, which additionally triggers IFN-γ production. The termination of TCR signaling allows the up-regulation of Il12Rβ2. Therefore, IL-12 can additionally activate STAT4, which together with STAT1, Hlx, Runx3, T-bet, AP-1, and NFAT, further induces IFN-γ transcription. In parallel, NFAT, together with STAT4 and NF-κB, binds to the promoter of the twist1 gene.
Th2 helper cells: NFAT transcriptional partners and targets
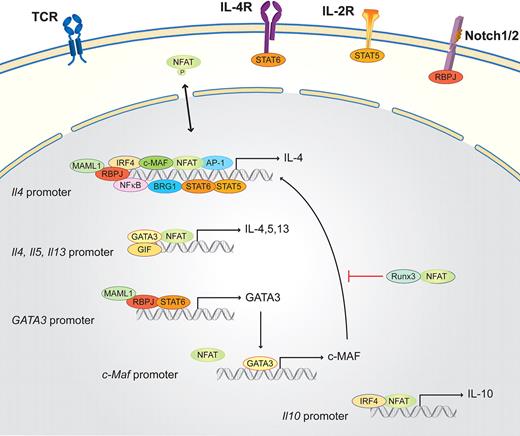
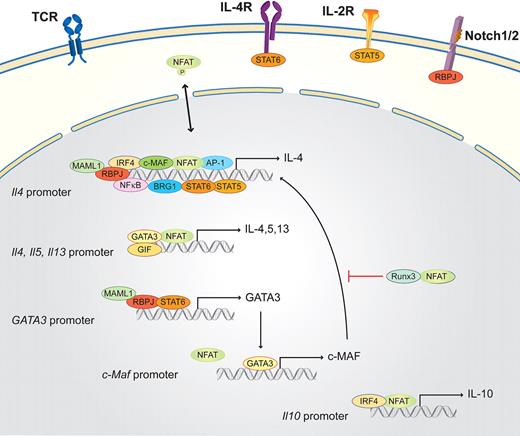
Th2 cells are essential in organizing the host defense against parasites and in inducing humoral responses in the form of helping B cells, which produce antibodies. Th2 cells produce IL-4, IL-5, IL-6, IL-9, IL-10, and IL-13.16,52,75 Briefly, in the presence of exogenous IL-4 (provided by mast cells, basophils, eosinophils, NKT cells, or previously differentiated Th2 cells), naive T cells, after TCR cross-linking, activate not only the TCR-dependent signaling machinery but also the IL-4 receptor (IL-4R) pathway (Figure 3). IL-4R, which is present on naive T cells, activates STAT6, which in turn, together with NFAT, AP-1, and NF-κB, drives Il4 transcription.19,76 Another downstream target of STAT6 is the master transcription factor in the Th2 subset, GATA3, which consequently induces the transcription of the long form of viral musculoaponeurotic fibrosarcoma oncogene homolog (c-MAF), which additionally helps to activate Il4 transcription. This activation results in a strong autocrine feedback loop that activates Il4, Il5, and Il13. Two STAT6-independent pathways have been discovered that promote Th2 differentiation: the Notch1/2 signaling pathway via RBPJ and MAML1 and the IL-2 receptor signaling pathway. The IL-2 receptor pathway boosts early Il4 transcription via STAT5.16,74,77,78 During Th2 differentiation, the whole Th2 cytokine locus undergoes a number of chromatin modifications. These mechanisms control the accessibility of DNA elements to transcription factors, such as NFATs, STATs, GATA3, and c-MAF. NFAT1 plays a pivotal role during the initiation process and reactivation as it binds to all 3 promoters (Il4, Il5, and Il13) and several enhancer elements within the Il4 locus, as the 3′Il4 enhancer in combination with GATA3.16 NFAT is also important for the rapid reactivation of resting Th2 cells to produce large amounts of IL-4. NFAT1 does not only activate the relevant genes for Th2 differentiation, but NFAT proteins have also been shown to mediate a negative feedback loop limiting the duration of Il4 transcription; therefore, the lack of NFAT1 results in a strong Th2 bias.16 The exact and nonredundant in vivo function of NFAT isotypes during Th2 differentiation remains complex and yet is not fully understood. NFAT1/NFAT4 double knockout mice submitted to airway inflammation display elevated Th2 cytokine production, higher IgE levels, and increased lung inflammation.32
Schematic of the NFAT interaction partners in Th2 cells. Th2 lineage is initiated by TCR and costimulatory signals, as well as the Th2 determining factor IL-4. IL-4 via the IL-4R together with IL-2 (via the IL-2R) and the Notch1/2/RBPJ/MAML1 signaling pathway activates the master transcription factor of the Th2 lineage, GATA3. GATA3, together with NFAT, NF-κB, AP-1, IRF4, c-MAF, STAT5, STAT6, BRG1, and RBPJ, binds the Il4 promoter region. GATA3 subsequently enhances, in combination with NFAT and GIF, the transcription of Il4, Il5, and Il13 via a positive feedback loop. GATA3 also induces the transcription of c-Maf. NFAT, in parallel with IRF4, binds the Il10 promoter and induces Il10 expression. NFAT and Runx inhibit Th2 differentiation.
Schematic of the NFAT interaction partners in Th2 cells. Th2 lineage is initiated by TCR and costimulatory signals, as well as the Th2 determining factor IL-4. IL-4 via the IL-4R together with IL-2 (via the IL-2R) and the Notch1/2/RBPJ/MAML1 signaling pathway activates the master transcription factor of the Th2 lineage, GATA3. GATA3, together with NFAT, NF-κB, AP-1, IRF4, c-MAF, STAT5, STAT6, BRG1, and RBPJ, binds the Il4 promoter region. GATA3 subsequently enhances, in combination with NFAT and GIF, the transcription of Il4, Il5, and Il13 via a positive feedback loop. GATA3 also induces the transcription of c-Maf. NFAT, in parallel with IRF4, binds the Il10 promoter and induces Il10 expression. NFAT and Runx inhibit Th2 differentiation.
One factor that could be involved in this regulation is the glycosylation-inhibiting factor (GIF); GIF secreted from naive CD4+ cells has been shown to maintain NFAT1 in the nucleus and represses IL-4 mRNA levels.79 In already committed Th2 cells, GATA3, NFAT, and c-MAF drive IL-4 secretion. Other important transcription factors that interact with NFAT are interferon regulatory factor 4 (IRF4), which potently synergizes with NFAT1 to specifically enhance NFAT1-driven transcriptional activation of the Il4 promoter. This function is dependent on the direct physical interaction of IRF4 with NFAT1. IRF4 synergizes with NFAT1 and the IL-4–inducing transcription factor c-MAF to augment Il4 promoter activity and to elicit significant levels of endogenous IL-4 production.32,33 NFAT1 and IRF4 have also been shown to synergistically enhance the Th2-specific enhancer activity of CNS-9, a cis-regulatory element 9 kb upstream of the Il10 gene loci.38
Runx1 inhibits Th2 differentiation by GATA3 repression. Runx3, together with T-bet, enhances IFN-γ in Th1 cells while simultaneously inhibiting the Il4 locus. In El-4 T cells, Runx3 physically interacts with NFAT, thereby suppressing the Il4 locus.34,35,38 PPARγ also inhibits Il4 promoter activation in a ligand-dependent manner at multiple NFAT sites.36 NFAT and STAT6 activity are also required for the recruitment of the Brahma-related gene-1 (BRG1) catalytic subunit of SWI/SNF-related complexes to the Th2 locus control region, and STAT6 associates with BRG1 in a differentiation-inducible manner.37
Th17 helper cells: NFAT transcriptional partners and targets
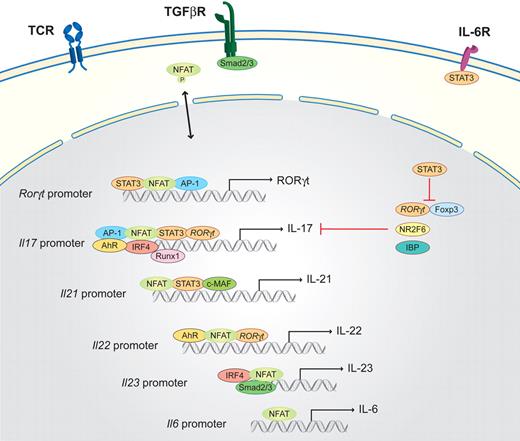
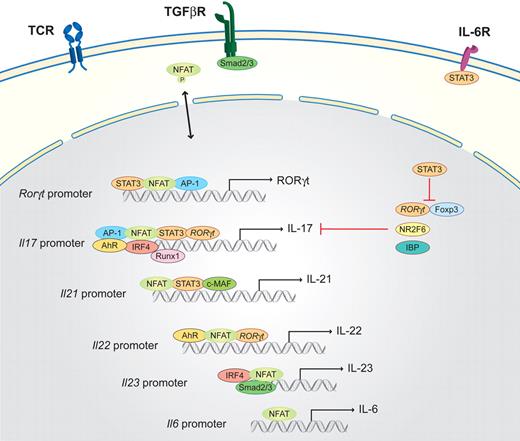
Th17 cells are important for the clearing of extracellular pathogens, especially at mucosal sites, but are also responsible for autoimmune diseases. Th17 cells have been extensively reviewed.42,80 Briefly, Th17 cells secrete the signature cytokines IL-17A, IL-17F, IL-21, and IL-22. NFAT is important in the induction of all of these cytokine genes (Figure 4).39,43 Furthermore, the crucial role of NFAT1 in Th17 differentiation has also been demonstrated by the analysis of NFAT1 knockout mice in an experimental colitis model because it plays a key regulatory role by controlling IL-6–dependent T-cell activation.48 Transforming growth factor-β (TGF-β), provided by dendritic and epithelial cells, inhibits Th1 and Th2 differentiation and favors Th17 and Treg differentiation, which is dependent on costimulatory signals and occurs in a concentration-dependent manner.81 If the cytokine milieu provides IL-6, which is mainly produced by monocytes, differentiation into the Th17 lineage is induced.6 The IL-6R activates STAT3, which subsequently induces the master transcription factor of the Th17 lineage, the retinoid-related orphan receptor, ROR (RORγt, as well as RORα).82 IL-6R activation results in the preferential induction of NFAT1 transcripts, leading to increased NFAT1 activity in response to TCR stimulation; this amplification loop further enhances IL-6 production.4 IL-6 has also been implicated in the induction of Il21 expression, which is NFAT dependent, and several NFAT-binding sites are located on the Il21 promoter.46,47
Schematic of the NFAT interaction partners in Th17 cells. Th17 differentiation is initiated by TCR-derived signals in combination with the activation of the IL-6R via IL-6 and the TGF-β signaling cascade (via the TGF-βR). STAT3 activated via the IL-6R together with NFAT and AP-1, binds to the promoter of RORγt, which is the master transcription factor of the Th17 lineage. Subsequently, RORγt, in combination with NFAT, AP-1, STAT3, AhR, IRF4, and Runx1, binds to the Il17 promoter region to induce transcription. In parallel, NFAT, together with STAT3 and c-MAF, activates the transcription of Il21 and, in combination with RORγt and AhR, the transcription of Il22. IL-21 and low concentrations of TGF-β (via Smad2/3) induce the expression of the IL-23R components (not shown); subsequently, NFAT, IRF4, and Smad2/3 drive the transcription of Il23.
Schematic of the NFAT interaction partners in Th17 cells. Th17 differentiation is initiated by TCR-derived signals in combination with the activation of the IL-6R via IL-6 and the TGF-β signaling cascade (via the TGF-βR). STAT3 activated via the IL-6R together with NFAT and AP-1, binds to the promoter of RORγt, which is the master transcription factor of the Th17 lineage. Subsequently, RORγt, in combination with NFAT, AP-1, STAT3, AhR, IRF4, and Runx1, binds to the Il17 promoter region to induce transcription. In parallel, NFAT, together with STAT3 and c-MAF, activates the transcription of Il21 and, in combination with RORγt and AhR, the transcription of Il22. IL-21 and low concentrations of TGF-β (via Smad2/3) induce the expression of the IL-23R components (not shown); subsequently, NFAT, IRF4, and Smad2/3 drive the transcription of Il23.
Il21 has been identified as one of the most highly induced genes after Th17 differentiation. Autocrine up-regulation of Il21 further enhances the expression of Il21 and Il23R.41 In vivo IL-23 signaling is required for the expression of Il22.40 Once Th17 differentiation is initiated and IL-23R, which is not expressed on naive T cells, is expressed, IL-23 plus TGF-β are capable of driving Il17 and Il23R expression in an amplification loop. In vivo, IL-23, which is mainly released by dendritic cells and macrophages and is an essential component for the maintenance but not the induction of Th17 cells, has been demonstrated to be very important80,83 ; and albeit still controversial, IL-22 also seems to be essential. Surprisingly, Il22-deficient mice are fully susceptible to experimental autoimmune encephalitis induction,84 which dismisses IL-22 as an essential and nonredundant pathogenic player in the development of autoimmune central nervous system inflammation.
Taken together, multiple proinflammatory cytokines might therefore be able to drive Th17 differentiation in combination with TGF-β in vivo, such as IL-1, IL-6, IL-21, or IL-22.40,48 The subsequently activated STAT3 pathway is required for RORγt and IL-17 induction.47,85 RORγt, together with Runx1, binds to the Il17a promoter and inhibits Foxp3 expression.45 RORγt also directly binds to Foxp3; therefore, Foxp3 can inhibit its transcriptional activities.41 This inhibition is relieved by the presence of the proinflammatory cytokines that drive Th17 differentiation by a posttranslational mechanism. Another transcription factor essential for Th17 differentiation is IRF-4, which was already discussed in Th2 differentiation. IRF-4 regulates Il21 and Il23R expression and is inhibited by the IRF-4–binding protein.44 IRF-4 is up-regulated after TCR activation and is therefore not Th-dependent,33,44,46 suggesting that IL-21, which is produced by Th17 cells in an autocrine manner and involved in the positive feedback loop, is transcriptionally regulated by cooperation between IRF-4 and NFAT. The aryl hydrocarbon receptor (AhR), which is highly expressed in the Th17 subset,80,86,87 is necessary for Il22 and, to a lesser extent, Il17 expression. Zhou and Littman40 reported that AhR cooperates with RORγt to induce maximal expression of Il17 and Il22. Recently, ICOS and c-MAF were shown to be essential in Th17 cell differentiation; the induction of c-MAF by ICOS induces IL-21 production. In Th17 cells, differentiated and activated ex vivo, the loss of Nr2f6 results in amplified NFAT DNA-binding capabilities at the Il17 promoter and subsequently increases Il17 transcription. Nr2f6-deficient mice consistently have hyperreactive lymphocytes and develop a late-onset immune pathology. These mice are also more susceptible to the Th17-dependent model of experimental autoimmune encephalomyelitis.68
iTregs: NFAT transcriptional partners and targets
Regulatory T cells (CD4+CD25+/Foxp3+) seem to be essential for limiting autoimmune responses and have been divided into several “flavors”: the natural regulatory T cells (nTreg) developing in the thymus and several inducible regulatory T-cell subsets that differentiate from naive peripheral CD4+ T cells as Foxp3+ iTreg, the type 1 regulatory cells (Tr1),21 the Th3,88,89 and the HOZOT cytotoxic Tregs.90 Significant differences have been found between the development of nTregs in the thymus and iTregs in the periphery6 ; therefore, it has been suggested that they might have nonoverlapping functions in the regulation of immune responses. nTregs could be responsible for the regulation of T-cell tolerance under resting conditions, whereas iTregs could function during antigen-specific responses as T helper cells.6
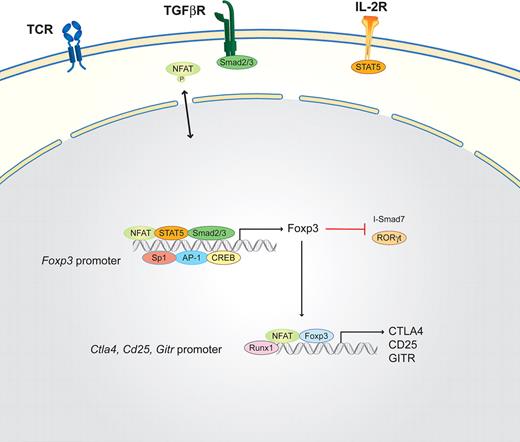
In the presence of TGF-β and IL-2/IL-15, naive T cells induce STAT5, Smad, and non-Smad signaling after T-cell receptor stimulation. If exposed to low-affinity antigen without costimulation, however, the NFAT signaling cascade, but not the mitogen-activated protein kinase pathway (and subsequently AP-1), is activated. This combination of transcription factors induces the master transcription factor of inducible regulatory T cells, Foxp3, which consequently suppresses I-Smad7 expression.91,92 In the absence of AP-1, NFAT up-regulates Cbl-b,93 further enhancing Foxp3 expression and resulting in a positive feedback loop for the expression of Foxp3.6,94 The Foxp3 promoter region has been analyzed in much detail to elucidate how TGF-β signaling can influence Foxp3 expression and Treg differentiation.49 Tone et al49,50 revealed that, in addition to the Foxp3 promoter, which is activated by the combined binding of NFAT, AP-1, Sp1, and STAT5, additional enhancer regions exist (Figure 5). Enhancer region 1 is activated by NFAT/(p)Smad3 cooperation, whereas (p)Smad3 binding is important only during the early induction of TGF-β. NFAT binding is required throughout at least a 24-hour period.49 Another transcription factor discovered to be important in the differentiation of Tregs is Runx1 as it physically and functionally interacts with Foxp3.51 This interaction is different from the interaction between NFAT and Foxp3 because they interact through regions distinct from their DNA-binding domains on widely separated sites.51 In addition to its essential role in the induction of Foxp3 transcription, NFAT additionally cooperates with Foxp3 to up-regulate CTLA-4 and CD25, 2 highly expressed surface markers of Tregs.50 Chromatin-immune precipitations confirmed the binding of NFAT and Foxp3 to the promoters of Il2, Ctla4, and Cd25.23 Foxp3 competes with NFAT1 to bind to the Nfat2 promoter to suppress the amplification loop of NFAT transcription, thereby potentially affecting the activation of the many NFAT-mediated cytokine genes downstream.22 In contrast to naive T cells where Runx1 cooperates with NFAT/AP-1 on the Il2 promoter to activate Il2 expression after TCR stimulation, Runx1 cooperates with the NFAT/Foxp3 complex in Tregs to suppress Il2 and activate Ctla4, Cd25, and Gitr expression.23,95,96
Schematic of the NFAT interaction partners in iTreg cells. iTregs are induced by high TGF-β concentrations via the TGF-βR in combination with low TCR signals. IL-2 via the IL-2R induces STAT5 that, in combination with NFAT, the Smad2/3 complex, Sp1, CREB, and AP-1, binds to the Foxp3 promoter region. Foxp3, the master transcription factor of regulatory T cells, subsequently inhibits I-Smad7 and RORγt and, together with NFAT and Runx1, induces the expression of CTLA4, CD25, and GITR.
Schematic of the NFAT interaction partners in iTreg cells. iTregs are induced by high TGF-β concentrations via the TGF-βR in combination with low TCR signals. IL-2 via the IL-2R induces STAT5 that, in combination with NFAT, the Smad2/3 complex, Sp1, CREB, and AP-1, binds to the Foxp3 promoter region. Foxp3, the master transcription factor of regulatory T cells, subsequently inhibits I-Smad7 and RORγt and, together with NFAT and Runx1, induces the expression of CTLA4, CD25, and GITR.
Two other regulatory T cells have been described, namely, the Th3 and Tr1 cells. The Th3 cells have been cloned from orally tolerized mice, which produce large amounts of TGF-β but small amounts of IL-10 and IL-4.88 In contrast, the Tr1 identified by Chen et al97 arise in the periphery when naive CD4+ T cells are activated by tolerogenic antigen-presenting cells in the presence of IL-10. These resulting Tr1 cells regulate immune responses by secreting Il-10 and TGF-β and have the capacity to suppress both naive and memory T-cell responses in vivo and in vitro.98,99 The HOZOT cytotoxic Tregs are FOXP3+CD4+CD8+CD25+ and produce high levels of IL-10 resulting from the activation of STAT5, NFAT, and NF-κB.98,100
In conclusion, NFAT plays a key role in the regulation of immune effector functions, and understanding the underlying cellular mechanisms at the molecular level in the fast-evolving field of T helper subsets could be helpful in the treatment of immune diseases, the use of cancer immunotherapy, and increasing vaccination effectiveness. Here, we reviewed the recent progress in our understanding of the molecular nature and regulation of the Ca2+/NFAT signaling pathway in the determination of lymphocyte response. Not only is the NFAT pathway involved in the signaling response to antigenic stimulation, but it regulates in promoter context-dependent transcriptional complexes the activation of the different Th master/lineage-specific transcription factors, as well as signature cytokines and their cytokine receptors. Future challenges we face are (1) to establish the biologic relevance of this recently accumulated molecular and biochemical knowledge in the complex context of animal disease models and (2) to translate this fundamental knowledge into the design of sound therapeutic approaches for immune diseases, all the way to their application in clinically relevant situations.
Acknowledgments
This work was supported by the FWF Austrian Science Fund (grants SFB-021 and P19505-B05), the Tyrolian Science Fund (grant TWF-2008-1-563), the Swiss Kamillo Eisner-Foundation (grant CH-6052 Hergiswil), the European Community Seventh Framework Program (grant agreement no. HEALTH-F4-2008-201106), and the Austrian Federal Ministry of Science and Research (BMWF-651.423/0001-11/2/2009).
The authors apologize to those colleagues whose work could not be cited because of space restrictions.
Authorship
Contribution: N.H.-K. and G.B. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Natascha Hermann-Kleiter, Schöpfstraße 41, A-6020 Innsbruck, Austria; e-mail: natascha.kleiter@i-med.ac.at.