To the editor:
Recently, Hauser and coworkers reported that a single nucleotide polymorphism (SNP) of the lactase persistence gene (LCT) at position 13910 of the donor influenced patient outcome after allogeneic hematopoietic stem cell transplantation.1 They reported in a study of 111 recipients/donor pairs that median overall survival (OS), death in remission, and relapse rate were significantly improved when recipients received transplantations from donors homozygous for CC at 13910 in the LCT gene. They argued that lactose malabsorption changes the composition of colonic microflora, which could be caused by genetic polymorphisms leading to nonpersistence of lactase phlorizin hydrolase, a β-galactosidase, expressed exclusively in the small intestine.1 Changes in the composition of colonic microflora and in the gut-associated immune system might influence the outcome of transplantation as reported for genetic variants of NOD2/CARD15 genes, which are associated with an increase in incidence and severity of acute graft-versus-host disease (GVHD) after transplantation.2,3
We studied the influence of the gene variant of LCT-13910 on transplantation in HLA-identical sibling donors of a homogeneous group of 123 patients, all diagnosed with acute myeloid leukemia and all receiving transplantations of non-T cell–depleted grafts after myeloablative conditioning.
The median age of our patients was 47 years (range, 16-68 years) and that of donors was 46 years (range, 15-72 years). Conditioning regimens included total body irradiation (68%) or chemotherapy alone (32%). The GVHD prophylaxis in 119 patients (96%) was cyclosporine A (CsA) and methotrexate (MTX) and in 5 patients (4%) CsA and mycophenolate mofetil. Forty-nine percent of the patients received a transplantation in first complete remission and 51% of the patients in more advanced disease.
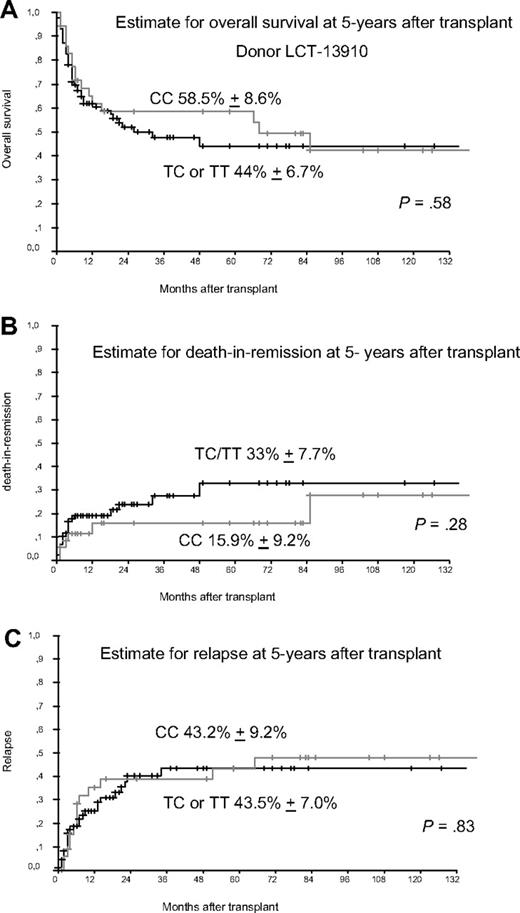
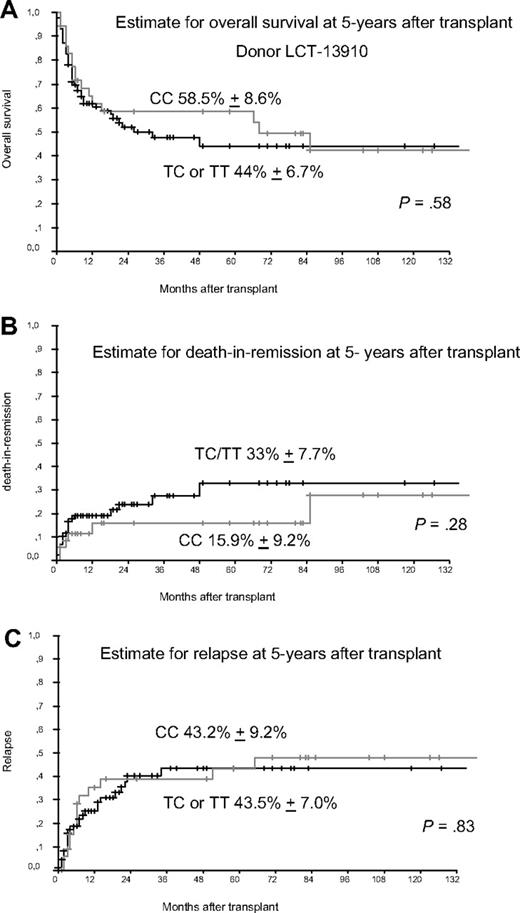
We preformed genotyping of LCT-13910 by real-time polymerase chain reaction and melting curve analysis as published earlier.4,5 The detected allele frequencies were found in Hardy Weinberg equilibrium (P > .05). We detected the homozygous C allele in 36 of 123 donors (29.3%), whereas 87 donors (70.7%) had CT or TT genotype, which is comparable to frequencies published in the study by Hauser et al. Although we included more patients than Hauser et al we did not find a statistically significant longer median OS rate in patients who received transplantations from donors with CC gene variant of LCT-13910 as shown in Figure 1. The estimated rate of death in remission was slightly better with 33% versus 15.9% but failed to achieve statistical significance. Furthermore, we found no difference in the estimated relapse rate at 5 years after transplantation. The different results reported by Hauser et al might be caused in part by a more heterogenous study population with regard to type of donor (HLA-identical sibling donors, mismatched donors, unrelated donors), types of diseases, and the use of various conditioning regimen (myeloablative and nonmyelablative). Gene polymorphisms of NOD2, TLR9 or IL23R might have influence on the outcome of transplantation as reported earlier,2,5,6 but not the SNP of LCT as analyzed here. Once again it seems to be crucial to analyze more homogenous groups of patients to reliably assess the role of gene polymorphisms in the transplant setting.
Donors with gene variant of LCT-13910. (A) Estimate for overall survival (OS) in patients receiving transplantations from donors with gene variants of LCT 13910. Patients receiving transplantations from donors with the homozygous CC gene variant of LCT at amino acid position 13910 compared with TC/TT gene variants showed no significantly different 5-year OS. (B) Estimate for death in remission (TRM) at 5 years after transplant. Patients receiving transplantations from donors with the homozygous CC gene variant of LCT at amino acid position 13910 compared with TC/TT gene variants had a slightly improved 5-year TRM (not significant). (C) Estimate for relapse 5 years after transplant, which shows no differences for patients receiving transplantations from donors with the homozygous CC gene variant of LCT at position 13910 compared with donors with TC or TT gene variants.
Donors with gene variant of LCT-13910. (A) Estimate for overall survival (OS) in patients receiving transplantations from donors with gene variants of LCT 13910. Patients receiving transplantations from donors with the homozygous CC gene variant of LCT at amino acid position 13910 compared with TC/TT gene variants showed no significantly different 5-year OS. (B) Estimate for death in remission (TRM) at 5 years after transplant. Patients receiving transplantations from donors with the homozygous CC gene variant of LCT at amino acid position 13910 compared with TC/TT gene variants had a slightly improved 5-year TRM (not significant). (C) Estimate for relapse 5 years after transplant, which shows no differences for patients receiving transplantations from donors with the homozygous CC gene variant of LCT at position 13910 compared with donors with TC or TT gene variants.
Authorship
Acknowledgments: The authors thank Stefanie Lorenzen, Roberta Freesa, Silke Gottwald, Christiane Schary (all Essen, Germany) for their excellent technical assistance with genotyping analyses.
This work was supported by grants from the Deutsche Krebshilfe 70-3093-El4 and Kulturstiftung Essen (05 032 Elmaagacli).
Contribution: A.H.E. designed and performed research and wrote the paper; and N.S., M.D., Y.H., H.O., R.T., and D.W.B. collected data and approved the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ahmet H. Elmaagacli, University Hospital of Essen, Hufelandstr 55, 45122 Essen, Germany; e-mail: ahmet.elmaagacli@uni-duisburg-essen.de.