Abstract
We have developed a dual bioluminescent reporter system allowing noninvasive, concomitant imaging of T-cell trafficking, expansion, and activation of nuclear factor of activated T cells (NFAT) in vivo. NFAT activation plays an important role in T-cell activation and T-cell development. Therefore we used this system to determine spatial-temporal activation patterns of (1) proliferating T lymphocytes during graft-versus-host disease (GVHD) and (2) T-cell precursors during T-cell development after allogeneic hematopoietic stem cell transplantation (HSCT). In the first days after HSCT, donor T cells migrated to the peripheral lymph nodes and the intestines, whereas the NFAT activation was dominant in the intestines, suggesting an important role for the intestines in the early stages of alloactivation during development of GVHD. After adoptive transfer of in vitro-derived T-cell receptor (TCR) H-Y transgenic T-cell precursors into B6 (H-2b) hosts of both sexes, NFAT signaling and development into CD4+ or CD8+ single-positive cells could only be detected in the thymus of female recipients indicating either absence of positive selection or prompt depletion of double-positive thymocytes in the male recipients. Because NFAT plays an important role in a wide range of cell types, our system could provide new insights into a variety of biologic processes.
Introduction
Nuclear factor of activated T cells (NFAT) is a major regulator of gene expression in the immune system and also comprises important biologic functions in a variety of other tissues.1 NFAT is a family of transcription factors encompassing 5 members, NFAT1-5.2 The DNA binding region of all NFAT members is a conserved sequence and located centrally within the protein. The activity of NFAT proteins is tightly regulated by the calcium/calmodulin-dependent phosphatase calcineurin. NFAT can form complexes with transcription factors of the AP-1 family (such as Fos and Jun). NFAT:AP-1 binding sites can be found in the regulatory regions of many genes that are involved in the regulation of the immune system.1 NFAT activation can be blocked by cyclosporine A (CsA) and FK506, which inhibit calcineurin.3
In T cells, NFAT is a prominent transcription factor downstream of the T-cell receptor (TCR)/CD3 signal cascade.2 Stimulation of the TCR is coupled to calcium mobilization and triggers activation of many intracellular enzymes including calcineurin, which dephosphorylates NFAT. Within minutes of TCR stimulation, dephosphorylated NFAT proteins translocate from the cytoplasm to the nucleus. This process has been implicated in the regulation of several important genes associated with T-cell activation, including interleukin-2 (IL-2), interferon-γ (IFN-γ), tumor necrosis factor-α (TNF-α), and Fas ligand (FasL).1 During T-cell development calcineurin/NFAT signaling initiates ERK hypersensitivity, which allows weak signals to induce positive selection.4,5 Inactivation of the B1 regulatory subunit of calcineurin (Cnb1) eliminates all calcineurin activity, which results in impaired positive selection, but does not affect negative selection of Cnb1-deficient double-positive (DP) thymocytes.4,5
Bioluminescence imaging (BLI) enables noninvasive in vivo monitoring of cells. A variety of luciferase enzymes with unique substrate requirements and spectral characteristics are available. Noninvasive imaging technologies provide important information on the biodistribution, proliferation, and persistence of cells.
In this study, we successfully combined a constitutive and NFAT-activation inducible reporter in a single system suitable for in vivo studies of primary cells by BLI. While others have developed NFAT-inducible reporter systems,6,7 we demonstrate for the first time the combination of NFAT-inducible and constitutive reporters. This system allowed noninvasive, in vivo imaging of primary cells, and we applied it to monitor the behavior of donor T cells during graft-versus-host disease (GVHD) and T-cell precursors during T-cell development and selection studying their trafficking, expansion, and activation status.
Methods
Synthesis of plasmid-encoded lentiviral vector
A plasmid encoding the recombinant lentiviral vector was produced using standard molecular biology techniques. The external Gaussia Luciferase (extGLuc) cDNA was previously published.8 The Click beetle red Luciferase (CBRLuc) cDNA was purchased from Promega. The NFAT-responsive enhancer consists of 4 consecutive repeats of the human NFAT consensus binding sequence 5′-GGAGGAAAAACTGTTTCATACAGAAGGCGT-3′. Separating the NFAT-responsive enhancer and the start codon of CBRLuc is a minimal promoter derived from the human β-globin promoter: 5′-GAATTCagggctgggcataaaagtcagggcagagccatctattgcttaca-tttgcttctgacacaactgtgttcactagcaacctcaaacagacaCC-3′. The NFAT-responsive enhancer and β-globin promoter DNA sequences were cloned using polyacrylamide gel electrophoresis–purified oligodeoxynucleotides purchased from Sigma-Genosys.
Cell lines
The temperature-sensitive gene for simian virus 40 (SV40) T-antigen was inserted into 293T (293tsA1609neo, no. CRL-11 268; ATCC), a human embryonic kidney epithelial cell derived from 293 (HEK-293).9 EL4 is a murine T-cell lymphoma cell line (no. TIB-39; ATCC), and 3T3 is a murine embryonic fibrobast cell line (no. CCL-92; ATCC). The human T-cell leukemia Jurkat cell line was a gift of Dr James W. Young (Memorial Sloan-Kettering Cancer Center).
Preparation of lentiviral vectors and transduction of primary cells
Lentiviral vectors were prepared by tripartite transfection of 293T cells, followed by viral vector harvest, concentration, and resuspension. The 293T cells were seeded on 10-cm plates. When the cells were approximately 70% confluent, the medium was replaced with 8 mL Dulbecco modified Eagle medium/fetal bovine serum (DMEM/FBS) and transfected approximately 1 hour later with 3 plasmids: pUC.MDG (cytomegalovirus promoter-driven vesicular stomatitis virus glycoprotein [VSV-G]), ΔR8.91 (cytomegalovirus promoter-driven gag and pol), and the recombinant lentiviral vector using 293T lipid-based TransIT-LT1 Transfection Reagent (Mirus Bio). Medium was replaced 14 to 16 hours later with 10 mL of DMEM/FBS. Viral vector was harvested 24 hours later and replaced with 10 mL of fresh DMEM/FBS. Viral vector was then cleared of particulates by centrifugation (466g for 5 minutes at 4°C) and concentrated by ultracentrifugation (13 000g overnight at 4°C). RPMI/FBS (50 μL) was added to the pellets followed by shaking 4 hours at 4°C. Resuspended pellets of viral vector were combined as needed, diluted in RPMI/FBS, and used for transduction.
Mice and bone marrow transplantation protocol
C57BL/6 (B6, H-2b) female mice (The Jackson Laboratory) between 6 and 8 weeks of age were used as donors. Bone marrow transplantation (BMT) protocols were approved by the Memorial Sloan-Kettering Cancer Center Institutional Animal Care and Use Committee. Bone marrow (BM) was taken aseptically from femurs and tibias of mice and prepared in 1 of 3 ways: (1) BM cells were T cell–depleted by incubation with anti–Thy-1.2 for 40 minutes at 4°C followed by incubation with Low-TOX-M rabbit complement at 37°C; (2) BM for lentiviral transduction was lineage-depleted (lin−) using negative magnetic cell separation (Miltenyi Biotec) according to manufacturer's instructions; or (3) sorted for lin−Sca-1+c-kit+ (LSK) cells after lineage depletion (EasySep; StemCell Technologies). We purified donor T cells from spleens and lymph nodes using positive selection with CD5+ magnetic beads. Typical purities were greater than 90% by flow cytometry with anti–CD3-fluorescein isothiocyanate (FITC). BM and T cells were resuspended in DMEM (Life Technologies) and transplanted by tail vein infusion into lethally irradiated mice. Before transplantation, recipient mice received total body irradiation (137Cs source) of 11Gy (C57BL/6) or 8.5Gy (BALB/c) as a split dose with 3 hours in between doses to reduce gastrointestinal toxicity. BM and CD5+ T cells from chimeric mice were isolated as previously described. Cells were stained for extGLuc and sorted. Subsequent generations of chimeric mice were created using sorted BM cells. T cells were isolated from the spleen and lymph nodes and used for in vivo studies. TCR H-Y transgenic mice (H-2b) were purchased from Taconic (model no. 004051).
Generation of chimeric mice
LSK cells or lin− BM were transduced with the NFAT vector and transplanted into lethally irradiated host C57BL/6 (B6, H-2b) mice. Lin− BM cells were expanded after transduction by culturing for 4 days with following cytokines: stem cell factor (10 ng/mL), hTPO (100 ng/mL), IGFII (20 ng/mL), and FGFI (10 ng/mL). Cells were stained on day 4 for extGLuc and then transplanted into syngeneic recipients. After immune reconstitution, transgenic primary T cells were stained for extGLuc, and the sorted cells were used without unnecessary manipulation by direct transduction, which might have resulted in activation.
In vitro T-cell stimulation and blocking of TCR signaling
Primary T cells were stimulated with phorbol 12-myristate acetate (PMA; 100 ng/mL)/ionomycin (1500 ng/mL) or CD3/CD28 for 16 hours in the presence or absence of CsA (300 ng/mL). TCR signaling was blocked by SRC Tyrosine Kinase inhibitor PP2 (10 μg/ml).
In vitro T-cell precursor expansion
OP9-DL1, a BM stromal cell line of (C57BL/6 × C3H)F2-op/op origin transduced with Delta-like1 (DL1). T-cell precursors were developed and expanded in vitro from sorted hematopoietic progenitor cells by OP9-DL1 coculture as described.10 Cell culture medium consisted of MEM (GIBCO) supplemented with 20% heat-inactivated FBS, 100 U/mL penicillin, 100 g/mL streptomycin, 10 ng/mL mouse IL-7, and 10 ng/mL mouse Flt3L. Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2.
In vivo BLI
Mice were anesthetized and then imaged 10 minutes after intraperitoneal injection of D-luciferin (6 mg/mouse; Caliper Life Sciences) for CBRLuc imaging or immediately after intravenous injection of coelenterazine (100 μg/mouse; Nanolight) for extGLuc imaging. Mice were placed into the light-tight chamber of an IVIS-200 BLI system (Caliper Life Sciences). Photographs of the mice were acquired first for anatomical reference, followed by extended exposures (seconds to minutes) to acquire the BLI signal. Luminescence data were displayed in pseudocolor, superimposed on the photographs, and calibrated to units of radiance (photons s−1sr−1cm−2). Analysis and quantification were performed using the Living image software 2.5 (Caliper Life Sciences).
Analysis of BLI
When analyzing the BLI data, the maximum radiance within a region placed over the organ of interest was chosen as being representative of the concentration of luciferase activity in that organ. Here, we assume that the activity concentration is homogenous within the organ and that the maximum radiance corresponds to the most superficial parts of the organ. This depth was measured by magnetic resonance imaging (MRI) to be approximately 1 mm for all 3 organs, and therefore we were able to compare signal magnitude across organs with minimal concern for depth related attenuation differences.
We constructed a presumptive constitutive image as the pixel-by-pixel max of the measured constitutive and NFAT-activation induced images, following the normalization of each to its own max. The resultant presumptive constitutive image rendered on a blue color scale was then blended with the measured NFAT-activation image rendered in red. Equal blends of blue and red are seen as magenta. These manipulations were conducted with ImageJ Software.
Tomographic 3D reconstruction
The 3D reconstruction depicted in Figure 6B was based upon multispectral data involving 4 wavelengths (580, 600, 620, and 640 nm) each acquired from both anterior and posterior views of the mouse. Acquisition from the 2 views was facilitated by a custom mouse holder11 that maintained the mouse in a rigid position and allowed registration to an X-ray computed tomography image set, for anatomical reference and tissue segmentation purposes. The light propagation within the mouse was calculated using a Simplified Spherical Harmonics (specifically the SP3) solution to the radiative transfer model.12 These solutions have been shown to be more accurate for small geometries (such as a mouse) and short wavelengths (< 600 nm) than the more simplistic and commonly used diffusion model.
Luciferase activity analysis
Cells were seeded in 24-well plates (HEK293T cells, 7 × 104 cells/well; NIH3T3 cells, 3 × 104 cells/well; Jurkat cells, 1 × 105 cells/well). Cells were transfected within the next 24 hours with plasmid DNA and lipid-based TransIT-LT1 Transfection Reagent (Mirus, WI) according to the manufacturer's instructions. Forty-eight hours after transfection, cells were treated with PMA (c = 100 ng/mL) plus ionomycin (c = 1500 ng/mL; Sigma-Aldrich) with or without a 30-minute pretreatment of cyclosporin A (CsA, c = 300 ng/mL). Six to 16 hours after treatments, cell lysates were prepared and measured to discriminate the 2 types of luciferase activity by using the Dual-Glo Luciferase Assay System (Promega) in a Clarity Luminescence Microplate Reader (BioTek) as specified by the manufacturers. The expression of CBRLuc was normalized with the expression of extGLuc reporter. Fold increase of normalized CBRLuc activity in treated cells was calculated over the values from untreated cells. The assays were performed in triplicates for each condition in each cell line.
Statistics and software
Calculations were performed in GraphPad Prism version 5.0 and SPSS 12.0 for Windows. Comparisons were made with the Mann-Whitney U test or Wilcoxon matched pairs test. Unless specified, P value less than or equal to .05 was considered significant.
Results
Generation of the dual-reporter system
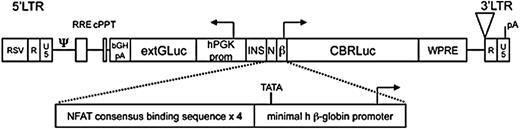
Our dual-reporter system consists of a bidirectional lentiviral vector construct encoding a constitutively expressed membrane-anchored Gaussia luciferase (extGLuc)8,13 and an NFAT-activation inducible Click-beetle red luciferase (CBRLuc; Figure 1). NFAT-dependent transcription of CBRLuc was accomplished using a synthetic enhancer/promoter consisting of a minimal promoter downstream of 4 NFAT DNA-binding elements.7 The NFAT DNA-binding element is a conserved binding site and is therefore expected to be sensitive to all NFAT family members. The distinct substrate requirements for extGLuc and CBRLuc bioluminescence (coelenterazine and luciferin, respectively) enable their differentiation. The membrane-anchored extGLuc also functions as a selection marker for successfully transduced cells.
Dual reporter lentiviral construct. Schematic diagram of the dual reporter, bidirectional lentiviral vector. LTR indicates long terminal repeat; RSV, respiratory syncytial virus; R, R element of retroviral LTR; U3, U3 element of retroviral LTR; ψ, packaging signal; RRE, rev response element; cPPT, central polypurine tract; bGH pA, bovine growth hormone polyadenylation signal; hPGK prom, human phosphoglycerate kinase-1 promoter; INS, insulator core derived from the chicken CHS4 element; N, nuclear factor of activated T cells (NFAT)-responsive enhancer; β, minimal human β-globin promoter; CBR Luciferase, click beetle red Luciferase; WPRE, woodchuck posttranscriptional regulatory element; pA, polyadenylation signal; and TATA, TATA box.
Dual reporter lentiviral construct. Schematic diagram of the dual reporter, bidirectional lentiviral vector. LTR indicates long terminal repeat; RSV, respiratory syncytial virus; R, R element of retroviral LTR; U3, U3 element of retroviral LTR; ψ, packaging signal; RRE, rev response element; cPPT, central polypurine tract; bGH pA, bovine growth hormone polyadenylation signal; hPGK prom, human phosphoglycerate kinase-1 promoter; INS, insulator core derived from the chicken CHS4 element; N, nuclear factor of activated T cells (NFAT)-responsive enhancer; β, minimal human β-globin promoter; CBR Luciferase, click beetle red Luciferase; WPRE, woodchuck posttranscriptional regulatory element; pA, polyadenylation signal; and TATA, TATA box.
The dual-reporter system is functional in cell lines
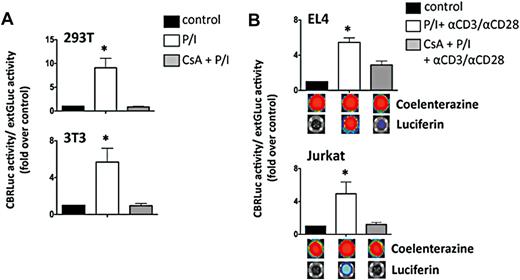
To validate our construct, we determined the transgene expression and function in transduced EL4 and Jurkat cell lines. ExtGLuc expression was confirmed by luciferase assay, BLI and flow cytometry. NFAT-activation with PMA/ionomycin or αCD3/αCD28 induced a robust CBRLuc signal activity that was abrogated by CsA-induced inhibition of calcineurin, thereby verifying NFAT-dependence (Figure 2).
In vitro functionality of NFAT-inducible lentiviral vector. (A) 293T and 3T3 cell lines were transfected with the NFAT vector before stimulation with PMA (100 ng/mL)/ionomycin (1500 ng/mL) for 6 hours in the presence or absence of CsA (300 ng/mL). NFAT-CBRLuc activity normalized to extGLuc activity was measured using a dual luciferase assay and expressed as a fold induction above unstimulated cells. Data shown represent averages ± SEM. 293T, n = 8; 3T3, n = 6. *P < .05, stimulated cells vs control. (B) EL4 and Jurkat cells were transduced with the NFAT vector. Three days after transduction cells were stimulated overnight with PMA (100 ng/mL)/ionomycin (1500 ng/mL) and αCD3/CD28 for 16 hours in the presence or absence of CsA (300 ng/mL). Luciferase activity was detected by using the Dual-Glo Luciferase Assay System (Promega) in a Clarity Luminescence Microplate Reader (BioTek) or using the IVIS bioluminescence imaging system (Xenogen). Data shown represent averages ± SEM. EL4, n = 6; Jurkat, n = 5. *P < .05, stimulated cells vs control.
In vitro functionality of NFAT-inducible lentiviral vector. (A) 293T and 3T3 cell lines were transfected with the NFAT vector before stimulation with PMA (100 ng/mL)/ionomycin (1500 ng/mL) for 6 hours in the presence or absence of CsA (300 ng/mL). NFAT-CBRLuc activity normalized to extGLuc activity was measured using a dual luciferase assay and expressed as a fold induction above unstimulated cells. Data shown represent averages ± SEM. 293T, n = 8; 3T3, n = 6. *P < .05, stimulated cells vs control. (B) EL4 and Jurkat cells were transduced with the NFAT vector. Three days after transduction cells were stimulated overnight with PMA (100 ng/mL)/ionomycin (1500 ng/mL) and αCD3/CD28 for 16 hours in the presence or absence of CsA (300 ng/mL). Luciferase activity was detected by using the Dual-Glo Luciferase Assay System (Promega) in a Clarity Luminescence Microplate Reader (BioTek) or using the IVIS bioluminescence imaging system (Xenogen). Data shown represent averages ± SEM. EL4, n = 6; Jurkat, n = 5. *P < .05, stimulated cells vs control.
Generation of chimeric mice
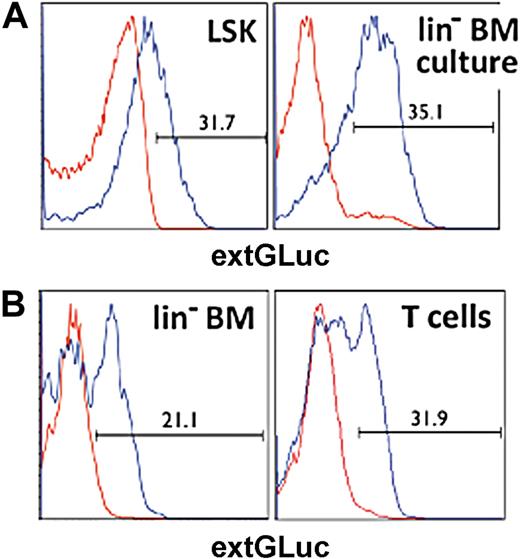
To obtain naive T cells that carry our dual-reporter vector, we isolated LSK hematopoietic precursor cells (HPCs) from murine BM and transduced these cells with our vector. We used these transduced HPCs to generate chimeric mice (Figure 3A). After 6 to 8 weeks, we were able to sort out the genetically engineered naive T cells by staining for extGLuc (Figure 3B).
Generation of chimeric mice. (A) Histograms display the transduction efficiency for LSK and lin− BM cells determined by flow cytometric analysis of extGLuc expression. Positive cell numbers are shown as percentage of live (DAPI−) cells. Nontransduced control cells, red. Transduced cells, blue. (B) BM and T cells were harvested from chimeric B6 mice 6-8 weeks after BMT. ExtGLuc expression was determined for lin− BM and CD5+ T cells by flow cytometry analysis. Nontransduced control cells are indicated in red, transduced cells in blue.
Generation of chimeric mice. (A) Histograms display the transduction efficiency for LSK and lin− BM cells determined by flow cytometric analysis of extGLuc expression. Positive cell numbers are shown as percentage of live (DAPI−) cells. Nontransduced control cells, red. Transduced cells, blue. (B) BM and T cells were harvested from chimeric B6 mice 6-8 weeks after BMT. ExtGLuc expression was determined for lin− BM and CD5+ T cells by flow cytometry analysis. Nontransduced control cells are indicated in red, transduced cells in blue.
The dual-reporter system is functional in primary cells ex vivo
We then tested the functionality of the dual-reporter system in primary T cells ex vivo. ExtGLuc expression was confirmed by Luciferase assay, BLI, and flow cytometry. NFAT-activation with PMA/ionomycin or αCD3/αCD28 induced robust CBRLuc signal activity that was abrogated by CsA-induced inhibition of calcineurin, thereby verifying NFAT-dependence (Figure 4A-B). The extGLuc signal activity was similar in all conditions, excluding a significant nonspecific effect on cell viability by CsA (Figure 4A).
In vitro functionality of NFAT-inducible lentiviral vector. (A) Sorted extGLuc+ and extGLuc− primary T cells from chimeric B6 mice were stimulated with PMA/Ionomycin or αCD3/CD28 in the presence or absence of Cyclosporine A (CsA). ExtGLuc and NFAT-induced CBRLuc activity were assessed using the IVIS bioluminescence imaging system. A representative image from 1 of 2 identical experiments is shown. (B) Activation marker (CD25, CD44, and CD62L) expression were determined for primary transgenic T cells after stimulation with PMA (100 ng/mL)/ionomycin (1500 ng/mL) and αCD3/CD28 for 16 hours in the presence or absence of CsA (300 ng/mL) and analyzed by flow cytometry. (C) ExtGLuc+ T cells from chimeric B6 were challenged with wild-type BALB/c (allogeneic) or B6 (syngeneic) antigen presenting cells plus αCD28 antibody (c = 2 μg/mL) for 72 hours. The extGluc and CBRLuc signal activity (photons/s) were quantified. Data shown represent average of 2 identical experiments.
In vitro functionality of NFAT-inducible lentiviral vector. (A) Sorted extGLuc+ and extGLuc− primary T cells from chimeric B6 mice were stimulated with PMA/Ionomycin or αCD3/CD28 in the presence or absence of Cyclosporine A (CsA). ExtGLuc and NFAT-induced CBRLuc activity were assessed using the IVIS bioluminescence imaging system. A representative image from 1 of 2 identical experiments is shown. (B) Activation marker (CD25, CD44, and CD62L) expression were determined for primary transgenic T cells after stimulation with PMA (100 ng/mL)/ionomycin (1500 ng/mL) and αCD3/CD28 for 16 hours in the presence or absence of CsA (300 ng/mL) and analyzed by flow cytometry. (C) ExtGLuc+ T cells from chimeric B6 were challenged with wild-type BALB/c (allogeneic) or B6 (syngeneic) antigen presenting cells plus αCD28 antibody (c = 2 μg/mL) for 72 hours. The extGluc and CBRLuc signal activity (photons/s) were quantified. Data shown represent average of 2 identical experiments.
Flow cytometric analyses confirmed transgene expression as well as activation of the T cells. To determine the postactivation kinetics of CBRLuc, we treated activated primary T cells with the Src tyrosine kinase inhibitor PP2, to block TCR signaling. We found that CBRLuc activity within viable cells decreased by 75% within 10 hours (supplemental Figure 2, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In the next step, we assessed the NFAT-driven T-cell bioluminescence in response to alloreactive antigen presenting cells. Sorted extGLuc+ primary T cells from chimeric B6 mice were mixed together with wild-type BALB/c (allogeneic) or wild-type B6 (syngeneic) antigen-presenting cells plus αCD28 antibody for 72 hours. A robust CBRLuc signal activity in the T cells was induced by alloreactive antigen-presenting cells (Figure 4C).
Dual in vivo imaging of T cells in GVHD
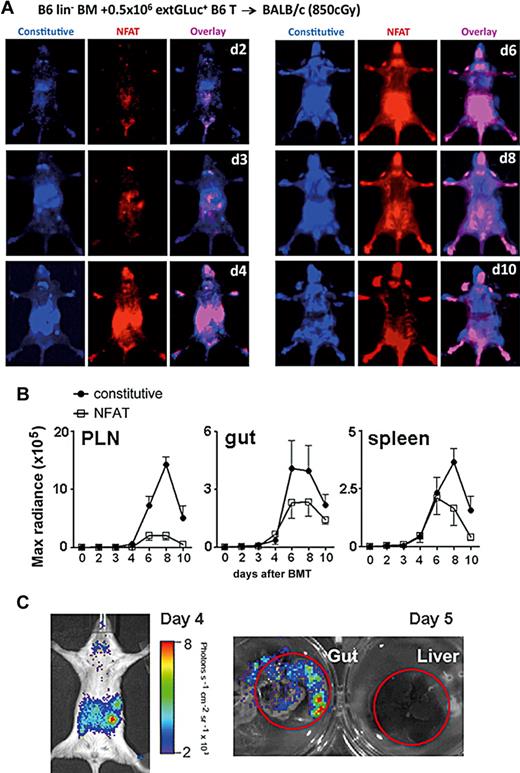
We then tested this NFAT reporter system in a mouse model of GVHD, a major complication of allogeneic hematopoietic stem cell transplantation (HSCT). In these experiments, we transplanted lethally irradiated animals with lineage-depleted BM and donor T cells (Figure 5A-B). The trafficking pattern of alloreactive transgenic donor T cells (extGLuc activity) was comparable with previously published data using firefly-luciferase or enhanced green fluorescent protein (eGFP) transgenic splenocytes.14,15 In the first 2 to 3 days after BMT, donor T cells migrated to the peripheral lymph nodes (PLN) and the intestines (constitutive extGluc signals representing the number of cells), but NFAT activation was predominantly seen in the intestines, and not in the PLN (Figure 5A-B and supplemental Figure 1A). In the following days, 4 to 8 the NFAT activity was comparable; however, there were significant'y more T cells in the PLN than in the gut, resulting in a higher percentage of activated T cells overall in the gut (Figure 5A-C and supplemental Figure 1A). Subsequently, the NFAT activation to constitutive signal ratio (CBRLuc-to-extGLuc) was higher in the gut at each time point. This observation suggests either more activation per cell or a relatively higher percentage of activated cells in the gut. In addition, the difference of the ratio CBRLuc-to-extGLuc between days 2 and 8 was significantly higher in the gut (Figure 5A-B and supplemental Figure 1B). Interestingly, both the constitutive and induced signals from the alloreactive T cells dropped precipitously after day +8 (Figure 5A-B). The decline of alloreactive T cells might represent a contraction phase involving activation-induced cell death (AICD).16
Distinct trafficking and activation kinetics of donor T cells in GVHD. (A) Donor B6 transgenic CD5+ T cells (0.5 × 106) were followed in BALB/c recipients (8.5 Gy) at serial time points, days (B) 2-10 after BMT. T-cell migration was monitored by the derived-constitutive extGLuc activity (blue) and T-cell activation by the NFAT-inducible CBRLuc activity (red). The overlay areas are displayed in magenta. Representative images from 1 of 5 experiments are shown. (B) Kinetics of extGLuc and CBRLuc signal intensities were quantified for PLN, gut, and spleen. Data shown represent averages ± SEM, n = 8. (C) NFAT-inducible CBRLuc activity is displayed on day 4 in vivo and on day 5 liver and gut ex vivo. Representative images from 1 of 5 experiments are shown.
Distinct trafficking and activation kinetics of donor T cells in GVHD. (A) Donor B6 transgenic CD5+ T cells (0.5 × 106) were followed in BALB/c recipients (8.5 Gy) at serial time points, days (B) 2-10 after BMT. T-cell migration was monitored by the derived-constitutive extGLuc activity (blue) and T-cell activation by the NFAT-inducible CBRLuc activity (red). The overlay areas are displayed in magenta. Representative images from 1 of 5 experiments are shown. (B) Kinetics of extGLuc and CBRLuc signal intensities were quantified for PLN, gut, and spleen. Data shown represent averages ± SEM, n = 8. (C) NFAT-inducible CBRLuc activity is displayed on day 4 in vivo and on day 5 liver and gut ex vivo. Representative images from 1 of 5 experiments are shown.
Dual in vivo imaging of T-cell precursors during T-cell development and selection
In previous studies we showed that adoptive transfer of ex vivo–generated T-cell precursors markedly enhanced T-cell reconstitution and graft-versus-tumor activity after HSCT.10,17 Here we used these ex vivo–generated T-cell precursors and our dual in vivo imaging system to assess T-cell development and selection during immune reconstitution. We adoptively transferred CD4−CD8−CD25+CD44+ (DN2) and CD4−CD8−CD25+CD44− (DN3) T cell precursor cells derived from an OP9-DL1 coculture of transduced LSK cells into lethally irradiated recipients of allogeneic lineage-depleted BM cells. NFAT activation was first detectable in the thymus between 11 to 14 days after the transfer of transduced T-cell precursors (Figure 6A). We acquired bioluminescence data in a manner suitable for tomographic reconstruction and calculated the 3D distribution of the NFAT-activation signal.12 The result clearly showed a localization of activated cells in the region of the thymus (Figure 6B), which was confirmed by subsequent ex vivo measurements.
Adoptively transferred T precursor cells migrate and develop primarily in the thymus during immune reconstitution. (A) 2 × 105 BALB/c lin− BM plus 0.5-1 × 106 transgenic B6 DN2/3 T cell precursor cells were transplanted into BALB/c recipients. Images display CBRLuc activity (NFAT signaling) from the thymus, day 12 in vivo and day 21 ex vivo. The CBRLuc activity quantification was performed in 10 recipients. The dot plot displays CD45+ extGLuc+ cells and percentages of DP, SP-CD4, and SP-CD8. (B) Tomographic reconstruction of the bioluminescence source distribution superimposed upon orthogonal slices through a registered computed tomography (CT) image set. Slices were selected so as to pass through the center of the most intense region of bioluminescence and correspond to the thymus.
Adoptively transferred T precursor cells migrate and develop primarily in the thymus during immune reconstitution. (A) 2 × 105 BALB/c lin− BM plus 0.5-1 × 106 transgenic B6 DN2/3 T cell precursor cells were transplanted into BALB/c recipients. Images display CBRLuc activity (NFAT signaling) from the thymus, day 12 in vivo and day 21 ex vivo. The CBRLuc activity quantification was performed in 10 recipients. The dot plot displays CD45+ extGLuc+ cells and percentages of DP, SP-CD4, and SP-CD8. (B) Tomographic reconstruction of the bioluminescence source distribution superimposed upon orthogonal slices through a registered computed tomography (CT) image set. Slices were selected so as to pass through the center of the most intense region of bioluminescence and correspond to the thymus.
In another in vivo experiment, we used the TCR H-Y transgenic mouse model18 to analyze the development of adoptively transferred T-cell precursors. In this model, the cells are homozygous for the TCR H-Y transgene and express a αβTCR specific for a minor-histocompatibility antigen (H-Y), which is encoded on the Y chromosome and only expressed in male mice. Thymocytes from these TCR H-Y transgenic mice are positively selected in female, but deleted in male mice. When we adoptively transferred TCR H-Y transgenic DN2/DN3 T cell precursors into B6 (H-2b) hosts of both sexes, we detected transferred T-cell precursors in the thymus of both male and female mice, whereas NFAT signaling and development into CD4+ or CD8+ single-positive (SP) cells could be detected only in the thymus of female mice (Figure 7).
Selection of adoptively transferred TCR H-Y transgenic T cell precursor cells predominantly in female recipients. (A) 2 × 105 B6 lin− BM plus 0.5-1 × 106 TCR H-Y transgenic DN2/3 T cell precursor cells were transplanted into male (left) and female (right) B6 recipients (n = 10). Constitutive and NFAT signal activity were determined. Thymi were harvested to determine the absolute thymocyte counts. (B) The mean of thymic NFAT activity from 5 identical experiments was quantified in male and female recipients. *P ≤ .05 was considered significant. (C) Dot plots display CD45+ extGLuc+ cells and percentage of DP, SP-CD4, and SP-CD8.
Selection of adoptively transferred TCR H-Y transgenic T cell precursor cells predominantly in female recipients. (A) 2 × 105 B6 lin− BM plus 0.5-1 × 106 TCR H-Y transgenic DN2/3 T cell precursor cells were transplanted into male (left) and female (right) B6 recipients (n = 10). Constitutive and NFAT signal activity were determined. Thymi were harvested to determine the absolute thymocyte counts. (B) The mean of thymic NFAT activity from 5 identical experiments was quantified in male and female recipients. *P ≤ .05 was considered significant. (C) Dot plots display CD45+ extGLuc+ cells and percentage of DP, SP-CD4, and SP-CD8.
Discussion
We have generated a lentiviral vector for dual in vivo noninvasive imaging consisting of both an NFAT-inducible luciferase (Click beetle) and a constitutively expressed luciferase (Gaussia). This reporter system has the following advantages: (1) the constitutive marker allows the sorting of engineered cells; (2) the constitutive marker is detectable in vivo and therefore is suitable to monitor trafficking and expansion and can act as a normalizer against which the NFAT activation induced signal may be judged; and (3) NFAT activation can be detected in vivo and assessed independent of, but in close temporal proximity to, the constitutive signal measurement. After validation, we applied this system in 2 mouse models, a GVHD and an immune reconstitution model.
The migration pattern of alloreactive T cells in GVHD has been previously characterized.14 Analyses by Beilhack et al showed initial proliferation of donor T cells in secondary lymphoid organs with subsequent homing to the intestines, liver, and skin. Panoskaltsis-Mortari et al15 demonstrated also an early migration of donor T cells to the lymph nodes (by 24 hours after BMT), but found that this migration did not require recognition of host alloantigens most likely occurring by CCR7-independent and -dependent extravasation.19
Our findings indicate a dominant role for the intestines in the early stages of alloactivation during the development of GVHD, which is in accordance to data by Murai et al showing that gut Peyer patches (PPs) are required to activate antihost CTL responses and that mice deficient for PPs failed to develop acute GVHD.20 In contrast, Beilhack et al demonstrated that the secondary lymphoid organs are the primary site of alloactivation, so that blocking access to all secondary lymphoid organs can prevent the onset of acute GVHD.21 In our study, we cannot exclude the possibility that there might have been activation of alloreactive T cells in the PLN directly post-BMT (day 1), since measurements on day 1 post-BMT revealed no signal activity.
Interestingly, we also found a decrease in donor alloreactive T-cell numbers after day +8. This is in agreement to one GVHD study by Maeda et al22 and studies regarding T-cell activation, expansion, and contraction in response to other antigens as reviewed by Badovinac and Harty.23
Immune reconstitution studies by Kisielow et al18 showed that adoptively transferred hematopoietic stem cells from TCR H-Y transgenic female donor were mostly deleted in male recipients but positively selected in female recipients. DP thymocytes, deficient for the thymocyte-specific B1 regulatory subunit of calcineurin (Cnb1), were shown to have a deficit in positive selection and to be unable to progress to the SP stage.5 However, cell death through negative selection in Cnb1-deficient DP thymocytes was normal. Processes involved in the positive selection as the induction of CD69 expression on DP thymocytes, the shutdown of recombination activating gene 1 (RAG-1) and the down-regulation of CD4 and CD8 expression were inhibited by treatment with FK506 treatment, suggesting that the activation of calcineurin is required.24 In contrast to the effect on positive selection, negative selection in the thymus of DP thymocytes was not affected by the treatment with FK506.24 Interestingly, although we could detect DP thymocytes in the thymus of male recipients, we were unable to detect any NFAT activation. This is in agreement with previous studies showing that NFAT is not required for negative selection and suggests prompt NFAT-independent negative selection of double-positive thymocytes, which is consistent with data by Teh et al.25
To use the proposed reporter system most effectively, one has to consider the changes to the measured signals that are a function of depth. This is of particular importance when analyzing the relative signal magnitudes from reporters with differing emission spectra, as we have here. The spectrum for CBR peaks at 615 nm, whereas extGLuc peaks at 480 nm. The difference in attenuation between these wavelengths at typical depths within the mouse tissue can be orders of magnitude, complicating the generation of images attempting to depict regional relative NFAT-activation such as the overlay images of Figure 5A. Without resorting to tomographic reconstruction techniques, this type of overlay is only roughly accurate. In some regions, for example, there can be an NFAT-activation signal without the presence of a measurable constitutive signal. We mitigated this anomalous behavior to the extent possible by normalizing the constitutive and NFAT activation images based on MRI organ depth measurements.
Bioluminescence tomography (BLT) may allow more quantitative interpretation of these datasets, however, we felt that the quantitative accuracy of BLT has not yet been sufficiently validated. Furthermore, BLT generally requires more involved acquisition procedures, vitiating one of the key advantages of using a bioluminescence reporter system. Nevertheless, we consider BLT to be an important and promising approach to obtain more quantitative and spatially accurate bioluminescence data.
In conclusion, we have developed a novel dual-reporter system and used this system in studies of GVHD and immune reconstitution to noninvasively and concurrently analyze spatial-temporal T-cell trafficking, proliferation, and activation. To our knowledge, this is the first study showing simultaneous in vivo visualization of trafficking and functional NFAT signaling of primary cells. This reporter system should be applicable to study any tissue with NFAT activation and could provide new insights into a variety of biologic processes.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the staff of the Memorial Sloan-Kettering Cancer Center Research Animal Resources Center for excellent animal care, and the staff of the Molecular Cytology Core Facility, Comparative Pathology Laboratories and Small Imaging Facility for assistance with sample preparation, microscopy, and imaging. We also thank Odette M. Smith for her assistance and Andrea Stroux (Charité, Germany) for statistical analyses support.
This research was supported by National Institutes of Health (NIH) award numbers RO1-HL069929, RO1-CA107096, RO1-AI080455, PO1-CA33049, R01-HL095075 (all M.R.M.v.d.B.), PO1-CA059350 (M.S.), P50-CA86438 (R.G.B.), and R24-CA83084 (Jason Koutcher). Support was also received from the US Department of Defense United States Army Medical Research Acquisition Activity (USAMRAA) Award W81XWH-09-1-0294 (M.R.M.v.d.B.), the Ryan Gibson Foundation, the Elsa U. Pardee Foundation, the Byrne Foundation, the Emerald Foundation, and The Experimental Therapeutics Center of Memorial Sloan-Kettering Cancer Center funded by Mr. William H. Goodwin and Mrs. Alice Goodwin, the Commonwealth Foundation for Cancer Research, The Bobby Zucker Memorial Fund (M.R.M.v.d.B.), and The Lymphoma Foundation.
I.K.N. and A.G. are both supported by the Deutsche Krebshilfe, Mildred-Scheel-Stiftung; A.M.H. is supported by NIH grant T32 AIO7621 and is a Starr Stem Cell Scholar Fellowship Awardee; J.C.M. was supported by NIH Medical Scientist Training Program (MSTP) grant GM07739 and a Cancer Research Institute Pre-Doctoral Fellowship.
This manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
National Institutes of Health
Authorship
Contribution: I.K.N. designed and performed experiments, analyzed data, and wrote the paper; J.C.M. generated the vector, analyzed data, and wrote the paper; J.J.T. performed experiments and analyzed data; N.L.Y., A.M.H., A.G., U.K.R., and M.T.S. performed experiments; B.J.B. designed experiments, analyzed and interpreted data, and wrote the paper; A.D.K. analyzed and interpreted data; I.S., E.B.S., R.J.B., and R.G.B. provided reagents and designed experiments; R.G.B. edited the manuscript; and M.S. and M.R.M.v.d.B. designed experiments, supervised the study, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marcel R. M. van den Brink, Department of Medicine, Memorial Sloan-Kettering Cancer Center (MSKCC), 1275 York Ave, New York, NY 10021; e-mail: vandenbm@mskcc.org.
References
Author notes
J.C.M. and J.J.T. contributed equally to this work.