To the editor:
It has recently been reported that the majority of patients with essential thrombocythemia (ET) bear a JAK2 V617F mutation on both parental alleles.1 However, a prior report using different methodology indicated that biallelic JAK2 mutations are infrequent (< 5% of MPN patients).2 We have undertaken a systematic investigation to clarify this discrepancy and elucidate the true prevalence of biallelic JAK2 mutations in ET.
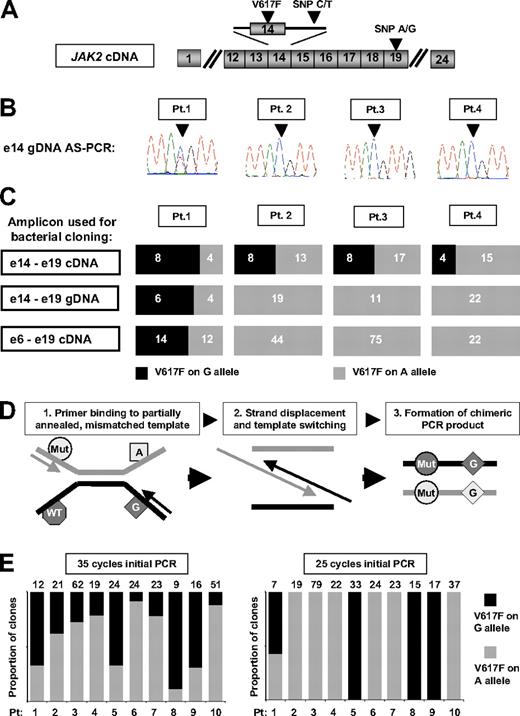
Initial studies focused on reproducing the methodology and results of both previous reports.1,2 First, granulocyte genomic DNA (gDNA) from V617F-positive ET patients was analyzed by allele-specific polymerase chain reaction (AS-PCR),3 which amplifies only the V617F-mutant allele and generates an amplicon for sequencing which includes an informative intron 14 SNP (Figure 1A). Using this approach, biallelic JAK2 mutations were detected in 2 of 30 patients (eg, Figure 1B patient 1), in keeping with Olcaydu and colleagues, who used similar methodology.2 In the report of Lambert and colleagues,1 the frequency of biallelic JAK2 mutations was determined by PCR amplification of granulocyte cDNA and genotyping by restriction enzyme digestion. Given the difficulty of ensuring complete enzymatic digestion, bacterial cloning was also performed. We therefore used granulocyte cDNA to generate a similar amplicon (encompassing the V617F mutation and an informative exon 19 SNP; Figure 1A) for bacterial cloning and genotyping. In contrast to the AS-PCR/sequencing assay, this methodology suggested the presence of biallelic JAK2 mutations in all 4 patients studied (Figure 1C), consistent with Lambert and colleagues.1 Thus, our data reproduced the findings of both previous reports and demonstrated that the 2 methodologies give conflicting results.
Detection of biallelic JAK2 mutations in patients with essential thrombocythemia by allele-specific PCR and bacterial cloning of gDNA or cDNA PCR products. (A) Schematic representation of JAK2 cDNA showing the position of the V617F mutations and the intron 14 and exon 19 single nucleotide polymorphisms (SNPs) used in these studies. (B) Granulocyte genomic DNA (gDNA) from patients 1-4 was amplified using a JAK2 V617F mutation-specific and common reverse primer to generate an amplicon encompassing an informative intron 14 SNP (arrowhead), with sequencing of the amplicon showing a JAK2 mutation on both the C- and T-alleles in patient 1, and a JAK2 mutation on the C allele only in patients 2-4. (C) Amplicons generated from cDNA or gDNA were column-purified, ligated into pGem-T Easy (Promega) or TOPO XL (Invitrogen) vectors and used for transformation of competent Escherichia coli bacteria per the manufacturer's instructions. Individual bacterial colonies were then picked and genotyped by sequencing with M13 primers. Analysis of a cDNA amplicon, encompassing JAK2 exons 14 to 19, by PCR amplification and bacterial cloning suggested the presence of biallelic JAK2 mutations in all 4 patients; however, similar analysis of both a gDNA amplicon encompassing JAK2 exons 14 to 19 and a cDNA amplicon encompassing JAK2 exons 6 and 19 indicated the presence of biallelic JAK2 mutations in patient 1 only. The number of bacterial clones with each genotype is indicated on the individual bar charts. (D) Model to explain the formation of chimeric PCR products in a mixed template reaction due to the extension of a partially annealed, mismatched template, giving rise to the artifactual appearance of biallelic mutations. (E) Analysis by bacterial cloning and genotyping of an exon 14-19 cDNA amplicon generated by either 35 or 25 cycles of initial PCR amplification: analysis of the 35-cycle amplicon suggested biallelic JAK2 mutations in all 10 patients, whereas reducing the initial PCR step to 25 cycles indicated biallelic mutations only in patient 1, concordant with the allele-specific PCR/sequencing assay. These data indicate that apparent biallelic JAK2 mutations are likely to reflect generation of chimeric DNA molecules during the later stages of PCR amplification. The number of individual bacterial clones genotyped is recorded above the graph. All ET patients were diagnosed according to criteria recently published by the British Committee for Standards in Haematology.4
Detection of biallelic JAK2 mutations in patients with essential thrombocythemia by allele-specific PCR and bacterial cloning of gDNA or cDNA PCR products. (A) Schematic representation of JAK2 cDNA showing the position of the V617F mutations and the intron 14 and exon 19 single nucleotide polymorphisms (SNPs) used in these studies. (B) Granulocyte genomic DNA (gDNA) from patients 1-4 was amplified using a JAK2 V617F mutation-specific and common reverse primer to generate an amplicon encompassing an informative intron 14 SNP (arrowhead), with sequencing of the amplicon showing a JAK2 mutation on both the C- and T-alleles in patient 1, and a JAK2 mutation on the C allele only in patients 2-4. (C) Amplicons generated from cDNA or gDNA were column-purified, ligated into pGem-T Easy (Promega) or TOPO XL (Invitrogen) vectors and used for transformation of competent Escherichia coli bacteria per the manufacturer's instructions. Individual bacterial colonies were then picked and genotyped by sequencing with M13 primers. Analysis of a cDNA amplicon, encompassing JAK2 exons 14 to 19, by PCR amplification and bacterial cloning suggested the presence of biallelic JAK2 mutations in all 4 patients; however, similar analysis of both a gDNA amplicon encompassing JAK2 exons 14 to 19 and a cDNA amplicon encompassing JAK2 exons 6 and 19 indicated the presence of biallelic JAK2 mutations in patient 1 only. The number of bacterial clones with each genotype is indicated on the individual bar charts. (D) Model to explain the formation of chimeric PCR products in a mixed template reaction due to the extension of a partially annealed, mismatched template, giving rise to the artifactual appearance of biallelic mutations. (E) Analysis by bacterial cloning and genotyping of an exon 14-19 cDNA amplicon generated by either 35 or 25 cycles of initial PCR amplification: analysis of the 35-cycle amplicon suggested biallelic JAK2 mutations in all 10 patients, whereas reducing the initial PCR step to 25 cycles indicated biallelic mutations only in patient 1, concordant with the allele-specific PCR/sequencing assay. These data indicate that apparent biallelic JAK2 mutations are likely to reflect generation of chimeric DNA molecules during the later stages of PCR amplification. The number of individual bacterial clones genotyped is recorded above the graph. All ET patients were diagnosed according to criteria recently published by the British Committee for Standards in Haematology.4
We pursued several experimental strategies to explain these discrepant results. First, an 8-kb gDNA amplicon encompassing exons 14 to 19 was studied by bacterial cloning. This approach gave results concordant with AS-PCR/sequencing (Figure 1C), excluding insensitivity of the AS-PCR/sequencing assay or genotyping of a proximal versus distant SNP as explanations for our results. Next, a 2-kb cDNA amplicon encompassing exons 6 to 19 was similarly studied. This approach also gave results concordant with AS-PCR/sequencing (Figure 1C), excluding the use of cDNA compared with gDNA as an explanation.
Finally, we investigated template-switching as a source of discrepancy. This is an assay-specific phenomenon, reflecting differences in template, primers, and/or PCR conditions, which may occur when mixed templates (eg, heterozygous SNP and/or mutant/wild-type alleles) are amplified5,6 (Figure 1D). As the likelihood of template-switching increases in the later stages of the PCR reaction,5,6 we studied granulocyte cDNA from 10 of our 30 ET patients and compared results obtained using 25 and 35 PCR cycles, with PCR products analyzed quantitatively by bacterial cloning and sequencing (Figure 1E). Using 35 cycles, apparent biallelic mutations were present in all 10 patients. In marked contrast, using 25 cycles, biallelic mutations were found only in patient 1, who also had biallelic mutations detected by AS-PCR/sequencing.
These data indicate that apparent biallelic JAK2 mutations in ET patients frequently reflect assay-specific generation of chimeric DNA molecules during PCR amplification. Our study suggests that the true prevalence of biallelic JAK2 mutations in ET is approximately 5% to 10%.
Authorship
Acknowledgments: We thank Prof Nick Cross for helpful comments on the manuscript and the Addenbrooke's Hematology Disorders Sample Bank for processing and managing patient samples. Work in the authors' laboratories is supported by the United Kingdom Medical Research Council, Leukaemia and Lymphoma Research, the Kay Kendall Leukaemia Fund, the National Institute for Health Research Cambridge Biomedical Research Center, and the Leukemia & Lymphoma Society of America.
Contribution: P.A.B. designed and performed experiments, analyzed data, and cowrote the manuscript; C.A.O. and P.J.C. contributed to experimental design and reviewed the manuscript; and A.R.G. directed the research and cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Prof Tony Green, Cambridge Institute for Medical Research, Hills Rd, Cambridge CB2 0XY, United Kingdom; e-mail: arg1000@cam.ac.uk.