Abstract
Inactivation of p15INK4b, an inhibitor of cyclin-dependent kinases, through DNA methylation is one of the most common epigenetic abnormalities in myeloid leukemia. Although this suggests a key role for this protein in myeloid disease suppression, experimental evidence to support this has not been reported. To address whether this event is critical for premalignant myeloid disorders and leukemia development, mice were generated that have loss of p15Ink4b specifically in myeloid cells. The p15Ink4bfl/fl-LysMcre mice develop nonreactive monocytosis in the peripheral blood accompanied by increased numbers of myeloid and monocytic cells in the bone marrow resembling the myeloproliferative form of chronic myelomonocytic leukemia. Spontaneous progression from chronic disease to acute leukemia was not observed. Nevertheless, MOL4070LTR retrovirus integrations provided cooperative genetic mutations resulting in a high frequency of myeloid leukemia in knockout mice. Two common retrovirus insertion sites near c-myb and Sox4 genes were identified, and their transcript up-regulated in leukemia, suggesting a collaborative role of their protein products with p15Ink4b-deficiency in promoting malignant disease. This new animal model demonstrates experimentally that p15Ink4b is a tumor suppressor for myeloid leukemia, and its loss may play an active role in the establishment of preleukemic conditions.
Introduction
The INK4a/ARF/INK4b locus covers a 40-kb region located on the human chromosome 9. This locus encodes for 3 genes (p15INK4b, p14ARF, and p16INK4a) with essential roles in regulation of the cell cycle. Whereas ARF regulates the p53 pathway, 2 other proteins (p16INK4a and p15INK4b) are important inhibitors of cyclin-dependent kinases (CDKs) CDK4 and CDK6, which play a critical role in regulation of the G1-to-S transition of the cell cycle.1 The entire locus is very frequently deleted in a variety of human tumors, including leukemias.1 In the hematopoietic system, deletions of the entire locus are mainly associated with malignancies of immature lymphocytes, most frequently in T-cell acute lymphocytic leukemia.2 Interestingly, specific inactivation of p15INK4b, via increased methylation of the CpG island in the 5′ end of the gene, is frequently observed in leukemias of myeloid origin. Indeed, the hypermethylation-associated repression of the p15INK4b expression has been reported in almost 80% of all patients with acute forms of myeloid leukemia.3,4 Specific inactivation of the gene has also been frequently detected in 50% of patients diagnosed with myelodysplastic syndrome and almost 60% of patients with myeloproliferative disorders, respectively.5,6 More importantly, hypermethylation of p15INK4b has been linked with poor prognosis in patients.7-9 These clinical results established the repression of p15Ink4b expression as the most common epigenetic abnormality in myeloid leukemia and suggest that p15Ink4b is a potent suppressor of myeloid disease. Thus far, however, only minimal evidence has been produced to support this hypothesis. Embryonal knockout of both alleles of p15Ink4b gene results in a very mild lymphoproliferative disorder and a subtle predisposition to spontaneous development of nonhematopoietic tumors.10 In addition, although p15Ink4b homozygous knockout mice have an increased frequency of early myeloid progenitors in the bone marrow (BM),11 they did not show an increased propensity over wild-type mice to develop retrovirus-induced myeloid leukemia.12 These results demonstrated that the complete embryonic removal of p15Ink4b does not provide a model system that is indicative of the human condition, where there is a direct link between p15INK4b inactivation and myeloid leukemia.
Considering the limitations of embryonal knockout, we decided to prepare a new animal model system that would have the potential to more closely recapitulate development of myeloid diseases in humans. We used the promoter for the myeloid-specific LysM gene13 to drive the expression of the Cre recombinase and conditionally delete exon 2 of p15Ink4b in myeloid cells. This mouse model closely mirrors the situation in human myeloid diseases, such as myelodysplastic syndrome and myeloproliferative disorders, where methylation of the p15INK4b CpG island represses expression of this gene without any obvious effect on regulation of p16INK4a and ARF expression.5,6
Here, we describe the p15Ink4b-LysMcre mouse and used this animal model to address whether myeloid-specific inactivation of p15Ink4b can be a causal event in premalignant myeloid disorders and the development of leukemias. Our results show that mice with targeted deletion of p15Ink4b in myeloid cells develop nonreactive monocytosis in peripheral blood (PB) accompanied by increased numbers of myeloid and monocytic cells in the BM compared with wild-type animals. The spontaneous progression from this chronic type of myeloproliferative disease to acute leukemia was not observed, even in aged animals; however, injection of neonatal mice with the MOL4070LTR retrovirus resulted in statistically significant development of myeloid leukemia in p15Ink4bfl/fl-LysMcre mice with 60% penetrance. These results strongly support the hypothesis that inactivation of p15INK4b can initiate preleukemic disease and provide the first clear experimental evidence that p15Ink4b can function as a myeloid tumor suppressor.
Methods
Generation of transgenic mice
A targeting construct for conditional knockout was generated on the backbone of the pEL14 targeting vector used for embryonal knockout and described earlier.10 Briefly, a vector containing p15loxP-exon 2-loxPNeo was constructed by insertion of a loxP site upstream of exon 2 of p15Ink4b into the BamHI site and by insertion of the neomycin cassette flanked by 2 loxP sites downstream of p15Ink4b exon 2 into the XhoI site. The DNA fragment with a floxed exon 2 and a floxed neomycin cassette was used to replace the neomycin cassette in the targeting vector pEL14 using BamHI and XhoI restriction sites to generate the targeting vector p15cKO. The targeting vector p15cKO was electroporated into the CJ7 embryonic stem (ES) cell line.14 Resistant ES clones were screened by Southern analysis for homologous recombination of the p15Ink4b allele using genomic probes located at the 5′ and 3′ end of the gene.10 Correctly targeted ES cell clones were injected into C57BL/6 blastocysts to generate chimeras that transmitted the mutated allele to the progeny. Mice positive for the floxed p15Ink4b allele were obtained by breeding and crossed to each other on a C57BL/6–129SvJ background. The offspring was screened for homozygosity and then crossed to LysMcre mice (The Jackson Laboratory).
Genotyping was performed on DNAs isolated from tails using primers specific for the intron of the p15Ink4b gene: pg1(ATACCAAGGAAACAAGATCCTC) and pg2 (AGGACAGCCAGGGCTATACAG; Figure 1A); and primers specific for Cre: Cre1 (CGATGCAACGAGTGATGAGG) and Cre2 (GCATTGCTGTCACTTGGTCCT).
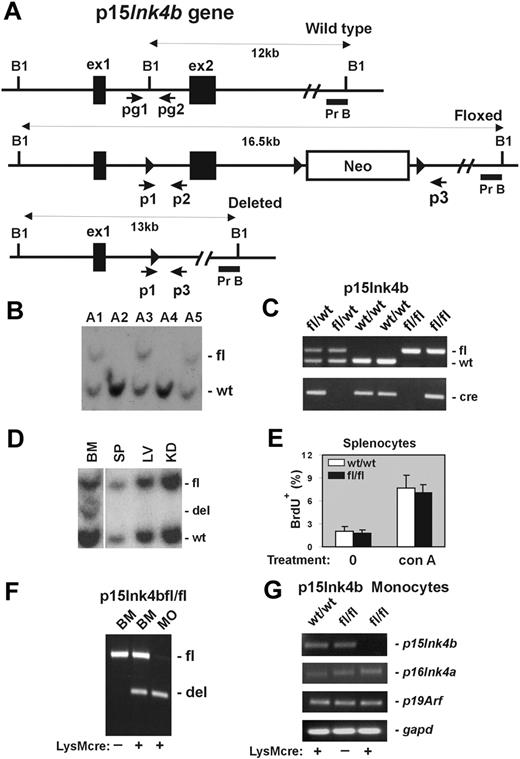
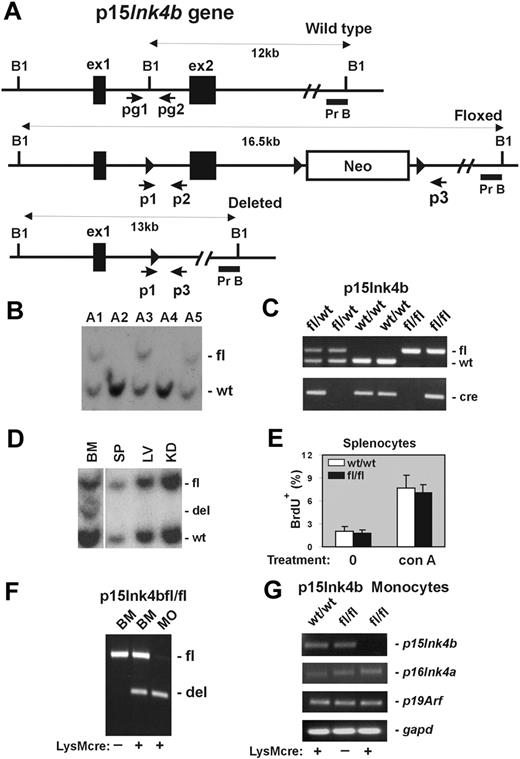
Design and generation of the p15Ink4b conditional null allele. (A) Schematic of the mouse p15Ink4b gene (Wild type), targeted (Floxed), and Cre-driven deleted (Deleted) p15Ink4b alleles. Genomic sequences encoding 2 p15Ink4b exons (ex1 and ex2), a neomycin resistance cassette (Neo), and 3 loxP sites (▶). The size of DNA fragments resulting from BamHI (B1) digestion and the probe used for Southern blot analyses (Pr B) are shown. Positions of primers pg1 and pg2 used for genotyping of experimental mice and primers p1, p2, and p3 used for evaluation of Cre-driven efficiency of recombination by PCR are also illustrated. (B) Southern blot analysis of genomic DNA isolated from the tails of the p15Ink4b chimeras used for generation of the p15Ink4bfl/fl-LysMcre and p15ink4bfl/wt-LysMcre mice. (C) PCR genotyping of mice carrying floxed p15Ink4b and LysMcre alleles using pg1 and pg2 primers and the Cre-specific primer set. The picture shows PCR products from mice carrying the p15Ink4b alleles indicated on top of the lanes. (D) Southern blot analyses of LysMcre-driven recombination of floxed p15Ink4b. Genomic DNAs were isolated from BM, spleen (SP), liver (LV), and kidney (KD) of p15Ink4bfl/wt-LysMcre mouse, digested with BamH1 and hybridized with genomic probe B (Pr B). (E) Proliferation of splenocytes isolated from p15Ink4bfl/fl-LysMcre and p15Ink4bwt/wt-LysMcre mice. Untreated (0) and concanavalin A (conA, 10 μg/mL; 2 days)-treated splenocytes were analyzed by flow cytometry for BrdU incorporation. Data are mean ± SD (error bars) for BrdU incorporations. (F) Triple-primer PCR assay with primers p1, p2, and p3 (positions in the p15Ink4b gene are shown in panel A). Targeted p15Ink4b alleles produce a 700-bp band as a PCR product of primers p1 and p2. Extension cycles of the PCR program were chosen such that a large PCR product of primers p1 and p3 with an expected length of 5.5 kb was not amplified. After the deletion of exon 2, the priming site for primer p2 is lost and primer p3 is close to p1, which together amplify a 500-bp PCR product. The ratio of the band intensities was used to determine the approximate recombination efficiency in DNAs isolated from BM and purified BM monocytes (MO). DNA isolated from the BM of the p15Ink4bfl/fl mouse without (−) the LysMcre transgene was used as a negative control for Cre-driven recombination. (G) Reverse-transcribed PCR analyses for p15Ink4b, p16Ink4a, p19Arf, and gapd expression. Total RNAs were isolated from BM-derived monocytes purified from p15Ink4bwt/wt and p15Ink4bfl/fl mice with (+) or without (−) the LysMcre transgene.
Design and generation of the p15Ink4b conditional null allele. (A) Schematic of the mouse p15Ink4b gene (Wild type), targeted (Floxed), and Cre-driven deleted (Deleted) p15Ink4b alleles. Genomic sequences encoding 2 p15Ink4b exons (ex1 and ex2), a neomycin resistance cassette (Neo), and 3 loxP sites (▶). The size of DNA fragments resulting from BamHI (B1) digestion and the probe used for Southern blot analyses (Pr B) are shown. Positions of primers pg1 and pg2 used for genotyping of experimental mice and primers p1, p2, and p3 used for evaluation of Cre-driven efficiency of recombination by PCR are also illustrated. (B) Southern blot analysis of genomic DNA isolated from the tails of the p15Ink4b chimeras used for generation of the p15Ink4bfl/fl-LysMcre and p15ink4bfl/wt-LysMcre mice. (C) PCR genotyping of mice carrying floxed p15Ink4b and LysMcre alleles using pg1 and pg2 primers and the Cre-specific primer set. The picture shows PCR products from mice carrying the p15Ink4b alleles indicated on top of the lanes. (D) Southern blot analyses of LysMcre-driven recombination of floxed p15Ink4b. Genomic DNAs were isolated from BM, spleen (SP), liver (LV), and kidney (KD) of p15Ink4bfl/wt-LysMcre mouse, digested with BamH1 and hybridized with genomic probe B (Pr B). (E) Proliferation of splenocytes isolated from p15Ink4bfl/fl-LysMcre and p15Ink4bwt/wt-LysMcre mice. Untreated (0) and concanavalin A (conA, 10 μg/mL; 2 days)-treated splenocytes were analyzed by flow cytometry for BrdU incorporation. Data are mean ± SD (error bars) for BrdU incorporations. (F) Triple-primer PCR assay with primers p1, p2, and p3 (positions in the p15Ink4b gene are shown in panel A). Targeted p15Ink4b alleles produce a 700-bp band as a PCR product of primers p1 and p2. Extension cycles of the PCR program were chosen such that a large PCR product of primers p1 and p3 with an expected length of 5.5 kb was not amplified. After the deletion of exon 2, the priming site for primer p2 is lost and primer p3 is close to p1, which together amplify a 500-bp PCR product. The ratio of the band intensities was used to determine the approximate recombination efficiency in DNAs isolated from BM and purified BM monocytes (MO). DNA isolated from the BM of the p15Ink4bfl/fl mouse without (−) the LysMcre transgene was used as a negative control for Cre-driven recombination. (G) Reverse-transcribed PCR analyses for p15Ink4b, p16Ink4a, p19Arf, and gapd expression. Total RNAs were isolated from BM-derived monocytes purified from p15Ink4bwt/wt and p15Ink4bfl/fl mice with (+) or without (−) the LysMcre transgene.
Efficiency of Cre-mediated site-specific recombination of the loxP cassettes was evaluated by Southern blot hybridization and a triple primer polymerase chain reaction (PCR) method using primers p1 TGCCTCTCATCTACTCTCCAG, p2 GCTTCAGGTGTCTGATTTCTC, and p3 TCCGATCATATTCAATAACCC (Figure 1A). Mice with genotypes p15Ink4bfl/fl-LysMcre, p15Ink4bfl/wt-LysMcre, and p15Ink4bwt/wt-LysMcre were used in experiments. Handling and treatment of mice were performed in accordance with animal protocols approved by the National Institutes of Health Intramural Animal Care and Use Committee.
Cell lines and plasmid constructs
The murine myeloid cell line M1 was cultured and differentiated as described previously.15 The retroviral vector pMSCV-IRES-GFP (MIG) was kindly provided by Scott Kogan (University of California-Los Angeles, San Francisco). Retroviral vectors expressing c-Myb (MIG-cMyb) and Sox4 (MIG-Sox4) were constructed by subcloning murine c-Myb cDNA from pcDNAcMyb16 and murine Sox4 cDNA from pIRES-2-Sox4 (kindly provided by A. Perkins17 ) into Bgl II, XhoI and Bgl II, Hpa I sites of MIG, respectively.
Histologic and immunohistochemical analysis
Tissues processed for microscopic evaluation were fixed, embedded in paraffin, mounted on slides, and stained with hematoxylin and eosin. For immunohistochemistry, sections from fixed tissues were stained using conventional techniques with the following anti-mouse antibodies: CD3 (Dako North America), B220 (CD45R; PharMingen), myeloperoxidase (MPO; Dako North America), and F4/80 (Caltag) as described previously.12 Images were taken with a Zeiss Axiophot microscope (Carl Zeiss) using 5×, 20×, or 63× lenses (Carl Zeiss), acquired with a Photometrics Coolsnap HQ camera (Photometrics) using iVision Version 4.0.5 acquisition software (Biovision) and processed in Photoshop 3.0 (Adobe Systems).
Retrovirus-induced leukemia and phenotype analyses
Newborn mice were inoculated intraperitoneally with 4 × 104 infectious particles of the MOL4070LTR retrovirus as described previously.18 Mice were under daily observation for early signs of leukemia, including lethargy, labored breathing, enlarged lymph nodes, or abdominal masses. Moribund mice were killed, and tissues were harvested for analysis. BM cells were assessed microscopically as May-Grunewald-Giemsa–stained cytospin preparations. For immunophenotyping by flow cytometry, cells were stained with fluorescein isothiocyanate–conjugated anti–mouse Mac-1, F4/80, Gr-1, Thy-1, B220, Sca-1, c-Kit, and phycoerythrin-conjugated anti–mouse Mac-1 antibodies (BD Biosciences). Diseases were classified according to the Bethesda proposals.19,20
In vitro differentiation assay
Progenitor c-Kit+ cells were enriched from the BM of p15Ink4bwt/wt-LysMcre and p15Ink4bfl/fl-LysMcre mice using EasySep Mouse CD117 Selection Cocktail kit (StemCell Technologies). Cells were transduced with MSCV-IRES-GFP-based retroviruses expressing empty vector (MIG), c-Myb, or Sox4 in the presence of 10 μg/mL of polybrene (Sigma-Aldrich) by spinoculation at 1700g for 60 minutes. Twenty-four hours after infection, green fluorescent protein (GFP)+ cells were sorted by fluorescence-activated cell sorter (FACS) and plated in triplicate in methylcellulose media containing murine stem cell factor, murine interleukin-3, and murine interleukin-6 (StemCell Technologies) at a final concentration of 5 × 102 cells/mL. The averages of duplicate platings, performed in triplicate for each genotype, were graphed. A minimum of 4 mice for each genotype and time point were used to generate the mean myeloid colony number for each group.
Proliferation assay and cell-cycle analyses
Targeted and wild-type mice were injected intraperitoneally with 100 mg/kg of bromodeoxyuridine (BrdU; BD Biosciences) and killed 2 hours after injection. BM cells were stained with phycoerythrin-conjugated anti–mouse Mac-1, fixed, and stained using the BrdU Flow Cytometry kit (BD Biosciences) before analysis by flow cytometry for incorporation of BrdU and for cell-cycle distribution using FlowJo Version 7.2.5 software (TreeStar).
Results
Generation of p15Ink4b conditional knockout mice
To investigate the possible consequences of the P15INK4B repression in human acute myeloid leukemia (AML), we decided to generate a mouse model with myeloid-specific deletion of the gene. For this purpose, we constructed mice in which the p15Ink4b gene could be conditionally targeted. A targeting vector was prepared by inserting a loxP site upstream of exon 2 and neomycin-resistant gene cassette flanked by 2 loxP sites downstream of exon 2 (Figure 1A). After transfection of the vector into CJ7 ES cell line,14 clones resistant to selection medium were screened for correct targeting by Southern blot analysis (data not shown). Chimeric founder mice, generated by microinjecting targeted ES cell clones into C57BL/6 blastocysts, were analyzed by Southern blot hybridization using genomic probe B (Figure 1B). Positive chimeras were crossed with C57BL/6 mice to produce offspring heterozygous for the floxed allele (p15Ink4bfl/wt). These mice were further bred with LysMcre mice to induce deletion of p15Ink4b in myeloid cells. Mice with Cre expression, driven via the LysM promoter, have been shown to undergo efficient deletion of floxed genes, especially in macrophages and neutrophils.13 The p15Ink4bfl/wt-LysMcre mice were bred to produce p15Ink4bwt/wt-, p15Ink4bfl/wt-, and p15Ink4bfl/fl-LysMcre offspring that were genotyped for the presence of the p15Ink4b floxed allele and the LysMcre transgene using PCR (Figure 1C). These offspring were born at the expected Mendelian ratio and did not exhibit any obvious morphologic or behavioral abnormalities. Southern blot analyses demonstrated deletion of floxed allele in less than 50% of cells isolated from total BM, roughly corresponding to the proportion of myeloid cells in this tissue. No obvious Cre-driven deletion was detected in DNAs isolated from other tissues, such as spleen, liver, and kidney (Figure 1D). In contrast to embryonal knockout, we did not observe a lymphoproliferative disorder in mice that have myeloid specific inactivation of p15Ink4b (Figure 1E). To evaluate recombination efficiency, we developed a triple-primer PCR assay with primers p1, p2, and p3 annealing to the p15Ink4b gene (Figure 1A). Using this PCR approach, we confirmed approximately 40% efficiency of Cre-driven recombination of the floxed p15Ink4b in DNA isolated from total BM cells and identified more than 90% efficiency of recombination in DNA isolated from BM-derived monocytes (Figure 1F). No spontaneous recombination was detected in BM DNA isolated from p15Ink4bfl/fl mice lacking the LysMcre transgene. High efficiency of LysMcre-derived recombination was confirmed by semiquantitative reverse-transcribed PCR where expression of the p15Ink4b gene in BM-derived monocytes isolated from p15Ink4bfl/fl-LysMcre mice was reduced by more than 95% (Figure 1G). Furthermore, the expression levels of p16Ink4a, and p19Arf located in the same locus, were not affected. Thus, the p15Ink4b-LysMcre mouse model shows a near-complete and specific loss of p15Ink4b expression in myeloid cells with no effect on the expression of neighboring genes within the same locus, closely mimicking the situation in AML patients.
Adult p15Ink4bfl/fl-LysMcre mice develop monocytosis
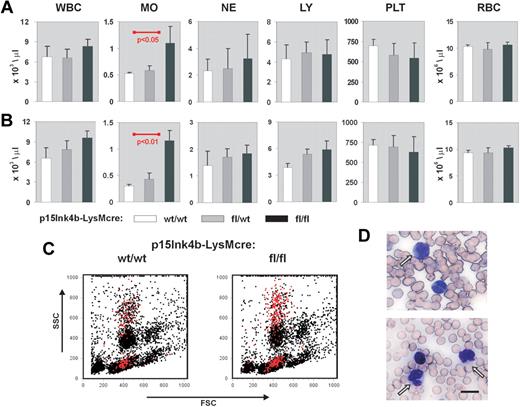
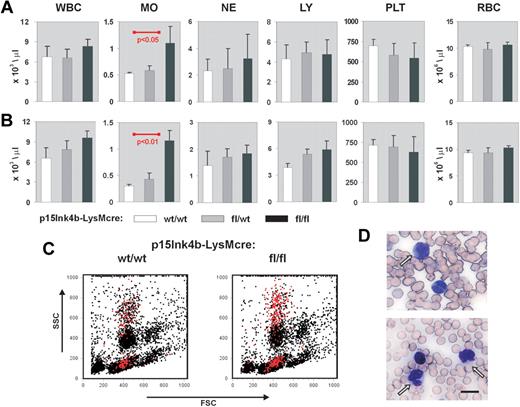
Myeloid disease and leukemias result in a pronounced increase in circulating white blood cells, particularly those of myeloid origin. To monitor disease development in p15Ink4bfl/fl-LysMcre mice, we performed white cell counts on circulating blood from young (3-4 months), juvenile (5-7 months), and aged (> 8 months) targeted and wild-type animals. Monocytes, neutrophils, lymphocytes, platelets, and red blood cells were also enumerated at each age range. We did not observe any statistically significant alterations in the number of hematopoietic cells circulating in PB at 3 to 4 months (data not shown). However, p15Ink4bfl/fl-LysMcre mice that were to 5 to 7 months of age showed a 2-fold increase in the numbers of circulating monocytes (t test, P < .05) compared with p15Ink4bwt/wt-LysMcre control mice (Figure 2A). Monocytosis was maintained in targeted mice beyond 8 months of age, whereas wild-type mice showed a marked decrease in monocytes resulting in even greater significance in the statistical comparisons (t test, P < .01; Figure 2B). Flow cytometry and May-Grunewald-Giemsa staining confirmed the increased numbers of mature/maturing monocytes in PB (Figure 2C-D). We did not observe any dysplastic cells in the smears or any increased number of red blood cells containing Howell-Jolly bodies that would indicate an impaired splenic function. These results suggest that targeted deletion of the p15Ink4b gene in myeloid cells causes a nonreactive monocytosis in the PB of experimental animals.
Nonreactive monocytosis in p15Ink4bfl/fl-LysMcre mice. Blood cell counts in p15Ink4bwt/wt-LysMcre mice (wt/wt, n = 5), in p15Ink4bfl/wt-LysMcre mice (fl/wt, n = 5), and in p15Ink4bfl/fl-LysMcre mice (fl/fl, n = 6) at 5 to 7 months (A) and 8 to 10 months (B). WBC indicates white blood cells; MO, monocytes; NE, neutrophils; LY, lymphocytes; PLT, platelets; and RBC, red blood cells. Data are mean ± SD (error bars) for cell concentrations. P values were calculated using the Student t test and only shown where statistically significant. (C) Representative flow cytometric analysis of PB nucleated cells. Monocytes (Gr-1−/lo/Mac-1+) are highlighted in red. (D) May-Grunewald-Giemsa staining of PB from p15Ink4fl/fl-LysMcre mice with nonreactive monocytosis confirmed increased numbers of mature and maturing mononuclear cells. Scale bar represents 10 μm
Nonreactive monocytosis in p15Ink4bfl/fl-LysMcre mice. Blood cell counts in p15Ink4bwt/wt-LysMcre mice (wt/wt, n = 5), in p15Ink4bfl/wt-LysMcre mice (fl/wt, n = 5), and in p15Ink4bfl/fl-LysMcre mice (fl/fl, n = 6) at 5 to 7 months (A) and 8 to 10 months (B). WBC indicates white blood cells; MO, monocytes; NE, neutrophils; LY, lymphocytes; PLT, platelets; and RBC, red blood cells. Data are mean ± SD (error bars) for cell concentrations. P values were calculated using the Student t test and only shown where statistically significant. (C) Representative flow cytometric analysis of PB nucleated cells. Monocytes (Gr-1−/lo/Mac-1+) are highlighted in red. (D) May-Grunewald-Giemsa staining of PB from p15Ink4fl/fl-LysMcre mice with nonreactive monocytosis confirmed increased numbers of mature and maturing mononuclear cells. Scale bar represents 10 μm
Expansion of myelomonocytic cells in the BM of p15Ink4bfl/fl-LysMcre mice
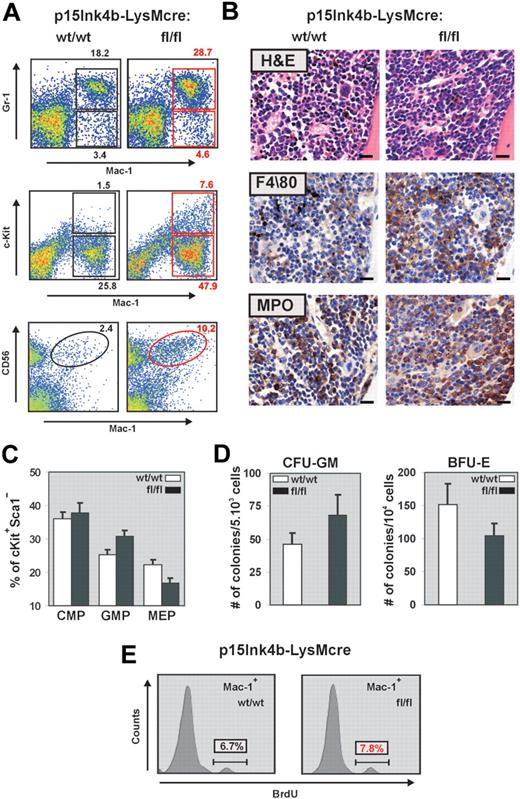
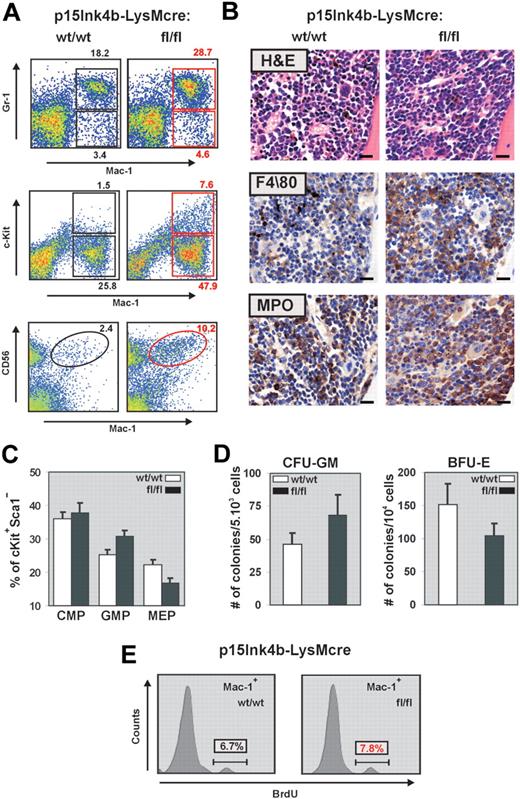
During myelopoiesis, monocytes are formed from immature progenitors in the BM through a stepwise process of differentiation involving a myelomonocytic intermediate. To determine whether the increased frequency of monocytes detected in the PB correlated with changes in the number of myeloid cells and progenitors, we examined BM cells by flow cytometry for the expression of Gr-1, Mac-1, and c-Kit. BM cells from p15Ink4bfl/fl-LysMcre mice demonstrated a significant increase in both mature myeloid (Gr-1+/Mac-1+) and monocytic (Gr-1−/lo/Mac-1+) cells (Figure 3A). This increase correlated with a significantly higher proportion of immature myeloid (Mac-1+/lo/c-Kit+) cells in the BM (7.6% in floxed vs 1.5% in wild-type mice). It has been reported that an increase in the number of CD56+ myelomonocytic cells distinguishes nonreactive monocytosis from the pathogen induced.21 FACS analyses of BM cells with anti–Mac-1 and anti-CD56 revealed an increase in Mac-1+/CD56+ cells in targeted mice and provide additional evidence for nonreactive myeloproliferative diseases in association with myeloid-specific deletion of p15Ink4b. Increased numbers of myeloid and monocytic cells in the BM were further supported by hematoxylin and eosin staining and by immunohistochemistry using monocytic (F4/80) and myeloid (MPO) markers (Figure 3B).
Myelomonocytic and monocytic cell expansion in the BM of p15Ink4bfl/fl-LysMcre mice. (A) Representative flow cytometric analysis of BM single-cell suspensions for Gr-1 and Mac-1 highlights expanded mature myeloid (Gr-1+/Mac-1+) and monocytic (Gr-1−/lo/Mac-1+) populations as well as immature myeloid cells (Mac-1+/lo/c-Kit+) in p15Ink4bfl/fl-LysMcre mice. Increased population of Mac-1+/CD56+ cells detected in targeted mice confirms a nonreactive myeloproliferative disease in targeted mice. (B) Histopathologic sections of BM from representative p15Ink4bwt/wt-LysMcre and p15Ink4bfl/fl-LysMcre animals. Hematoxylin and eosin-stained and immunohistochemistry using anti F4/80 and MPO antibodies of paraffin-embedded bone sections. Scale bars represent 20 μm. (C) Frequencies of myeloid progenitors in BM cells isolated from targeted and wild-type mice. Lineage-depleted BM cells were purified by SpinSep (StemCell Technologies) and stained with fluorochrome-conjugated monoclonal antibodies to CD34, c-Kit, interleukin-7 receptor (IL-7R), Sca-1, and FcγRII as described previously.11 Myeloid progenitors (CMPs, CD34+FγcRIIlo, GMPs, CD34+FcγRIIhi, and MEP CD34−FcγRIIlo) were identified by flow cytometry within the Lin−IL-7R−Sca-1−c-Kit+ BM cell population. Data are mean ± SD (error bars) from 3 independent analyses. (D) In vitro differentiation of FACS-purified CMP progenitors. Lineage-depleted BM cells were purified by SpinSep (StemCell Technologies) and stained with fluorochrome-conjugated monoclonal antibodies to CD34, c-Kit, interleukin-7 receptor (IL-7R), Sca-1, and FcγRII as described previously.11 CMP (CD34+FγcRIIlo) purified by FACS from the Lin−IL-7R−Sca-1−c-Kit+ BM cell population using FACSAria cell sorter (BD Biosciences) were expanded for 3 days in MyeloCult medium supplemented with SCF (50 ng/mL) and IL-11 (100 ng/mL). Cells were plated in triplicate in methylcellulose media (MethoCult M03434, StemCell Technologies). Burst-forming units–erythroid and granulocyte-macrophage colony-forming cells colonies were counted on days 7 and 12, respectively. The averages of duplicate platings performed in triplicate for each genotype, and SD values (error bars) are shown. (E) Flow cytometric analysis of BrdU incorporation into Mac-1+ BM cells indicates an increased proliferation of this population in p15Ink4bfl/fl-LysMcre mice.
Myelomonocytic and monocytic cell expansion in the BM of p15Ink4bfl/fl-LysMcre mice. (A) Representative flow cytometric analysis of BM single-cell suspensions for Gr-1 and Mac-1 highlights expanded mature myeloid (Gr-1+/Mac-1+) and monocytic (Gr-1−/lo/Mac-1+) populations as well as immature myeloid cells (Mac-1+/lo/c-Kit+) in p15Ink4bfl/fl-LysMcre mice. Increased population of Mac-1+/CD56+ cells detected in targeted mice confirms a nonreactive myeloproliferative disease in targeted mice. (B) Histopathologic sections of BM from representative p15Ink4bwt/wt-LysMcre and p15Ink4bfl/fl-LysMcre animals. Hematoxylin and eosin-stained and immunohistochemistry using anti F4/80 and MPO antibodies of paraffin-embedded bone sections. Scale bars represent 20 μm. (C) Frequencies of myeloid progenitors in BM cells isolated from targeted and wild-type mice. Lineage-depleted BM cells were purified by SpinSep (StemCell Technologies) and stained with fluorochrome-conjugated monoclonal antibodies to CD34, c-Kit, interleukin-7 receptor (IL-7R), Sca-1, and FcγRII as described previously.11 Myeloid progenitors (CMPs, CD34+FγcRIIlo, GMPs, CD34+FcγRIIhi, and MEP CD34−FcγRIIlo) were identified by flow cytometry within the Lin−IL-7R−Sca-1−c-Kit+ BM cell population. Data are mean ± SD (error bars) from 3 independent analyses. (D) In vitro differentiation of FACS-purified CMP progenitors. Lineage-depleted BM cells were purified by SpinSep (StemCell Technologies) and stained with fluorochrome-conjugated monoclonal antibodies to CD34, c-Kit, interleukin-7 receptor (IL-7R), Sca-1, and FcγRII as described previously.11 CMP (CD34+FγcRIIlo) purified by FACS from the Lin−IL-7R−Sca-1−c-Kit+ BM cell population using FACSAria cell sorter (BD Biosciences) were expanded for 3 days in MyeloCult medium supplemented with SCF (50 ng/mL) and IL-11 (100 ng/mL). Cells were plated in triplicate in methylcellulose media (MethoCult M03434, StemCell Technologies). Burst-forming units–erythroid and granulocyte-macrophage colony-forming cells colonies were counted on days 7 and 12, respectively. The averages of duplicate platings performed in triplicate for each genotype, and SD values (error bars) are shown. (E) Flow cytometric analysis of BrdU incorporation into Mac-1+ BM cells indicates an increased proliferation of this population in p15Ink4bfl/fl-LysMcre mice.
Similar to our previous results in embryonal knockout,11 FACS analyses of Lin−/IL7R−/Sca−/cKit+ BM revealed an increased frequency of granulocyte-monocyte progenitors (GMPs) and a decreased frequency of megakaryocyte-erythroid progenitors (MEPs) in targeted mice (Figure 3C). FACS purified common myeloid progenitors (CMPs) from floxed p15Ink4b mice also had an increased propensity to differentiate to granulocyte-macrophage colony-forming cells at the expense of burst-forming units–erythroid in a methylcellulose assay (Figure 3D). These results suggest that LysMcre-driven inactivation of p15Ink4b expression occurs early in myeloid lineage development. Indeed, the PCR assay detected the LoxP-LysMcre-mediated partial excision of p15Ink4b in both GMP and, to a lesser extent, CMP populations (data not shown).
In addition to an early role in regulation of myeloid differentiation, the p15Ink4b protein blocks the cell cycle by arresting cells in early G1. To determine whether the increased numbers of myelomonocytic cells in p15Ink4fl/fl-LysMcre mice can also be the result of increased proliferation of mature myeloid cells, we performed a BrdU incorporation assay. There was no difference in proliferation of total BM cells (data not shown); however, we detected a small but reproducible increase of BrdU incorporation into the Mac-1+ cells isolated from the BM of targeted mice (Figure 3E).
Overall, these data show that myeloid-specific loss of p15Ink4b results in the expansion of monocytes, myelomonocytic cells, and early progenitors in the BM. These effects may be partially explained by one or both of 2 mechanisms: enhanced differentiation of CMP to GMP at the expense of MEP progenitors and increased proliferation of maturing Mac-1+ cells.
Analyses of myelomonocytic cells in spleen tissue
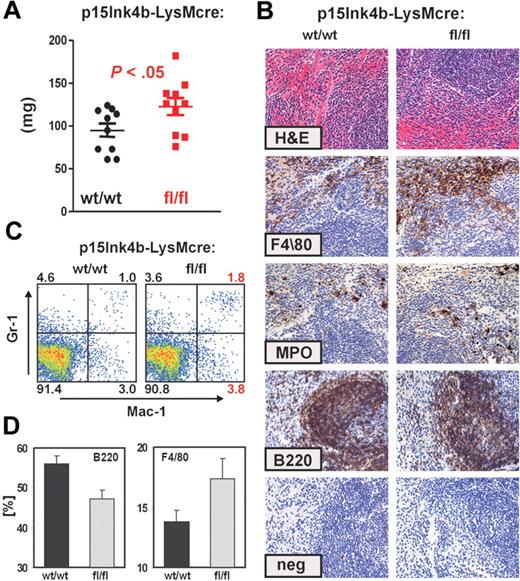
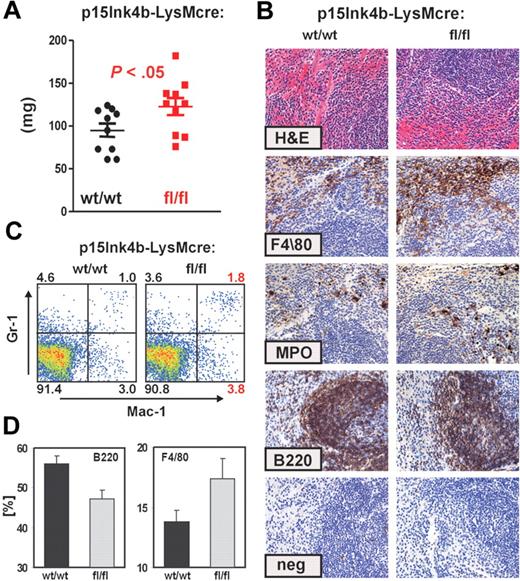
To investigate whether myelomonocytic cells from the BM expanded into other organs, we performed histopathologic analyses of spleens harvested from age-matched (6- to 8-month-old) mice. Spleens from p15Ink4bfl/fl-LysMcre were larger than p15Ink4bwt/wt-LysMcre mice (t test, P < .05; Figure 4A). Analysis of the spleen architecture (hematoxylin and eosin staining) did not reveal any obvious differences between targeted and wt mice. However, immunohistochemical staining of fixed splenic tissue with monocytic (F4/80), myeloid (MPO), and B-cell (B220) markers revealed a small increase in the myelomonocytic cell populations in the red pulp of the p15Ink4b-targeted mice (Figure 4B). To quantify differences in myeloid cell populations, we performed FACS analyses. Consistent with immunohistochemistry, we detected approximately a 25% increase in monocyte populations (Gr-1−/lo/Mac-1+) and a 2-fold greater percentage of mature myeloid (Gr-1+/Mac-1+) cells in p15Ink4bfl/fl-LysMcre mice (Figure 4C). We also detected a decreased population of B220+ cells in spleens isolated from p15Ink4bfl/fl-LysMcre mice (Figure 4D). These results suggest that inactivation of p15Ink4b in myeloid cells leads to statistically significant enlargement of spleens because of the increased population of myelomonocytic cells. However, the results are not as dramatic as observed in BM.
Increased myelomonocytic cell population in enlarged spleens of p15Ink4bfl/fl-LysMcre mice. (A) Composite spleen weights from age-matched mice of indicated genotypes demonstrate splenomegaly in p15Ink4bfl/fl-LysMcre mice. Data are mean ± SD. P value was calculated using the Student t test and shown (p15Ink4bwt/wt-LysMcre n = 10; p15Ink4bfl/fl-LysMcre, n = 10). (B) Hematoxylin and eosin and immunohistochemistry staining of paraffin-embedded spleen sections from wild-type (wt/wt) and floxed (fl/fl) mice. Scale bars represent 20 μm. (C) Gr-1 and Mac-1 FACS analysis of representative single-cell spleen suspensions confirmed expansion of mature myeloid (Gr-1+/Mac-1+) and monocytic (Gr-1−/lo/Mac-1+) cell populations. (D) The frequency of B220 and F4/80+ cells in the spleen of p15Ink4bwt/wt- (n = 4) and p15Ink4bfl/fl-LysMcre (n = 4) mice presented as a percentage of the total nucleated cells.
Increased myelomonocytic cell population in enlarged spleens of p15Ink4bfl/fl-LysMcre mice. (A) Composite spleen weights from age-matched mice of indicated genotypes demonstrate splenomegaly in p15Ink4bfl/fl-LysMcre mice. Data are mean ± SD. P value was calculated using the Student t test and shown (p15Ink4bwt/wt-LysMcre n = 10; p15Ink4bfl/fl-LysMcre, n = 10). (B) Hematoxylin and eosin and immunohistochemistry staining of paraffin-embedded spleen sections from wild-type (wt/wt) and floxed (fl/fl) mice. Scale bars represent 20 μm. (C) Gr-1 and Mac-1 FACS analysis of representative single-cell spleen suspensions confirmed expansion of mature myeloid (Gr-1+/Mac-1+) and monocytic (Gr-1−/lo/Mac-1+) cell populations. (D) The frequency of B220 and F4/80+ cells in the spleen of p15Ink4bwt/wt- (n = 4) and p15Ink4bfl/fl-LysMcre (n = 4) mice presented as a percentage of the total nucleated cells.
Increased penetrance of retrovirus-induced myeloid leukemia in p15Ink4bfl/fl mice
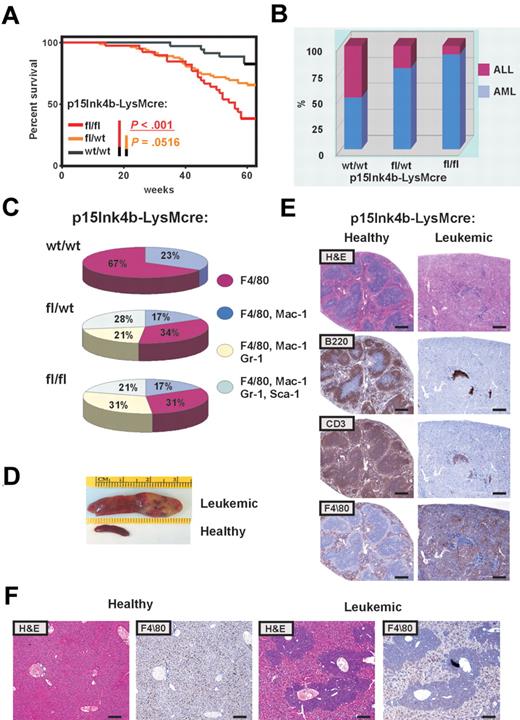
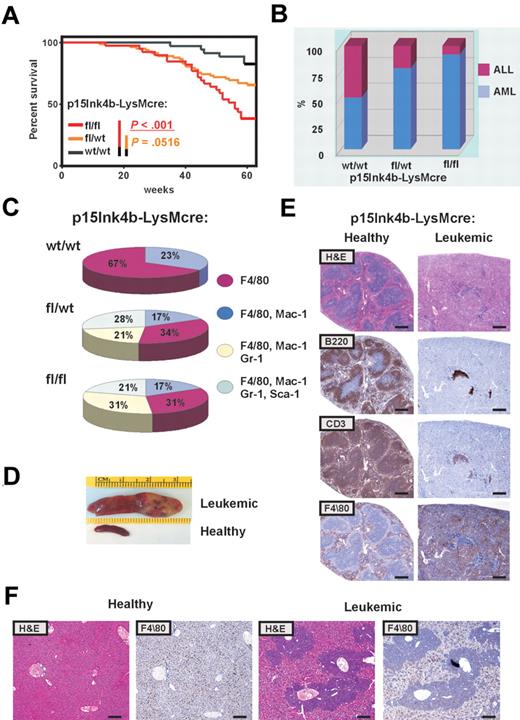
Inactivation of p15Ink4b in myeloid cells promoted a mild preleukemic myeloproliferative-like disease with nonreactive monocytosis in targeted mice. Three of 42 mice spontaneously progressed to a form of disease with leukemic symptoms and were killed. A significantly increased number of mature circulating myeloid cells in PB and cKit+ progenitors in BM of all 3 animals confirmed myeloproliferative-like disease that most closely resembles the advanced form of chronic myelomonocytic leukemia (CMML; supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). However, none of the targeted mice progressed to an acute form of leukemia over the period of 15 months. These results argue that inactivation of the p15Ink4b gene without an additional genetic/epigenetic hit is not sufficient to cause acute leukemia. To identify genetic changes that could cooperate with the loss of p15Ink4b in leukemia development, we used retrovirus-induced mutagenesis. One day after birth, mice were inoculated intraperitoneally with MOL4070LTR retrovirus18 and monitored for 15 months for signs of disease development. Complete blood counts were performed when leukemic symptoms were identified. Moribund mice were killed and histopathologically examined, and tissues were harvested for molecular and histologic analysis. Leukemias were classified according to Bethesda proposals for classification of hematopoietic neoplasms in mice.19,20 Control mice (p15Ink4bwt/wt-LysMcre) develop leukemia with low penetrance (6 of 35; Figure 5A). Low incidence of retrovirus-induced leukemia in p15Ink4bwt/wt-LysMcre may be explained by the mixed C57BL/6–129sv genotype of experimental mice, as it was shown previously that mice with the C57BL/6 background have limited spread of virus.22 The incidence of retrovirus-induced leukemia was statistically highly significant for p15Ink4bfl/fl-LysMcre (24 of 39, P < .001) and marginally significant for p15Ink4bfl/wt-LysMcre mice (28 of 82, P < .052, log-rank test; P < .038; Gehan-Breslow-Wilcoxon test), compared with wt littermates (Figure 5A). Furthermore, FACS analyses of tumor cells demonstrated that the majority of leukemias that developed in p15Ink4bfl/fl-LysMcre (92%) and p15Ink4bfl/wt-LysMcre mice (79%) were of myeloid origin. In contrast, there was an equal distribution of myeloid and lymphoid tumors in control mice (Figure 5B). Myeloid tumors were mostly monocytic (F4/80+, F4/80+/Mac1+) and myelomonocytic (F4/80+/Mac1+/Gr-1+; Figure 5C). We also detected 28% and 21% of myeloid tumors with maturation in p15Ink4bfl/wt-LysMcre and p15Ink4bfl/fl-LysMcre mice, respectively (Figure 5C). Necropsy revealed an aggressive form of AML with markedly enlarged pale spleens infiltrated with tumor cells (Figure 5D). Histologic examination of leukemic spleens revealed aggressive infiltration of monocytic leukemia into the spleen accompanied by a decrease in B (B220) and T (CD3) lymphoid cells (Figure 5E). Perivascular infiltrations of F4/80-positive monocytic leukemic cells were detected in the enlarged liver of mice with retrovirus-induced myeloid leukemia (Figure 5F). Control experiments did not reveal any obvious difference in propagation and integration of MOL4070LTR retrovirus in experimental mice (supplemental Figure 1), confirming a crucial role for the loss of p15Ink4b in predisposition of targeted mice to retrovirus-induced leukemia development. The tumorigenic potential of myeloid leukemia was further evaluated in transplantation experiments. Leukemic cells isolated from enlarged spleens of diseased mice (2 × 106/0.5 mL) were injected intravenously into healthy syngeneic recipients. Mice with transplanted tumor cells developed myeloid leukemia phenotypically identical to original tumors (data not shown). These results experimentally demonstrate, for the first time, that p15Ink4b clearly functions as a tumor suppressor for myeloid leukemia development.
High penetration of retrovirus-induced myeloid leukemia in p15Ink4bfl/fl-LysMcre mice. (A) Kaplan-Meier survival curves for p15Ink4bfl/fl-LysMcre (n = 39), p15Ink4bfl/wt-LysMcre (n = 82), and p15Ink4bwt/wt-LysMcre (n = 35) mice infected with the MOL4070LTR retrovirus. P values were calculated using the log-rank test. (B) Proportion of myeloid and lymphoid leukemia in wild-type (wt/wt) and targeted mice (fl/wt and fl/fl) infected with the retrovirus. (C) Distribution of myeloid leukemia subtypes in targeted and wt mice based on immunophenotype. (D) Representative pictures of a spleen from a clinically healthy mouse and an enlarged leukemic spleen with infiltrated leukemic cells. (E) Histologic analysis of paraffin-embedded sections of clinically healthy and leukemic spleens. Hematoxylin and eosin and immunohistochemical staining using anti-B220 (B220), anti-CD-13 (CD13), and anti-F4/80 (F4/80) are shown. Scale bar represents 400 μm. (F) Histologic analysis of paraffin-embedded sections of clinically healthy and leukemic liver stained with hematoxylin and eosin and anti-F4/80. Scale bar represents 200 μm.
High penetration of retrovirus-induced myeloid leukemia in p15Ink4bfl/fl-LysMcre mice. (A) Kaplan-Meier survival curves for p15Ink4bfl/fl-LysMcre (n = 39), p15Ink4bfl/wt-LysMcre (n = 82), and p15Ink4bwt/wt-LysMcre (n = 35) mice infected with the MOL4070LTR retrovirus. P values were calculated using the log-rank test. (B) Proportion of myeloid and lymphoid leukemia in wild-type (wt/wt) and targeted mice (fl/wt and fl/fl) infected with the retrovirus. (C) Distribution of myeloid leukemia subtypes in targeted and wt mice based on immunophenotype. (D) Representative pictures of a spleen from a clinically healthy mouse and an enlarged leukemic spleen with infiltrated leukemic cells. (E) Histologic analysis of paraffin-embedded sections of clinically healthy and leukemic spleens. Hematoxylin and eosin and immunohistochemical staining using anti-B220 (B220), anti-CD-13 (CD13), and anti-F4/80 (F4/80) are shown. Scale bar represents 400 μm. (F) Histologic analysis of paraffin-embedded sections of clinically healthy and leukemic liver stained with hematoxylin and eosin and anti-F4/80. Scale bar represents 200 μm.
Identification of genes that may cooperate with the loss of p15Ink4b in myeloid leukemia development
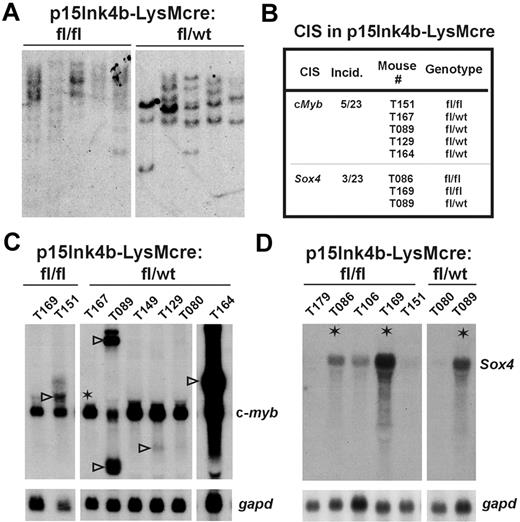
High penetrance of retrovirus-induced myeloid leukemia in targeted mice implies that genes activated by retroviruses may cooperate with the loss of p15Ink4b in malignant transformation of myeloid cells. Southern blot analysis was performed to visualize integrated MOL4070LTR proviruses in leukemic tissues isolated from p15Ink4bfl/fl-LysMcre and p15Ink4bfl/wt-LysMcre mice. As shown in Figure 6A, tumors isolated from either p15Ink4bfl/fl-LysMcre or p15Ink4bfl/wt-LysMcre mice contained from 2 to 8 retrovirus insertions per tumor. We observed differences in the intensity of bands in some tumors, suggesting that tumors in many cases are not monoclonal but at least biclonal or oligoclonal. Using 2 different PCR strategies, inverse PCR and ligation-mediated PCR,23 we have analyzed 26 genomic DNAs isolated from myeloid leukemia (9 p15Ink4bfl/fl-LysMcre, 14 p15Ink4bfl/wt-LysMcre, 3 p15Ink4bwt/wt-LysMcre) and recovered more than 100 insertion sites (supplemental Table 2). Two of these sites with recurrent virus integrations were referred to as common integration sites (CISs) for myeloid leukemia developed in the p15Ink4b-targeted mice. Retroviruses in these genomic sites integrated close or within genes encoding 2 transcriptional regulators c-Myb and Sox4 (Figure 6B). To confirm deregulation of both genes, we analyzed expression of c-myb and Sox4 mRNAs in total RNAs isolated from leukemias. For controls, we used total RNAs from leukemias that did not have retrovirus integrated in the vicinity of these genes, which will provide a more accurate comparator than normal spleen or BM cells (Figure 6C-D). Aberrant c-myb transcript was detected in 4 leukemias (Figure 6C open arrowheads) with retrovirus integrated directly within gene intron1 (T151), exon 8 (T089), intron 7 (T129), and intron 3 (T164). The expression level of the aberrant form of c-myb in tumor T129 is lower than the level of endogenous c-myb, suggesting that this particular clone was minor in the leukemic tumor mass population. In one case, we have also detected integration of virus 23 kb upstream of c-myb (Figure 6C, T167 asterisk). In this case, we did not observe up-regulation of c-myb expression compared with other leukemias. This genomic locus, named Mml1, was previously found to be the site of frequent integration of retroviruses in promonocytic leukemia; however, no up-regulation of c-myb expression was observed.24 The second CIS for our group of myeloid leukemias was identified in the genomic locus surrounding the intronless Sox4 gene. We have identified 3 retroviral insertions in this locus. Two of them were located at the 3′ end (T086 and T089) and one 5′ end (T169) of the gene (Figure 6B). Northern blot revealed high expression of the Sox4 gene in tumors T089 and T169 and moderately elevated expression in T086 (Figure 6D).
Identification of retrovirus CISs in myeloid leukemia of p15Ink4b-targeted mice. (A) Southern blot analysis of DNAs isolated from leukemic spleens of p15Ink4b-targeted (fl/fl and fl/wt), MOL4070LTR-infected mice. Genomic DNA for Southern blot analysis was isolated by phenol extraction, digested with EcoRI, separated on a 0.7% agarose gel, and transferred onto a nylon membrane. The blots were hybridized with a viral LTR probe labeled by the random priming method as described earlier.12 (B) Two CISs close to c-Myb and Sox4 encoding genes were identified in myeloid leukemias in p15INK4b-LysMcre mice by inverse and ligation-mediated PCR. (C) Northern blot analyses of c-myb expression in myeloid leukemias. (◁), aberrantly expressed c-myb mRNA in leukemias with retrovirus integrated within the gene. *Transcription in leukemia with retrovirus integrated 23 kb upstream of the c-myb gene. (D) Northern blot analyses of Sox4 expression in myeloid leukemias. *Leukemias with retrovirus integrated upstream or downstream of the Sox4 gene. Loading and integrity of analyzed RNAs were confirmed by hybridization with a gapd probe.
Identification of retrovirus CISs in myeloid leukemia of p15Ink4b-targeted mice. (A) Southern blot analysis of DNAs isolated from leukemic spleens of p15Ink4b-targeted (fl/fl and fl/wt), MOL4070LTR-infected mice. Genomic DNA for Southern blot analysis was isolated by phenol extraction, digested with EcoRI, separated on a 0.7% agarose gel, and transferred onto a nylon membrane. The blots were hybridized with a viral LTR probe labeled by the random priming method as described earlier.12 (B) Two CISs close to c-Myb and Sox4 encoding genes were identified in myeloid leukemias in p15INK4b-LysMcre mice by inverse and ligation-mediated PCR. (C) Northern blot analyses of c-myb expression in myeloid leukemias. (◁), aberrantly expressed c-myb mRNA in leukemias with retrovirus integrated within the gene. *Transcription in leukemia with retrovirus integrated 23 kb upstream of the c-myb gene. (D) Northern blot analyses of Sox4 expression in myeloid leukemias. *Leukemias with retrovirus integrated upstream or downstream of the Sox4 gene. Loading and integrity of analyzed RNAs were confirmed by hybridization with a gapd probe.
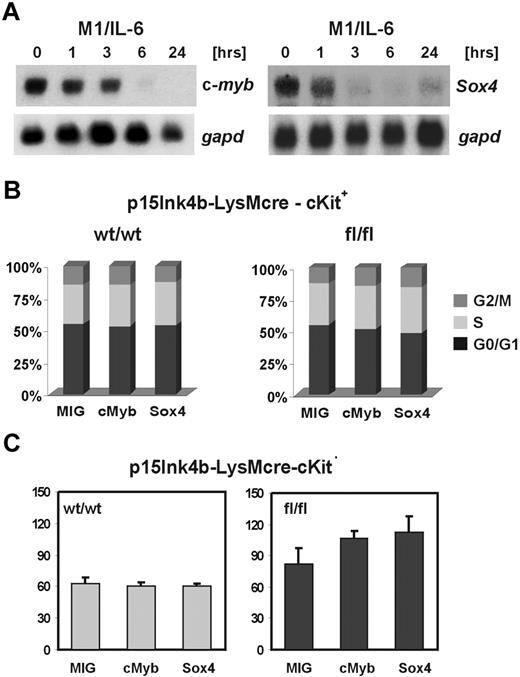
The transcription factors c-Myb and Sox4 play an important role in regulation of proliferation and/or differentiation of myeloid cells.17,25 Expression of both is rapidly repressed in myeloid cells as they differentiate along the monocytic pathway (Figure 7A). This suggests an incompatibility of their expression with terminal stages of monocytic differentiation. Indeed, enforced expression of either one prevents terminal differentiation of myeloid cells.17,26,27 We reasoned that the inability to down-regulate c-Myb and Sox4 expression during myeloid differentiation may cooperate with the loss of p15Ink4b in leukemia development. To investigate a possible cooperative role of c-Myb and Sox4 with the loss of p15Ink4b, we constructed retroviral vectors encoding murine c-Myb (MIG-cMyb) and Sox4 (MIG-Sox4) proteins. Hematopoietic progenitors enriched for c-Kit+ cells were purified from BM, infected with retroviruses, and GFP+ cells were sorted by FACS. Cell-cycle analyses revealed that enforced expression of either c-Myb or Sox4 did not affect cell-cycle distribution of infected cells (Figure 7B). Next, we investigated whether myeloid colony formation can be affected by deregulated expression of c-Myb or Sox4. When plated in methylcellulose cultures in the presence of growth factors supporting myeloid cells development, c-Kit+ cells isolated from p15Ink4bwt/wt-LysMcre mice infected with either empty vector (MIG) or vectors expressing c-Myb or Sox4 show no difference in plating efficiency. In contrast, we have observed an increase in myeloid colony numbers in c-Kit+ cells isolated from BM cells of p15Ink4bfl/fl-LysMcre mice ectopically expressing either c-Myb or Sox4 (Figure 7C). Myeloid colonies from cells expressing c-Myb and Sox4 were slightly smaller compared with colonies infected with empty vector. These results suggest that expression of c-Myb and Sox4 in the absence of p15Ink4b affects plating efficiencies rather than proliferation of progenitor cells.
Cooperation of deregulated c-Myb and Sox4 with the loss of p15Ink4b in myeloid colony formation. (A) Northern blot analyses of c-myb and Sox4 expression in M1 cells during interleukin-6 (IL-6)–induced monocytic differentiation. Loading and integrity of analyzed RNAs were confirmed by hybridization with the gapd probe. (B) Cell-cycle distribution of cKit+ cells purified from p15Ink4bwt/wt- and p15Ink4bfl/fl-LysMcre BM infected with empty retrovirus (MIG), or retrovirus expressing c-Myb (cMyb), or Sox4 (Sox4) transcription factors. (C) Myeloid colony formation of cKit+ BM cells purified from targeted (fl/fl) or wt (wt/wt) mice infected with empty vector (MIG) or transcription factors-expressing (cMyb or Sox4) retroviruses. GFP+ cells were sorted and cultured in methylcellulose with myeloid-specific cytokines. Data are mean ± SD (error bars) from 2 independent experiments, each performed in triplicate.
Cooperation of deregulated c-Myb and Sox4 with the loss of p15Ink4b in myeloid colony formation. (A) Northern blot analyses of c-myb and Sox4 expression in M1 cells during interleukin-6 (IL-6)–induced monocytic differentiation. Loading and integrity of analyzed RNAs were confirmed by hybridization with the gapd probe. (B) Cell-cycle distribution of cKit+ cells purified from p15Ink4bwt/wt- and p15Ink4bfl/fl-LysMcre BM infected with empty retrovirus (MIG), or retrovirus expressing c-Myb (cMyb), or Sox4 (Sox4) transcription factors. (C) Myeloid colony formation of cKit+ BM cells purified from targeted (fl/fl) or wt (wt/wt) mice infected with empty vector (MIG) or transcription factors-expressing (cMyb or Sox4) retroviruses. GFP+ cells were sorted and cultured in methylcellulose with myeloid-specific cytokines. Data are mean ± SD (error bars) from 2 independent experiments, each performed in triplicate.
Discussion
Experimental evidence is provided here that the lineage-specific silencing of p15Ink4b plays a role in early leukemia development. Mice with the LysMcre-driven deletion of p15Ink4b in myeloid tissues spontaneously developed nonreactive monocytosis in the PB that was accompanied by increased numbers of myelomonocytic cells and immature c-Kit+ myeloid progenitors in the BM and splenic tissues. Morphologic characteristics of p15Ink4bfl/fl-LysMcre mice most closely resemble the mild myeloproliferative form of CMML. In accord with our finding, silencing of p15INK4b through hypermethylation was reported previously in approximately 60% of CMML cases. The majority of patients in this study were diagnosed with class I CMML with less than 10% of blasts in the BM. However, even within this category, a higher proportion of methylation was detected in a more aggressive form of CMML-1, with 5% to 10% of blasts compared with patients with less than 5% of blasts in the BM.6 These results argue that myeloid-specific inactivation of p15Ink4b, which mimics suppression of its expression in CMML, is not just a passive result of a global change in DNA methylation frequently observed in precancerous lesions, but instead it may be of functional significance in the establishment of these preleukemic conditions.
Although LysMcre-driven inactivation of p15Ink4b triggers development of a premalignant condition similar to the myeloproliferative form of CMML, there is no spontaneous progression to acute leukemia, even in aged animals. This observation is in accord with the generally accepted hypothesis that no single event is sufficient for leukemia development.28 Inoculation of p15Ink4bfl/fl-LysMcre mice with MOL4070LTR retrovirus, however, resulted in the development of AML, with more than 60% frequency compared with only rare occurrences of the disease in control mice (Figure 5A). Strikingly, previous experiments with homozygous germline deletion of the gene show no increase in the retrovirus-induced leukemia development over wild-type littermates.12 Whether the global loss of p15Ink4b leads to compensation through deregulation of other Ink4 proteins or whether deletion of p15Ink4b outside of myeloid compartment negatively contributes to the development of myeloid leukemia is not clear at present. Interestingly, however, the lymphoproliferative disorder observed in embryonal knockout could result in an enhanced removal of retrovirus-expressing preleukemic cells by the immune system. Previously, tumor suppressor activity of p15Ink4b was demonstrated only in an animal model where both p16INK4a and p15INK4b genes were simultaneously inactivated. These animals developed a wide spectrum of tumors with prevalence of skin tumors and soft tissue sarcomas, but not myeloid leukemia.29 These results nicely correlated with frequent deletion of an entire locus CDKN2A-CDKN2B in human carcinomas and sarcomas30,31 and suggested that p15INK4b may work as the backup for p16INK4a tumor suppressor in these diseases. Our work is the first demonstration that p15Ink4b can act as a tumor suppressor in the presence of a fully functional p16Ink4a locus. This finding correlates with the frequent and specific suppression of p15INK4b expression, via an epigenetic mechanism, in human myeloid disease and suggests that a key aspect of the role for p15INK4b in myeloid disease and leukemia is its specific inactivation in myeloid cells.
Acute leukemia is the consequence of genetic and/or epigenetic changes that generally result in an enhanced proliferation and survival of progenitor hematopoietic cells with impaired differentiation.32 Our work emphasizes the importance of multiple genetic aberrations in the development of AML. Specifically, we have identified more than 100 genes, through retroviral insertional mutagenesis, whose protein products may cooperate with the loss of p15Ink4b expression in the formation of AML. In our model, recurrent retrovirus integrations were detected within or in close proximity to 2 genes in particular, c-myb and Sox4, resulting in their frequent up-regulation in myeloid tumors. Both genes encode nuclear transcription factors known to play critical roles in hematopoietic cell proliferation and differentiation. A direct role for c-Myb overexpression in myeloid disease has been previously demonstrated by constitutive expression of c-Myb in hematopoietic cells, which blocks the differentiation of monocytic and granulocytic lineages.26,27 More recent results also established a role for deregulated c-MYB in human leukemia, where the amplification of c-Myb locus in T-cell acute lymphocytic leukemia33,34 and in myelomonocytic AML35 was reported. A critical role for c-MYB in MLL-ENL-driven myeloid leukemia is also evident from recent experiments.36 The present work adds to this body of evidence by identifying the cooperation of p15Ink4b inactivation and aberrant c-Myb expression for the induction of myeloid leukemia in mice.
The identification of a cooperative role for Sox4 in the development of myeloid leukemia is a novel finding of this study as there has been little evidence previously associating this gene with myeloid cells. Sox4 homozygous null mice display defective B-cell development, but they do not show any obvious abnormalities in myelomonocytic cells.37 Deregulated expression of SOX4 has been implicated in the genesis of several human cancers, but its possible role in leukemias has not been examined.38-40 To our knowledge, the only evidence previously demonstrating a role for Sox4 in AML was provided by Boyd et al who showed that Sox4 prevents granulocytic differentiation and is up-regulated by proviral insertion in myeloid tumors developed in AKXD-23 mice.17 Here we associate Sox4 down-regulation with monocyte differentiation and suggest a role for Sox4 up-regulation during the development of AML in mice, specifically through cooperation with myeloid-specific loss of p15Ink4b.
Frequent activations of both transcriptional regulators c-Myb and Sox4 by retroviruses in tumors derived from our mouse model system suggest that these genes may cooperate with the loss of p15Ink4b in the development of myeloid disease. Increased myeloid colony formation of p15Ink4bfl/fl-LysMcre progenitor cells transduced with vectors encoding either c-Myb or Sox4 compared with wt progenitor cells supports this assumption (Figure 7C). However, more experiments need to be done to confirm collaboration of these transcription factors with the loss of p15Ink4b in development of myeloid leukemia. Nevertheless, our data suggest that further investigation into the potential cooperation of p15INK4B suppression with c-MYB or SOX4 deregulation in human AML is warranted.
Down-regulation of p15Ink4b expression is one of the most frequent epigenetic aberrations in myeloid leukemia, but until now its role in preleukemic or leukemic cell development was not clear. Our new animal model confirms the role for the loss of p15Ink4b in the establishment of preleukemic conditions in myeloid cells and provides the first experimental evidence that p15Ink4b can clearly function as a tumor suppressor for myeloid leukemia development independently of p16Ink4a and p19Arf.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the National Cancer Institute, Center for Cancer Research Gene Targeting Facility for generation of chimeric mice, Chris Perella and Mary Albaugh (National Cancer Institute–Frederick, National Institutes of Health) for excellent animal care, Jinesh Gheeya for technical assistance, and Archibald Perkins for providing the mouse Sox4 cDNA construct.
This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
National Institutes of Health
Authorship
Contribution: J.B. designed experiments, performed research, analyzed and interpreted data, and wrote the paper; M.S., S.Z., J.F., and R.K. performed research; M.R.-M., performed research and edited the paper; and L.W. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Juraj Bies, Laboratory of Cellular Oncology, National Cancer Institute, National Institutes of Health, Bldg 37/4124, 37 Convent Dr, MSC-4263, Bethesda, MD 20892-4263; e-mail: biesj@mail.nih.gov; and Linda Wolff, Laboratory of Cellular Oncology, National Cancer Institute, National Institutes of Health, Bldg 37/4124, 37 Convent Dr, MSC-4263, Bethesda, MD 20892-4263; e-mail: wolffl@mail.nih.gov.