Abstract
The multiple myeloma SET domain (MMSET) protein is overexpressed in multiple myeloma (MM) patients with the translocation t(4;14). Although studies have shown the involvement of MMSET/Wolf-Hirschhorn syndrome candidate 1 in development, its mode of action in the pathogenesis of MM is largely unknown. We found that MMSET is a major regulator of chromatin structure and transcription in t(4;14) MM cells. High levels of MMSET correlate with an increase in lysine 36 methylation of histone H3 and a decrease in lysine 27 methylation across the genome, leading to a more open structural state of the chromatin. Loss of MMSET expression alters adhesion properties, suppresses growth, and induces apoptosis in MM cells. Consequently, genes affected by high levels of MMSET are implicated in the p53 pathway, cell cycle regulation, and integrin signaling. Regulation of many of these genes required functional histone methyl-transferase activity of MMSET. These results implicate MMSET as a major epigenetic regulator in t(4;14)+ MM.
Introduction
Multiple myeloma is an incurable malignancy of mature plasma cells, associated in approximately 40% of cases with recurrent chromosomal translocations that lead to overexpression of known and putative oncogenes.1,2 MMSET (WHSC1, NSD2) is linked to the immunoglobulin promoter/enhancer in t(4;14) translocations, found in 15%-20% of multiple myeloma.3 Chromosomal fusion leads to overexpression of MMSET and FGFR3 genes; however, approximately 30% of the patient samples overexpress only the MMSET gene, suggesting its pivotal role in the disease.4-6 The other nuclear receptor Su(var)3-9, Enhancer-of-zeste, Trithorax (SET) domain-containing (NSD) family members, NSD1 and NSD3, were both found to be rearranged as fusion proteins with NUP98 in rare cases of acute myeloid leukemia, and NSD3 is overexpressed in breast cancer,7,8 suggesting that deregulation of these proteins plays a causative role in malignancy. The MMSET gene undergoes complex alternative splicing and differential promoter usage, giving rise to a number of different transcripts from the locus, most of which are overexpressed in t(4;14) myelomas (Figure 1A).2,9,10 The protein domains found in full-length MMSET include 2 conserved Pro-Trp-Trp-Pro motif (PWWP) domains, 4 plant homeo domain fingers, and 1 SET domain, all of which are commonly found in transcriptional regulators.11,12 Our previous report suggested that MMSET may be part of a corepressor complex.13 SET domain-containing proteins can methylate lysine residues on histone tails.14 Methylation and other covalent modifications of histone tails, such as acetylation, phosphorylation, ubiquitination, or sumoylation, can alter gene expression depending on the residue altered, the type of the modification, and whether the modified histone residue is found in a gene promoter, enhancer, or the body of a gene.15
Promoters of actively transcribed genes are marked by the presence of H3K4me3, whereas the transcribed body of active genes is characterized by methylation at H3K36 (H3K36me3).16,17 By contrast, CpG islands are depleted of H3K36 methylation.18 Inactive and silenced genes show methylation at H3K27me3 and H3K9me3, respectively.16,17,19 Previous reports suggested promiscuous activity of the SET domains of the NSD family proteins. NSD1 was initially shown to methylate both H3 and H4 histones, and more recently its specificity has been narrowed down to lysine 36 on histone H3.8 Likewise, MMSET was able to methylate both H3 and H4 histones in vitro.13,20 A recent report showed that the histone methyl-transferase (HMT) activity of NSD proteins is substrate specific, helping explain these discrepancies.21 With regard to multiple myeloma, key questions have included the nature of the genes regulated by MMSET, the changes in chromatin that occur on such genes, and the relevance of MMSET overexpression to oncogenesis.
In this study, we report that overexpression of MMSET in myeloma cells leads to aberrantly high global levels of H3K36 dimethylation, accompanied by a striking decrease in H3K27 methylation. Alteration of MMSET expression leads to changes in cell growth, adhesion, and chromatin accessibility. However, these effects are greatly dependent on the isoform of MMSET that is being manipulated. Our data suggest that in myeloma cells, MMSET globally disrupts chromatin structure and function representing a potential oncogenic insult that contributes to development of disease.
Methods
Cell culture, plasmids, and mutagenesis
Multiple myeloma cell lines were cultured in RPMI with 10% fetal bovine serum, penicillin, and streptomycin. All the DNA fragments used for cloning were polymerase chain reaction (PCR) amplified using high-fidelity Phusion polymerase (Finnzymes). MMSET I was cloned into the XhoI and EcoRI sites of the retroviral MIGR1 vector. MMSET II and REIIBP fragments were inserted into NotI and BamHI sites of the pRetroX-IRES-DsRedExpress vector (Clontech). Site-directed mutagenesis of MMSET residues F1177 and Y1118 was performed using the QuickChange Site Directed Mutagenesis Kit (Stratagene).
Loss- and gain-of-function systems
MMSET-inducible knockdown cell lines were created first by infection of KMS11 with a plasmid encoding a Tet repressor. This clonal cell line was then infected with a retrovirus containing Tet operator-controlled shRNA (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) for MMSET or scrambled control as previously described.22 Cells were selected with puromycin 1 μg/mL for 2 weeks. For the induction of shRNA, cells were grown with doxycycline 50 ng/mL for 7 days. For the repletion system, KMS11 knockout cells23 were transduced with retroviral vectors harboring different MMSET wild-type and mutant isoforms. All retroviruses were produced by transfection of amphotropic 293T cells with appropriate plasmids and FuGENE 6 Transfection Reagent (Roche) according to the manufacturer's protocol. After infection, MMSET knockout cells were sorted using the DsRed (MMSET II and REIIBP) or green fluorescent protein (MMSET I) markers and expanded for further studies.
RNA extraction and microarray analysis
The inducible knockdown cells were cultured in triplicate in the presence or absence of doxycycline for 7 days. Wild-type or mutant MMSET-repleted knockout cells were cultured in triplicate and collected for RNA extraction. RNA was extracted using QIAGEN RNeasy Kit and hybridized to Sentrix Human-6_v3 Expression BeadChip (Illumina). Data were analyzed using GeneSpring GX Version 11 (Agilent) using the Student t test without multiple test correction. Gene ontology was assessed using the DAVID Ontology software Version 6.7 (http://david.abcc.ncifcrf.gov/) and Ingenuity Pathway Analysis Version 8.7 (http://www.ingenuity.com/). All microarray data are available on Gene Expression Omnibus under accession number GSE24746.
cDNA preparation and qRT-PCR
cDNA was synthesized from 1 μg of total RNA using the Iscript cDNA Synthesis Kit (BioRad). Real-time RT-PCR determinations were performed using predesigned TaqMan assays (Applied Biosystems).
Adhesion assay
Inducible shRNA cells were grown for 7 days in the presence of doxycycline (50 ng/mL) in tissue culture plates. Plates were washed with phosphate-buffered saline and stained with crystal violet solution for 15 minutes. Wild-type or mutant MMSET repleted knockout cells were grown for 4 days before staining. Pictures were taken with Axiovert 200 microscope (Zeiss), AxioCam HRC camera (Zeiss), and AxioVision 40 Software Version 4.5.0.0 (Zeiss).
MNase accessibility assay
Micrococcal nuclease (MNase) accessibility assay was performed as described previously.24 Briefly, living cells were permeabilized with lysolecithin 0.025% for 2 minutes and then treated with 3 U of MNase (Worthington) for 5 minutes. DNA was purified by phenol/chloroform extraction and 5 μg loaded on 2% agarose gels.
Cell cycle analysis and apoptosis assay
Cells were treated with BrdU (5-bromo-2-deoxyuridine) for 30 minutes, and the extent of BrdU incorporation was assayed using the APC BrdU Flow Kit (BD Pharmingen) for flow cytometric analysis using the manufacturer's protocols. Apoptosis was measured using Annexin V-Cy5 Apoptosis detection Kit (Biovision), following the manufacturer's protocol.
Mass spectrometry and HMT assay
Histones were obtained using acid extraction, fractionated concurrent with mass spectrometry (MS) profiling, and digested using endoprotease Glu-C, as previously described.25 The Glu-C products were fractionated, and the 1-50 N-terminal H3.1 peptide was selected for direct infusion and MS/MS analysis. K27 methylation levels were estimated using a system of inequalities derived from the K36 and K36+K27 data. The reported values fit the data with the smallest possible error term and were calculated using Wolfram Mathematica Version 7.
ChIP
Chromatin immunoprecipitation (ChIP) was performed according to the manufacture's protocol (ChIP kit, Millipore) with the following modifications: 107 cells were used per sample. After cell lysis, extracts were subjected to sonication with a 60 Sonic Dismembrator (Fisher Scientific) at 4°C for 20 cycles, cycling ON for 20 seconds and OFF for 1 minute at 4.5 power leading to a DNA average size of 300 bp. Immunoprecipitations were performed overnight at 4°C using 5 μg of antibody. Imunoprecipitated chromatin was extracted using QIAquick PCR Purification Kit (QIAGEN). A subset of primers across different genes was selected for validation by qPCR. qPCR was performed using the QuantiTect SYBR Green PCR Kit (QIAGEN) on the Mx3000P Real-Time PCR System (Agilent Technologies). Primer sequences can be found in supplemental Table 5. Enrichment was calculated as a percentage of total input DNA.
Results
Altered MMSET expression promotes changes in global histone methylation patterns and higher chromatin structure
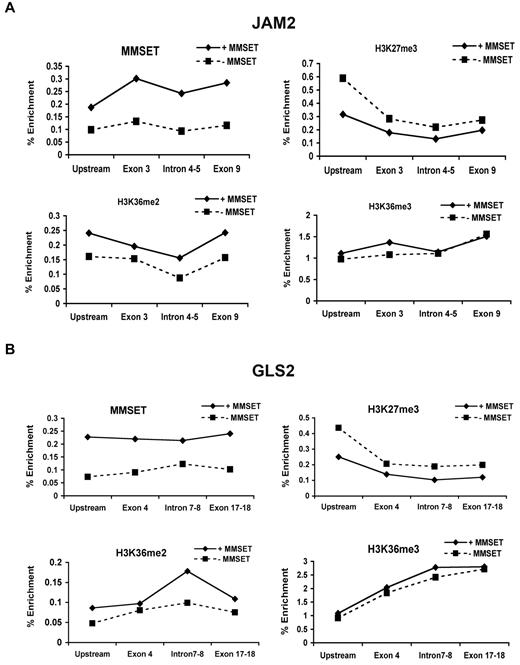
To determine whether changes in MMSET expression have global epigenetic effects, we used 2 different systems in which MMSET expression is lost in a t(4;14)+ multiple myeloma cell line. KMS11 cells were engineered to express doxycycline-inducible shRNAs, specific for various MMSET isoforms (Figure 1A). The 5′ shRNA efficiently knocks down MMSET I and II. The 3′ knockdown affects the SET domain-containing isoforms; however, our antibody does not permit the detection of the putative REIIBP protein (Figure 1A). The second loss-of-function (knockout) system was created by disruption of the overexpressed MMSET allele in KMS11 cells by homologous recombination (Figure 1C).23 The resulting cells express low levels of MMSET from the unrearranged allele, leading to decreased expression of both MMSET I and II (Figure 1C). Nuclear extracts from both systems were analyzed for changes in histone methylation following the loss of MMSET expression. In vitro experiments showed that MMSET can methylate a number of different residues on histones H3 and H413,20 (and data not shown). We did not detect any significant changes in H3K4 and H4K20 methylation (supplemental Figure 1A). However, we did observe a significant decrease of H3K36 dimethylation and trimethylation upon MMSET loss (Figure 1B-C). When MMSET was depleted using the 5′ shRNA, densitometric analysis showed an 80% decrease of H3K36me2 and a 60% decrease of H3K36me3. This was accompanied by a striking increase in H3K27 dimethylation and trimethylation. These changes were independent of the method of histone preparation (enzymatic or acid extraction). Similar results were obtained by interrogation of whole cell extracts using a reverse-phase protein microarray monitoring multiple histone modifications (supplemental Figure 1D). The shRNA directed against the C-terminal region of MMSET was able to increase H3K27me3 levels as well, but it had less effect on H3K36 methylation, leading to a 20% decrease of H3K36me3 and a 40% decrease of H3K36me2 (Figure 1B).
MMSET induces global changes in histone methylation. (A) Schematic representation of MMSET main isoforms showing the regions where the shRNA were designed. (Right) Cells containing the inducible shRNA were grown in the presence of doxycycline for 7 days. After 7 days, cell were washed and incubated for 7 more days without doxycycline to restore MMSET expression. Nuclear extracts were immunoblotted with anti-MMSET antibody. (B-C) Nuclear extracts from the knockdown and knockout systems were immunoblotted with the indicated antibodies. MMSET depletion induces methylation of H3K27me2/3 and decreases methylation of H3K36me2/3. (D) MMSET repletion induces global changes in chromatin. The MMSET knockout cell line was infected with retrovirus containing a vector control (lane 1), wild-type MMSET II (lane 2), or 2 different SET domain mutants (lanes 3-4). Nuclear extracts were immunoblotted with the indicated antibodies. Restoration of wild-type and active mutant of MMSET decreases methylation of H3K27me2/3 and increases methylation of H3K36me2/3. Repletion with the enzymatically dead mutant form of MMSET shows the same methylation pattern as the vector control. (E) MMSET expression affects chromatin structure. MNase assay in knockdown system. Lane 1: molecular weight marker; lanes 2-3: negative control with no enzyme. The presence of MMSET correlates with a more open chromatin state (lane 4). When MMSET is blocked, the chromatin closes, and it is not accessible for the digestion by MNase (lane 5). Re-expression of MMSET upon doxycycline removal opens the chromatin (lane 6).
MMSET induces global changes in histone methylation. (A) Schematic representation of MMSET main isoforms showing the regions where the shRNA were designed. (Right) Cells containing the inducible shRNA were grown in the presence of doxycycline for 7 days. After 7 days, cell were washed and incubated for 7 more days without doxycycline to restore MMSET expression. Nuclear extracts were immunoblotted with anti-MMSET antibody. (B-C) Nuclear extracts from the knockdown and knockout systems were immunoblotted with the indicated antibodies. MMSET depletion induces methylation of H3K27me2/3 and decreases methylation of H3K36me2/3. (D) MMSET repletion induces global changes in chromatin. The MMSET knockout cell line was infected with retrovirus containing a vector control (lane 1), wild-type MMSET II (lane 2), or 2 different SET domain mutants (lanes 3-4). Nuclear extracts were immunoblotted with the indicated antibodies. Restoration of wild-type and active mutant of MMSET decreases methylation of H3K27me2/3 and increases methylation of H3K36me2/3. Repletion with the enzymatically dead mutant form of MMSET shows the same methylation pattern as the vector control. (E) MMSET expression affects chromatin structure. MNase assay in knockdown system. Lane 1: molecular weight marker; lanes 2-3: negative control with no enzyme. The presence of MMSET correlates with a more open chromatin state (lane 4). When MMSET is blocked, the chromatin closes, and it is not accessible for the digestion by MNase (lane 5). Re-expression of MMSET upon doxycycline removal opens the chromatin (lane 6).
To corroborate these findings, we infected the MMSET knockout cells with retroviruses expressing MMSET isoforms. MMSET I, which lacks the SET domain, did not alter K36 or K27 methylation patterns (supplemental Figure 1B). REIIBP, which does contain the SET domain, also failed to modify chromatin, presumably due to its cytoplasmic localization in transduced KMS11 cells (supplemental Figure 1C). By contrast, re-expression of full-length MMSET II reversed the K36/K27 methylation pattern, with K36 becoming highly dimethylated and trimethylated, and K27 accumulating in the monomethyl state (Figure 1D and supplemental Figure 1D). Introduction of a mutation in the SET domain (Y1118A), which is expected to interfere with histone binding properties,13 prevented MMSET from altering histone modifications (Figure 1D and supplemental Figure 1D). However, a mutation predicted to expand the lysine-binding pocket of the SET (F1177A) domain showed a gain-of-function phenotype. When cells were transduced with this protein, most of the K36 was converted to a trimethylated state (Figure 1D and supplemental Figure 1D). In addition, we detected a modest but consistent increase in H3K27 acetylation in the presence of MMSET, possibly due to lower K27 methylation levels (Figure 1B-D). Collectively, these data show that the high levels of MMSET seen in multiple myeloma cause global changes in H3K36 and H3K27 methylation.
The unexpected global changes in histone modification in the KMS11 cells suggest that MMSET may affect chromatin structure. H3K27 methylation is associated with a more compact, repressed chromatin state, and its loss from histones may lead to global changes in the chromatin structure.26 To address this question, we performed a MNase accessibility assay24,27 on MMSET shRNA-inducible KMS11 cells (Figure 1E). Treatment of live and permeabilized cells with MNase led to a nucleosomal ladder of DNA fragments, suggesting an open chromatin structure. However, in the presence of doxycycline and loss of MMSET expression, there was a striking loss of MNase-mediated cutting of the chromatin, indicating a more closed structural state. This result is reversible, as removal of the doxycycline and re-expression of MMSET restores MNase sensitivity.
The H3K36/K27 methylation switch caused by MMSET is specific for t(4;14) myeloma cells
To investigate the potential clinical relevance of MMSET-associated chromatin changes, we investigated the global histone modification patterns in a panel of 9 t(4;14)+ and 7 t(4;14)− cell lines. All the t(4;14)+ cells expressed high levels of MMSET and H3K36me2 and low levels of H3K27me3. By contrast, t(4;14)− cells displayed low levels of H3K36 dimethylation and high H3K27 trimethylation (Figure 2A and supplemental Figure 2A). Other histone modifications, including H3K36me3, did not show consistent differences between the 2 groups of cells (supplemental Figure 2A-B).
t(4;14)-positive cells have a different histone methylation pattern than t(4;14)-negative cells. (A) Nuclear extracts from 9 t(4;14)-positive cells and 7 t(4;14)-negative cells were immunoblotted with the indicated antibodies. MMSET expression correlates with higher H3K36me2 and lower H3K27me3 marks. (B) Intact mass profiles of histones H3.1, H3.2, and H3.3 from 4 multiple myeloma cell lines. H929 and LP1 are t(4;14)+ and show a global increase in H3.1 and H3.2 methylation compared with the t(4;14)− KMS12 and MM.M1 cells. The [M+18H]18+ species are shown. Distinct peaks correspond to different numbers of methyl or acetyl groups (14 and 42 Da). The 6 methyl equivalent is labeled for each histone variant. (C) K36 methylation levels were determined from y174+, y184+, and y193+ from collision-induced dissociation (CID) of the N-terminal 1-50 [M+9H]9+ peptide of H3.1. (D) K36 dimethylation is reported on by y193+ ions (717.42 m/z) from CID of the N-terminal 1-50 [M+9H]9+ peptide of H3.1. Peak intensity (arbitrary units) is shown in the top right corner for each mass spectrum. (E) K27 methylation levels were determined from y244+, y274+, y356+, and y366+ for K36+K27 from CID of the N-terminal 1-50 [M+9H]9+ peptide of H3.1. Values for K27 methylation were deconvoluted from K36+K27 and K36 data using a system of inequalities (Mathematica). The methylation levels, with SEM, are shown in supplemental Table 1.
t(4;14)-positive cells have a different histone methylation pattern than t(4;14)-negative cells. (A) Nuclear extracts from 9 t(4;14)-positive cells and 7 t(4;14)-negative cells were immunoblotted with the indicated antibodies. MMSET expression correlates with higher H3K36me2 and lower H3K27me3 marks. (B) Intact mass profiles of histones H3.1, H3.2, and H3.3 from 4 multiple myeloma cell lines. H929 and LP1 are t(4;14)+ and show a global increase in H3.1 and H3.2 methylation compared with the t(4;14)− KMS12 and MM.M1 cells. The [M+18H]18+ species are shown. Distinct peaks correspond to different numbers of methyl or acetyl groups (14 and 42 Da). The 6 methyl equivalent is labeled for each histone variant. (C) K36 methylation levels were determined from y174+, y184+, and y193+ from collision-induced dissociation (CID) of the N-terminal 1-50 [M+9H]9+ peptide of H3.1. (D) K36 dimethylation is reported on by y193+ ions (717.42 m/z) from CID of the N-terminal 1-50 [M+9H]9+ peptide of H3.1. Peak intensity (arbitrary units) is shown in the top right corner for each mass spectrum. (E) K27 methylation levels were determined from y244+, y274+, y356+, and y366+ for K36+K27 from CID of the N-terminal 1-50 [M+9H]9+ peptide of H3.1. Values for K27 methylation were deconvoluted from K36+K27 and K36 data using a system of inequalities (Mathematica). The methylation levels, with SEM, are shown in supplemental Table 1.
Immunoblot analysis of histone modifications relies on the specificity and sensitivity of the antibody and is best at indicating relative and not absolute abundance of histone modifications under various conditions. MS on the other hand can differentiate unequivocally between different methylation states and can provide a detailed overview of histone modification profiles.25,28,29 We acquired mass spectrometric profiles of intact histones and carried out fragmentation of the N-terminal 1-50 H3 peptide to determine the site-specific histone modifications that occur on histone H3 in 2 t(4;14)-positive and 2 negative cell lines. Histones H3.1 and H3.2 from the t(4;14)+ cells showed an overall shift toward the heavier more methylated state in comparison to the histones from myeloma cells without the translocation (Figure 2B). This shift occurs on histones H3.1 and H3.2 but not on H3.3. Fragment ions containing lysine 36 showed that this residue is most commonly found in the unmethylated state in MMSET low cells (Figure 2C-D). By contrast, in the presence of pathological levels of MMSET, there is a dramatic increase in H3K36 dimethylation, with 60%-80% of H3K36 present in this state. There was a 2-3-fold increase in H3K36 monomethylation and trimethylation state as well, but the absolute amount of these modifications is much lower than H3K36me2. In t(4;14)-negative MMSET low cells, H3K27 was predominantly found in the dimethylated and trimethylated state, while in MMSET high t(4;14)-positive cells, there was a marked drop in H3K27 trimethylation and an accumulation in H3K27 monomethyl state (Figure 2E). Notably, there were few histone fragments that contained both K36me2 and K27me3, indicating that these modifications are not commonly found together (supplemental Table 3). All of these results were confirmed using the MMSET knockout system (supplemental Figure 3A-C and supplemental Table 4).
Loss of MMSET expression affects cell growth and apoptosis
Using the inducible shRNA system, we evaluated the effect of MMSET knockdown on cell cycle progression and proliferation. Loss of MMSET considerably inhibited the growth of KMS11 cells, particularly when the 3′ portion of the mRNA was targeted (Figure 3A). Knockdown of MMSET I and II with a shRNA targeted to the 5′ region of the mRNA that encodes both isoforms consistently led to increased cell adhesion, with cells growing in suspension changing shape and sticking to the tissue culture dish (Figure 3B). One of the inducible cell lines containing the 5′ shRNA also showed growth inhibition upon depletion of MMSET. Knockdown using the 3′ shRNA did not alter adhesion properties but significantly increased apoptosis of KMS11 myeloma cells (Figure 3C). Furthermore, MMSET knockdown using both 5′ and 3′ terminal shRNA led to decreased cell entry into S-phase as demonstrated by BrdU incorporation, with this effect more pronounced with the 3′ shRNA (Figure 3D). The differences in the phenotypes observed between the 2 shRNAs might be due to the persistence of MMSET isoforms unaffected by the different shRNA constructs: the 5′ shRNA leaving the REIIBP isoform intact and the 3' shRNA leaving MMSET I expression unchanged. We complimented these loss-of-function systems with a gain-of-function system, infecting KMS11 knockout cells with retroviruses expressing wild-type MMSET or forms of MMSET harboring mutations in the SET domain. Both the wild-type MMSET and the F1177A mutant, which retains HMT activity, stimulated growth of myeloma cells compared with the vector control. In distinct contrast, the HMT inactive Y1118A mutant failed to alter KMS11 growth (Figure 3E). Together, these data indicate that MMSET affects proliferation, apoptosis, and adhesion properties in multiple myeloma cells and that HMT activity of MMSET is critical for cell growth.
MMSET alters cell growth, cell cycle, and apoptosis in multiple myeloma cells. (A) MMSET affects growth of multiple myeloma cells. Cloned KMS11 cells harboring inducible shRNA were grown in the presence of doxycycline, and cells were counted at the indicated time points. Fresh media with or without doxycycline was added at days 7, 10, and 13 to allow continued cell growth. Two different clones are shown in the graphs. Knockdown using the C-terminal shRNA shows a drastic change in growth behavior. (B) MMSET alters adhesion of multiple myeloma cells. Knockdown cells where grown in the presence of doxycycline for 7 days, and pictures were imaged as described in “Adhesion assay.” (C) MMSET depletion induces apoptosis. Knockdown cells were grown in the presence of doxycycline, and annexin V staining was performed as described in “Cell cycle analysis and apoptosis assay.” (D) MMSET induces cell cycle arrest. Cell-cycle profiles of knockdown cells grown in the presence of doxycycline for 7 days were assessed after a 30-minute BrdU pulse as described in “Cell cycle analysis and apoptosis assay.” MMSET knockdown decreases the percentage of cells in S-phase, and this effect is more pronounced using the C-terminal shRNA. (E) MMSET induces proliferation. Knockout cells infected with vector control, wild-type, and mutants of MMSET were grown, and cell counts were performed over a period of 2 weeks. Cells containing wild-type and active mutant (F1177A) of MMSET proliferate faster than cells infected with vector control or the enzymatically dead MMSET (Y1118A).
MMSET alters cell growth, cell cycle, and apoptosis in multiple myeloma cells. (A) MMSET affects growth of multiple myeloma cells. Cloned KMS11 cells harboring inducible shRNA were grown in the presence of doxycycline, and cells were counted at the indicated time points. Fresh media with or without doxycycline was added at days 7, 10, and 13 to allow continued cell growth. Two different clones are shown in the graphs. Knockdown using the C-terminal shRNA shows a drastic change in growth behavior. (B) MMSET alters adhesion of multiple myeloma cells. Knockdown cells where grown in the presence of doxycycline for 7 days, and pictures were imaged as described in “Adhesion assay.” (C) MMSET depletion induces apoptosis. Knockdown cells were grown in the presence of doxycycline, and annexin V staining was performed as described in “Cell cycle analysis and apoptosis assay.” (D) MMSET induces cell cycle arrest. Cell-cycle profiles of knockdown cells grown in the presence of doxycycline for 7 days were assessed after a 30-minute BrdU pulse as described in “Cell cycle analysis and apoptosis assay.” MMSET knockdown decreases the percentage of cells in S-phase, and this effect is more pronounced using the C-terminal shRNA. (E) MMSET induces proliferation. Knockout cells infected with vector control, wild-type, and mutants of MMSET were grown, and cell counts were performed over a period of 2 weeks. Cells containing wild-type and active mutant (F1177A) of MMSET proliferate faster than cells infected with vector control or the enzymatically dead MMSET (Y1118A).
MMSET regulates expression of genes involved in cell cycle, apoptosis, DNA repair, and adhesion
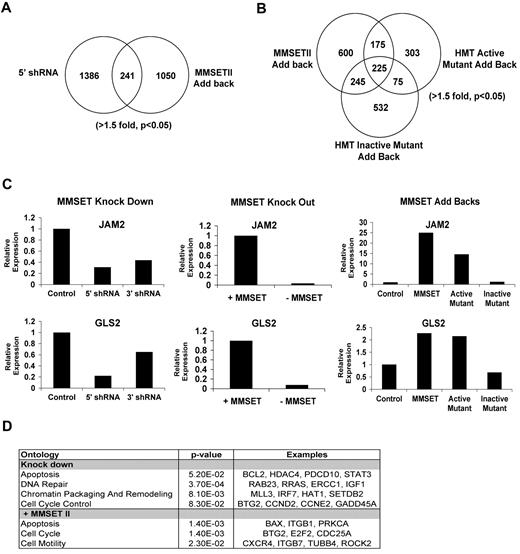
To determine the genes regulated by MMSET and the importance of histone methylation in MMSET action, we profiled gene expression in both the loss- and gain-of-function systems (Figure 4A-D). The N-terminal MMSET knockdown affected expression of 619 genes (fold change [FC] > 1.5, P < .05); 342 were up-regulated, and 279 had reduced expression levels. Re-expression of wild-type MMSET II in the knockout cell line led to increased expression of 677 genes and down-regulation of 652 genes. There were 241 genes regulated in both the loss- and gain-of-function systems (Figure 4A), with 88 of the genes showing reciprocal regulation in between the 2 systems (supplemental Table 5A). Of those, 31 genes were suppressed by MMSET expression, and 57 genes were activated by MMSET (supplemental Table 5A). The overlap of genes in the gain-of-function and loss-of-function lists generated by microarray analysis may represent excellent candidates of direct targets of MMSET. However, analysis of a group of genes identified by microarray analysis in only one of the systems shows that indeed many of these genes were regulated in both systems when validated by qRT-PCR (Figure 4C and supplemental Table 5B). Therefore, we analyzed the pathways implicated by genes regulated by MMSET in either the knockdown or repletion systems, using DAVID Ontology and Ingenuity Pathway Analysis software, and found enrichment of genes implicated in the regulation of cell death and the p53 pathway (eg, BAX, BCL2, and Caspase 6), the cell cycle (Cyclin E2, E2F2, TP53INP1, and CDC25A), genes for DNA repair (ATM, E2F2, and GADD45A), and integrin-mediated signaling (CDC42 and Integrin alpha-L; Figure 4D and data not shown). The effect of MMSET on integrin-related genes is of interest given that KMS11 cells, which grow in suspension, attach to the plastic when MMSET is depleted. The KMS11 knockout cell line, which does not adhere to plastic, becomes adherent when MMSET is added back (Figure 3B and supplemental Figure 4). Collectively, these data suggest that MMSET affects adhesion properties in multiple myeloma cells.
Identification of MMSET target genes by gene expression array. (A) Comparison of gene expression dataset from N-terminal knock down and MMSET II add back (t test paired; > 1.5-fold, P < .05). (B) Comparison of gene expression dataset from knockout cell line repleted with wild-type, active mutant, or dead mutant forms of MMSET (> 1.5-fold, P < .05). (C) Validation of MMSET target genes by real-time PCR. A representative experiment showing the regulation of JAM2 and GLS2 genes in the presence or absence of MMSET using the knockdown, knockout, and repletion systems. (D) Classification of MMSET target genes by Ontology analysis was performed using DAVID Ontology and Ingenuity Pathway Analysis. MMSET regulates genes mainly related with apoptosis, cell cycle, and DNA repair.
Identification of MMSET target genes by gene expression array. (A) Comparison of gene expression dataset from N-terminal knock down and MMSET II add back (t test paired; > 1.5-fold, P < .05). (B) Comparison of gene expression dataset from knockout cell line repleted with wild-type, active mutant, or dead mutant forms of MMSET (> 1.5-fold, P < .05). (C) Validation of MMSET target genes by real-time PCR. A representative experiment showing the regulation of JAM2 and GLS2 genes in the presence or absence of MMSET using the knockdown, knockout, and repletion systems. (D) Classification of MMSET target genes by Ontology analysis was performed using DAVID Ontology and Ingenuity Pathway Analysis. MMSET regulates genes mainly related with apoptosis, cell cycle, and DNA repair.
JAM2 and GLS2 were examples of genes regulated by MMSET. There was a decrease in the expression of JAM2 and GLS2 when MMSET was depleted using the knockdown and the knockout systems. Conversely, both genes were up-regulated when KMS11 knockout cells were repleted with wild-type MMSET and the active mutant F1177A but not when we overexpressed the HMT inactive Y1118A mutant (Figure 4C). This suggests that the HMT activity of MMSET was important for activation of those genes.
We extended the microarray analysis by comparison of gene expression changes after re-expression of wild-type MMSET and the catalytically inactive MMSET mutant (Y1118A). Strikingly, MMSET Y1118A, which failed to alter cell adhesion (supplemental Figure 4) and did not change bulk histone methylation (Figure 1D), still altered expression of 1077 genes, 44% overlapping with those regulated by wild-type MMSET (Figure 4B). When KMS11 cells were repleted with wild-type MMSET or the hyperactive F1117A MMSET mutant, cell cycle (E2F2, CDC25A, and ATM), adhesion (junctional adhesion molecule 3, Integrin, beta 1, and NRCAM), and p53 pathways were predominantly regulated (Figure 4D and data not shown). Comparing changes in gene expression mediated by wild-type versus mutant MMSET Y1118A showed that genes activated only by wild-type MMSET were stimulated 5-fold, while genes stimulated only by mutant MMSET Y1118A were activated by a factor of 2 (supplemental Figure 5A). Gene repression by wild-type and the Y1118A mutant was similar with genes inhibited by an average of 50% (supplemental Figure 5A-B). When the genes regulated in common by wild-type and mutant MMSET were compared, there was a correlation between gene regulation by the mutant and wild-type protein (supplemental Figure 5D), but wild-type MMSET with HMT activity intact activated this set of genes by an average factor of 4.1 and mutant MMSET affected gene expression by a factor of 2.7 (supplemental Figure 5C). Again, gene repression by the wild-type and mutant proteins was similar. These data suggest that gene activation by MMSET is closely linked to HMT activity.
Genes regulated only by the HMT inactive mutant (Y1118A) but not wild-type MMSET or the active mutant (F1177A) were enriched only in pathways such as biosynthesis of steroids or fructose metabolism. We also observed that CCND2, essential for the control of cell cycle, was up-regulated by wild-type MMSET but not the inactive mutant Y1118A, which may help explain the stimulated growth by only wild-type MMSET (Figure 3E). These results highlight the critical role of HMT activity of MMSET in multiple myeloma cell growth and link gene activation by MMSET with the control of cell growth.
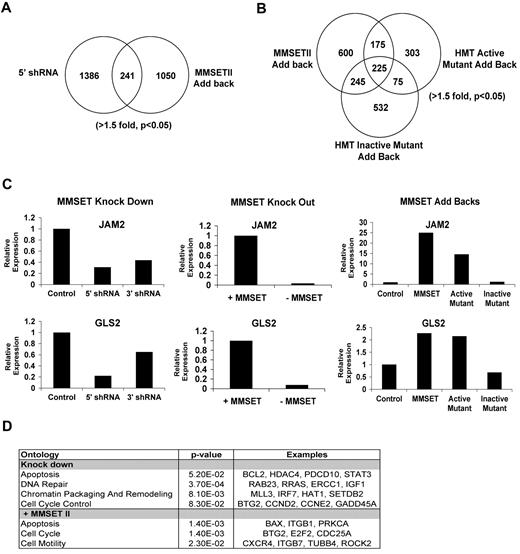
Genes bound by MMSET contain higher levels of H3K36 and lower levels of H3K27 methylation
As overexpression of MMSET led to global changes in histone modifications, we next determined how MMSET might regulate individual gene loci. To this end, we determined the state of chromatin of genes activated in the presence of overexpressed MMSET such as JAM2, a gene implicated in cellular adhesion, and GLS2, a p53-inducible regulator of glutamine metabolism and reactive oxygen species levels.30,31 ChIP using primers directed to widely spaced introns and exons of these genes shows MMSET binding at multiple points across these loci in KMS11 cells. MMSET binding was markedly decreased in KMS11 knockout (supplemental Figure 6A-B) and knockdown cells (Figure 5A-B). Depletion of MMSET binding to these loci was associated with a decrease in K36 dimethylation in knockdown cells and a decrease in K36 dimethylation and trimethylation in knockout cells. Accordingly, KMS11 knockout and knockdown cells showed increased H3K27 trimethylation in the promoter regions of the target genes and no significant change within the body of these genes (Figure 5A-B and supplemental Figure 6A-B). This is not true for every examined locus. For example, HOXC9, whose expression does not change in response to MMSET, is highly enriched by H3K27me3 and is not bound by MMSET (data not shown). Therefore, high level of expression of MMSET as found in t(4;14) myeloma cells affects global chromatin changes; however, only a subset of genes show significant alteration in expression.
MMSET binding across genes overlaps with high levels of H3K36me2 and low levels of H3K27me3. Chromatin of MMSET shRNA-inducible cells grown with or without doxycycline for 7 days was immunoprecipitated with anti-MMSET, anti-H3K27me3, anti-H3K36me2, or anti-H3K36me3 antibodies. Immunoglobulin G was used as negative control (not shown). Eluted DNA was PCR amplified with primers designed across JAM2 (A) and GLS2 (B) loci.
MMSET binding across genes overlaps with high levels of H3K36me2 and low levels of H3K27me3. Chromatin of MMSET shRNA-inducible cells grown with or without doxycycline for 7 days was immunoprecipitated with anti-MMSET, anti-H3K27me3, anti-H3K36me2, or anti-H3K36me3 antibodies. Immunoglobulin G was used as negative control (not shown). Eluted DNA was PCR amplified with primers designed across JAM2 (A) and GLS2 (B) loci.
Discussion
A growing amount of evidence suggests that a shift in histone methylation balance of a cell plays an important role in carcinogenesis. For example, chromosomal translocations of MLL, an H3K4 methyltransferase, lead to development of acute leukemias through recruitment of H3K79 methyl transferase Dot1.32-34 Overexpression of EZH2, an H3K27 methyltransferase, is found in prostate, breast cancer, and gastric cancer, while EZH2 mutations were recently identified in lymphoma.35-37 Inactivation of the SETD2 methyltransferase has been reported in renal carcinoma in association with loss of H3K36 trimethylation.38 In addition, inactivating mutations of histone demethylases, such as UTX, have also been implicated in oncogenesis.39,40
Elevated expression of a histone methyltransferase may alter normal cellular processes in a number of different ways. One possibility is that the high levels of the protein enhance binding and histone methylation of physiological target genes. Another possibility is that elevated expression of a histone methyltransferase allows for promiscuous interactions of the protein with chromatin, leading to changes in histone modifications on a novel set of target genes. Alternatively, increased amount of the HMT protein may sequester important cofactors, preventing their normal molecular function. As another possibility, the HMT could methylate cellular proteins, altering their biological activity. Finally, high levels of a HMT may have global effects on chromatin methylation and physical properties. Among these possibilities, our data suggest that at least one operant mechanism in cases of MMSET involves global changes in chromatin structure, accessibility, and function.
In this report, we show that overexpressed MMSET protein causes global changes in the chromatin modification state of t(4;14)+ myeloma cells. High levels of MMSET induce an increase in H3K36 dimethylation and trimethylation, which can be reversed upon MMSET knockdown. Surprisingly, cells overexpressing MMSET show a great reduction of H3K27 methylation, usually associated with gene repression. The MMSET-mediated increase in H3K36 methylation and concomitant loss of H3K27 methylation yields 2 chromatin marks favoring gene activation. While H3K36 methylation is dependent on the SET domain of MMSET, the mechanism by which overexpression of MMSET leads to loss of H3K27 methylation is uncertain. Previous reports41 and our own data indicate that H3K36me2 and H3K27me3 rarely exist on the same molecule, thus suggesting that the mere presence of abnormal amount of H3K36 methylation prevents K27 methylation from occurring. Alternatively, MMSET itself, or through an indirect mechanism, may sequester Polycomb proteins away from the DNA, thus leading to the loss of K27 methylation. Our previous report showed that MMSET can interact with LSD1, an H3K4 demethylase.13 In a similar manner, MMSET may be regulating expression or activity of H3K27 demethylases UTX and JMJD3; however, our microarray studies did not show any significant changes on the mRNA level of those proteins. Our group and others previously reported that MMSET can methylate histone H4 in vitro.13 While the presence of MMSET correlates consistently only with high levels of H3K36 and low levels of H3K27 methylation, we found that knockout cells repleted with MMSET have a slight increase in H4K20 methylation (supplemental Figure 1D). However, in a panel of t(4;14)+ cell lines, H4K20 methylation does not correlate with high MMSET expression (supplemental Figure 2B), suggesting that while MMSET may modulate H4K20 under some circumstances, methylation of this residue is not its primary biological function in multiple myeloma.
Despite the known genetic heterogeneity in myeloma cell lines (eg, immunoglobulin heavy chain-MYC translocations,42 c-MAF translocations,43,44 UTX mutations, J. Keats and P. L. Bergsagel, Mayo Clinic, written communication, August 25, 2010), the pattern of H3K36 and H3K27 methylation correlates perfectly with MMSET expression across the panel of t(4;14)+ and t(4;14)− cells. This is significant, because differences in chromosomal translocation breakpoints generate MMSET proteins of varying N-terminal sequences, suggesting that the absence of some domains of MMSET such as the N-terminal PWWP domain is not critical for the oncogenic action of MMSET. To our knowledge, no previous studies showed that simple overexpression of a histone methyltransferase had such a pronounced effect on histone modifications. NSD1, another family member, has been implicated in leukemogenesis through generation of a NUP98-NSD1 fusion protein.8,45 However, the oncogenic ability of this protein is possibly related to a new function of the fusion protein, rather that the higher levels of expression.
MMSET overexpression induced methylation changes on histone H3 variants H3.1 and H3.2 and did not affect methylation of the H3.3 variant. Histone H3.3 contains a serine at position 31 while H3.1/2 variants contain alanine.46 The proximity of serine 31 to lysine 36 in H3.3, particularly if the serine is phosphorylated, might prevent MMSET from properly interacting with or methylating H3.3 tails. Histone variant distribution across the genome is nonrandom. A recent study has shown that on a chromosomal scale map, H3.3 enrichment correlates with markers of active transcription (RNA polymerase II and H3K4me3).47 Similarly, MMSET distribution across the genome may not be uniform, which could explain why, despite global changes in histone methylation, MMSET only regulates a subset of genes. Further studies are needed to address this issue.
Loss of MMSET expression in a nonadhesive t(4;14)+ cell line lead to increased adhesion, slower cell growth, and induction of apoptosis. Many of the genes identified in both the gain- and loss-of-function microarray experiments fall into these categories. It is notable that MMSET depletion using the 3′ shRNA has a more pronounced effect on apoptosis. This shRNA is predicted to affect the REIIBP isoform not targeted by the 5′ shRNA. However, repletion of MMSET knockout cells with REIIBP failed to affect histone methylation (supplemental Figure 1), perhaps due to its cytoplasmic localization. However, it is possible that REIIBP plays a role in the cytoplasm by methylating other proteins and that the loss of REIIBP expression plays a significant role in induction of apoptosis. While MMSET I, which lacks the SET domain, is not found in all t(4;14)+ cells, KMS11 expresses high levels of both MMSET I and II (Figure 2A). The 3′ shRNA depletes only MMSET II, suggesting that persistence of MMSET I on its own may stimulate apoptosis. Loss of MMSET I and II together leads to compaction of chromatin, decreasing its accessibility. This may be partially explained by the global increase in H3K27 methylation, which is normally associated with repressed, more tightly wound chromatin.26 Further studies will evaluate whether chromosomes undergo gross physical changes in response to MMSET expression.
H3K36 methylation has been associated with actively transcribed genes, with higher enrichment of trimethylation at exon-intron boundaries and at the 3′ end of genes.48 In addition, trimethylation of H3K36 seems to correlate with the gene expression level as well as with gene length, with highly expressed and longer genes having the highest levels of this modification.49 It is possible that high levels of H3K36 methylation induced by overexpression of MMSET may lead to expression of certain alternatively spliced forms of genes over others. While much research has focused on H3K36me3, our data indicate that this modification is, on a molar basis, rare, and that most of the H3K36 in myeloma cells is found unmethylated in low MMSET cells or dimethylated in the presence of MMSET. Comparison of gene expression changes mediated by wild-type versus HMT inactive MMSET showed that while the extent of gene repression mediated by the 2 proteins was similar, gene activation was inhibited by the loss of HMT activity. HMT-inactive MMSET failed to mediate increased H3K36 dimethylation or trimethylation, failed to stimulate cell growth, and failed to activate CCDN2 and other genes related to cell cycle control. Collectively, this suggests that oncogenesis by MMSET is related to activation of gene expression. The mode by which increased H3K36 dimethylation activates gene expression remains to be fully understood.
Our study has identified pathological levels of MMSET as an important regulator of histone methylation and gene expression in t(4;14)+ multiple myeloma cells. The histone methyl transferase activity of MMSET appears critical for its biological function in promoting malignant cell growth. While the mechanism of the H3K36me2-H3K27me3 switch is not completely understood, our findings suggest that the inhibition of MMSET may be of benefit to patients with t(4;14) translocations. Furthermore, analysis of microarray datasets indicated that high MMSET levels are also found in advanced forms of solid tumors such as colon, lung, or prostate, implicating MMSET as a more broadly relevant therapeutic target.50
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Michael Kuehl for providing multiple myeloma cells and Josh Lauring for providing MMSET knockout cells, Russell Bandle and Hye-Jung Chung for protein array expertise, and Ari Melnick and Thomas Milne for critical reading of the manuscript.
This work was supported by a Multiple Myeloma Research Foundation (MMRF) award, a European Hematology Association fellowship, a fellowship from the Government of Navarra, Spain (E.M.-G.), Ruth Kirschstein National Research Service Awards T32CA070085 and F32HL099177 (R.P.), RO1CA123204, and a Leukemia & Lymphoma Society Specialized Center for Research Award and a Physical Sciences Oncology Center Grant 1U54CA143869-01 (J.D.L.).
National Institutes of Health
Authorship
Contribution: E.M.-G. and R.P. designed and performed experiments, analyzed data, and wrote the manuscript; D.-J.M. designed experiments and analyzed data; S.M.M.S., P.M.T., L.Z., A.H., and C.W. performed experiments and analyzed data; L.L. and L.M.S., designed and provided reagents; D.L.L. and N.L.K. designed experiments; and J.D.L. designed the project, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jonathan D. Licht, MD, Division of Hematology/Oncology, Northwestern University, Feinberg School of Medicine, Lurie 5-123, 303 E Superior St, Chicago, IL 60611; e-mail: j-licht@northwestern.edu.
References
Author notes
E.M.-G. and R.P. contributed equally to this manuscript.


![Figure 2. t(4;14)-positive cells have a different histone methylation pattern than t(4;14)-negative cells. (A) Nuclear extracts from 9 t(4;14)-positive cells and 7 t(4;14)-negative cells were immunoblotted with the indicated antibodies. MMSET expression correlates with higher H3K36me2 and lower H3K27me3 marks. (B) Intact mass profiles of histones H3.1, H3.2, and H3.3 from 4 multiple myeloma cell lines. H929 and LP1 are t(4;14)+ and show a global increase in H3.1 and H3.2 methylation compared with the t(4;14)− KMS12 and MM.M1 cells. The [M+18H]18+ species are shown. Distinct peaks correspond to different numbers of methyl or acetyl groups (14 and 42 Da). The 6 methyl equivalent is labeled for each histone variant. (C) K36 methylation levels were determined from y174+, y184+, and y193+ from collision-induced dissociation (CID) of the N-terminal 1-50 [M+9H]9+ peptide of H3.1. (D) K36 dimethylation is reported on by y193+ ions (717.42 m/z) from CID of the N-terminal 1-50 [M+9H]9+ peptide of H3.1. Peak intensity (arbitrary units) is shown in the top right corner for each mass spectrum. (E) K27 methylation levels were determined from y244+, y274+, y356+, and y366+ for K36+K27 from CID of the N-terminal 1-50 [M+9H]9+ peptide of H3.1. Values for K27 methylation were deconvoluted from K36+K27 and K36 data using a system of inequalities (Mathematica). The methylation levels, with SEM, are shown in supplemental Table 1.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/1/10.1182_blood-2010-07-298349/4/m_zh89991064330002.jpeg?Expires=1766013393&Signature=N7SN1WeL6Mjb1AZsM7T-02JTlKQIj5Eb8rZAlGSdsX03GTy0TqtsrVGSCSWw-1KG8-dM86lq4YUFqif02NHkK3mmtvzthR9q9cPT1GKUgI9BY2HIhVXjI6Sieg0qgXOjSJABVWmXmbAxKJEgres3mQ6lzmQCshoU2t0vargC1BoHtQABDcYPBhD4iw~ZDj7r-Ej0CMLTZ~NOtJwuYwlGZgSkMa1D4Frz~WaRJSZpXuqatw5-XfdjSI99mHHRZdKDLPFN5och9AqmUzjHLFMRFTW2US2zmfIcSfwvYzcrtdDp5g74RP5sgmZonqUAF4c5Vy7tRJCRrjigD31WUnQdoQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 2. t(4;14)-positive cells have a different histone methylation pattern than t(4;14)-negative cells. (A) Nuclear extracts from 9 t(4;14)-positive cells and 7 t(4;14)-negative cells were immunoblotted with the indicated antibodies. MMSET expression correlates with higher H3K36me2 and lower H3K27me3 marks. (B) Intact mass profiles of histones H3.1, H3.2, and H3.3 from 4 multiple myeloma cell lines. H929 and LP1 are t(4;14)+ and show a global increase in H3.1 and H3.2 methylation compared with the t(4;14)− KMS12 and MM.M1 cells. The [M+18H]18+ species are shown. Distinct peaks correspond to different numbers of methyl or acetyl groups (14 and 42 Da). The 6 methyl equivalent is labeled for each histone variant. (C) K36 methylation levels were determined from y174+, y184+, and y193+ from collision-induced dissociation (CID) of the N-terminal 1-50 [M+9H]9+ peptide of H3.1. (D) K36 dimethylation is reported on by y193+ ions (717.42 m/z) from CID of the N-terminal 1-50 [M+9H]9+ peptide of H3.1. Peak intensity (arbitrary units) is shown in the top right corner for each mass spectrum. (E) K27 methylation levels were determined from y244+, y274+, y356+, and y366+ for K36+K27 from CID of the N-terminal 1-50 [M+9H]9+ peptide of H3.1. Values for K27 methylation were deconvoluted from K36+K27 and K36 data using a system of inequalities (Mathematica). The methylation levels, with SEM, are shown in supplemental Table 1.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/1/10.1182_blood-2010-07-298349/4/m_zh89991064330002.jpeg?Expires=1766118620&Signature=U4o1pCNKOca-9CBoYFwxAisS-ut5knTxvLdKenfecSe77O2QEoEiEmM9O-qAIL3GVIr4qGY6sVBrnjKcImG0Aa3uCT147kSlV-eEDG2A3YJ-weJfnKM7447m6LtOY3WSQxvXvlscVpxiXW-NeoJJJpeNqum5jwWxVr3~FYn5MKtAt1P1b~avaQon7QwRh-Bo-yTR7vRsCC6zfcxD6XWmapOTfJY-iaR2p~HEPEUzkgg5DCjd9fiNkZS2NEhT4CaYtsPI7p5C8-xTLOK122HgRESiozoui-mG1WoBPn2usboNQhtxlmxFeym2nshuT6ERUrqY9ASuzXlLnTcxztV~Aw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)