Abstract
Elevated levels of microRNA miR-155 represent a candidate pathogenic factor in chronic B-lymphocytic leukemia (B-CLL). In this study, we present evidence that MYB (v-myb myeloblastosis viral oncogene homolog) is overexpressed in a subset of B-CLL patients. MYB physically associates with the promoter of miR-155 host gene (MIR155HG, also known as BIC, B-cell integration cluster) and stimulates its transcription. This coincides with the hypermethylated histone H3K4 residue and spread hyperacetylation of H3K9 at MIR155HG promoter. Our data provide evidence of oncogenic activities of MYB in B-CLL that include its stimulatory role in MIR155HG transcription.
Introduction
Transcriptional regulation of major hematopoietic oncogenes and tumor suppressor genes represents a critical step in tumor formation, tumor aggressiveness, and therapy resistance.1-3 In addition, a posttranscriptional inhibitory mechanism involving microRNA (miRNA) binding to the 3′-untranslated region of target mRNAs causes transcript degradation or interferes with the translation initiation and has been linked to tumorigenesis.4,5 Under physiologic conditions, miRNAs regulate developmental processes and cell fate decisions, and tight regulation of their levels represents an important factor in cell and tissue homeostasis.6 MiR-155, a well-studied miRNA, regulates hematopoietic cell development as documented by murine gene targeting experiments and also by other studies describing its function during immune B- and T-cell response, in production of cytokines and antibodies and in antigen presentation.7,8 Next, transgenic miR-155 overexpression in the mouse stimulates B-cell proliferation and frequent development of lymphomas.9 In humans, miR-155 up-regulation has been repeatedly reported in chronic B-cell lymphocytic leukemia (B-CLL), in its solid indolent form of a small lymphocytic lymphoma10-12 and also in aggressive types, including non-Hodgkin10,13,14 and Hodgkin lymphomas.13,15 Deregulation of several microRNAs was repeatedly described in B-CLL.16,17 B-CLL, the most common adult leukemia, is characterized by clonal accumulation of B cell–like mature-appearing elements (> 5000/μL)18 typically coexpressing the CD5, CD19, CD20, and CD23 surface markers. B-CLL represents a heterogeneous disease, the outcome of which may be predicted by the levels of surface protein CD38, intracellular tyrosine kinase ZAP70, or by a status of IgVH somatic hypermutation.18,19 Cytogenetic alterations of 2 loci that contain the p53 gene (deletion of 17p) and the ATM gene (deletion of 11q) are associated with poor prognosis, shorter duration of remission, and shortest overall survival,20 whereas normal karyotype or trisomy 12 is considered intermediate risk and the 13q14 deletion is considered a favorable mark. Subsets of B-CLL patients may progress to non-Hodgkin diffuse large B-cell lymphoma by a mechanism that remains largely unknown. Taken together, miR-155 appears to play a central role in B-cell function, and its up-regulation in lymphoproliferative disorders, including B-CLL, may lead to a block of differentiation and accumulation of lymphoid-like cells.
Recent studies brought evidence of a context-dependent transcriptional regulation of the MIR155HG. First, oncogenic properties of miR-155 have been demonstrated in breast cancer cells where MIR155HG is up-regulated by transforming growth factor-β/Smad pathway involving a Smad response element at the position −454 nt from the transcription start site (TSS).21 This regulatory pathway becomes disabled on inhibition of miR-155, resulting in derepression of miR-155 targets (including the RhoA protein) and in decreased cell migration and invasion.21 Second, the MIR155HG promoter region containing a conserved AP-1 site at position −40 nt appears critical for B-cell activation, as demonstrated in Epstein-Barr virus (EBV)–negative Burkitt lymphoma cell line (Ramos),22 whereby the transcription factors JunB and FosB (and possibly also c-Fos) bind to the AP-1 element on B-cell receptor cross-linking.22 Third, during the late response to Epstein-Barr virus infection, one of the virus-derived proteins (LMP1) efficiently activates miR-155 expression through the nuclear factor-κB (NF-κB) pathway. Notably, EBV infection is well known to be associated with some human lymphoproliferative disorders, including Burkitt, posttransplantation, and Hodgkin lymphomas. Two candidate NF-κB DNA recognition sites at positions −1150 and −1697 nt are essential for LMP1-dependent activation involving NF-κB (by facilitating binding of p65 to the MIR155HG promoter) and also p38/MAPK pathways that stimulate the MIR155HG promoter. The targets of up-regulated miR-155 apparently include hematopoietic transcription factor PU.1.23,24 Other studies found that down-regulation of PU.1 represents an important and critical step for both myeloid25 as well as lymphoid tumorigenesis26,27 by a mechanism involving transcriptional repression of the PU.1 gene. The reports summarized in this paragraph collectively document important regulatory elements upstream of the MIR155HG and emphasize the importance of miR-155 program in B cells that may be targeted on lymphoproliferation. In the current study, we add another mechanism involving the transcription factor MYB that binds and stimulates MIR155HG promoter, resulting in dysregulation of its epigenetic status and in aberrantly elevated levels of miR-155.
Methods
Patients and cells
The B-CLL patients (N = 113) and volunteers donated peripheral blood samples on informed consent during the years 2007 to 2010 in accordance with the Declaration of Helsinki. The Institutional Review Board for human subjects in Charles University approved this study. The diagnostic and prognostic evaluations were done as published elsewhere.28,29 Peripheral blood from B-CLL patients was fractionated by Ficoll gradient (GE Healthcare), or alternatively the RosetteSep Kit was used (StemCell Technologies). The controls included CD19+ peripheral blood lymphocytes separated by autoMACS Pro Separator (Miltenyi Biotec). The cell purity and viability exceeded 90% as determined by flow cytometry (FACSCanto II, BD Biosciences) and Trypan blue staining.
RNA expression
Cellular RNA was isolated by Trizol (Invitrogen), analyzed by 2100 Bioanalyzer (Agilent Technologies), and transcribed using High Capacity cDNA Reverse Transcription Kit with specific primers. Quantitative polymerase chain reaction (PCR; using the ABI 7900HT instrument) was run for 40 cycles (95°C for 15 seconds and 60°C for 1 minute) or alternatively (40 cycles, 95°C for 10 seconds, 60°C for 20 seconds, 72°C for 30 seconds) as TaqMan (Roche) or SYBR Green-based PCR. The converted CT values of specific (s) and control (c) amplicons calculated by 2−(CTc-CTs) equation were compared by Student t test.
Immunoblotting
Primary cells or cell lines (0.5-1 × 107) were lysed for 12 minutes in 200 μL RIPA buffer (50mM Tris-Cl, pH 8.0, 137mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, protease inhibitor cocktail P8340; Sigma) by vortexing on ice for 20 seconds 4 times followed by gentle sonication (50% amplitude, 3 cycles of 1 second, 5-second pause) on Branson Sonic Dismembrator (model 500) with a micro-tip. Denatured cell lysates (20 μg protein per lane) were resolved on 4% to 12% gradient Bis-Tris gel (NuPage; Invitrogen). The gels were dry-blotted by iBlot Gel Transfer System (Invitrogen). The membranes were blocked by 7.5% nonfat milk in phosphate-buffered saline (0.1% Tween-20). Antibodies used (diluted 1:600) were: (anti–v-Myb/c-myb [clone: EP769Y], ab45150 Abcam; anti-PU.1 [T-21], sc-352; Santa Cruz Biotechnology). Horseradish peroxidase-conjugated antibody was used to visualize bands using ECL Plus Western Blotting Detection System (GE Healthcare) on X-ray films. Anti-actin horseradish peroxidase-conjugated antibody (anti-actin [I-19], sc-1616; Santa Cruz Biotechnology) was used to determine sample loading.
ChIP
A total of 106 cells were processed as described previously.30 Antibodies used were: H3K9acetyl (07-353; Upstate Biotechnology), H3K4methyl (ab1012; Abcam), MYB (ab45150; Abcam), and a control antibody (NI01; Calbiochem). DNA from immunoprecipitates was quantified by PCR using standard curves and defined DNA copy numbers. Specific occupancy on DNA (“percentage of input”) was defined as a copy number of a specific DNA fragment in each immunoprecipitate compared with the copy number of that DNA fragment within 1/100 input dilution used for immunoprecipitation (1% input DNA). The control antibody values were subtracted from the values obtained using the specific antibodies.
Transfections
The MIR155HG promoter containing putative MYB binding site was subcloned into pGL4.17 plasmid (supplemental Data A and B, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) and transfected into HeLa cells (confluence 80% per 1.862 cm2, total plasmid content 1.5 μg) using jetPEI (Polyplus Transfection). The luciferase activity at 48 hours was determined by Steady-Glo Luciferase Assay System (Promega), and the values were normalized to the protein content relative to signal obtained from cells transfected with the reporter backbone. Primary B-CLL cells or the human Raji cell line (from ATCC) were transfected with MYB siRNA (sc-29855; Santa Cruz Biotechnology), hsa-miR-155 anti-miRNA inhibitor (AM12601; Ambion), hsa-miR-155 (AM17100; Ambion), negative control (AM17110; Ambion), MYB, and control plasmids (kindly provided by Dr M. Dvorak, Institute of Molecular Genetics, Prague) by either chemical transfection or nucleofection. Transfection of primary B-CLL cells (8 × 105 cells/1 mL OPTI-MEM/Iscove modified Dulbecco medium) was done by transfection reagent DMRIE-c (10459-014; Invitrogen). siRNA c-MYB (30 pmol) or anti–miR-155 (15 pmol) was mixed with 2 μL of DMRIE-c reagent (in 100 μL of OPTI-MEM/well) added to the cell suspension (500 μL of OPTIMEM/well) and incubated (at 5% CO2 in 37°C) for 5 hours. Next, an equal volume (500 μL) of Iscove modified Dulbecco medium (20% fetal calf serum, 1% of nonessential amino acids) was added to the culture. Cells were harvested after 48 hours (siRNA c-MYB) or 96 hours (anti–miR-155) and the RNA analysis performed (see “RNA expression”). Nucleofections were done by nucleofector Amaxa and Human B-cell nucleofection kit (VPA-1001; Lonza). A total of 2 million cells (in a final volume of 2 mL of Iscove modified Dulbecco medium, 1% of nonessential amino acids) were mixed with 100 μL of nucleofection solution after applying the nucleofection program M-013. Cells were harvested after 24 hours (hsa-miR-155 transfectants) or 48 hours (siRNA c-MYB, anti–miR-155, MYB transfectants), and RNA analysis was performed.
Microarray mRNA profiling
Total RNA was purified to prepare biotin-labeled cRNA target population according to Affymetrix 3′-IVT Express Kit protocol. cRNA probes were used for subsequent hybridization on the gene expression chips (Affymetrix Human Genome HG-U133 Plus 2.0 Array containing nearly 50 000 probe sets), followed by subsequent fluorescent staining, fluidics processing, and scanning according to the Affymetrix recommendations. We collected peripheral blood samples from B-CLL patients (N = 16) and healthy donors (N = 5) to perform genome-wide microarray analysis. We filtered out 4 B-CLL samples because of demands on clinical data quality and based on the GeneSpring Quality Control principal component analysis. The microarray data have been deposited in NCBI Gene Expression Omnibus (GEO, www.ncbi.nlm.nih.gov/geo) and are accessible through GEO Series accession number GSE26725.
Data analysis
High-throughput data analyses were performed using GeneSpring GX10 (Agilent Technologies) and TIGR MeV4 software Version 4.6.1.31 GeneSpring software was used for probe level data preprocessing and summarization (using RMA), quality control analysis based primarily on principal component analysis, and partially for statistical analysis of microarray data. MeV4 software was used for data visualization (creating heat maps). For the statistical analysis of quantitative PCR results, the software package R Version 2.12.0 (www./www.r-project.org), NET programming framework (C#), Microsoft MS SQL database, and Microsoft Excel were used.
GSEA
To determine the expression of miR-155 targets in B-CLL, we used the 6 following target prediction tools: PicTar (www.pictar.mdc-berlin.de), Targetscan (www.targetscan.org), Microrna (www.microrna.org), and Microcosm (www.ebi.ac.uk/enright-srv/microcosm); some are experimentally validated (Set Mir-155 Targets Diana, www.diana.cslab.ece.ntua.gr/tarbase, Gene Set Enrichment Analysis [GSEA] set, www.broadinstitute.org/gsea/msigdb/cards/AGCATTA,MIR-155.html). Set Mir-155 targets ALL represents union of all 6 sets (www.dnasuite.com/gsea). To determine the patterns of expression of the MYB targets, we used 3 different lists of MYB targets. Set MYB TARGETS GENEGO represents MYB targets from database www.genego.com. Set MYB TARGETS RULAI.CSHL.EDU is based on data from Transcriptional Regulatory Element Database at www.rulai.cshl.edu/cgi-bin/TRED. Set MYB TARGETS LITERATURE is based on a literature search. Set MYB TARGETS ALL represents union of all 3 sets (www.dnasuite.com/gsea).
To show that a notable part of the MYB and miR-155 downstream targets are differentially regulated, we used GSEA (www.broadinstitute.org/gsea).32 GSEA determines the significance of a gene list in the expression data of all measured genes ordered by a significance measure (ie, t test P value of a gene expression between 2 phenotypes or some other measure of differential regulation). The significant lists are those in which notable part of genes lies at the beginning or at the end of the ranked whole genome list (core enrichment). GSEA ranks lists of genes by enrichment scores and normalized enrichment scores. The primary measure of significance of a list is the false discovery rate (FDR) q value. The default GSEA settings mark as significant those lists with the FDR q value of ≤ 0.25. For GSEA, we custom compiled gene lists of MYB and miR-155 target genes based on databases and data described below in the text and also derived from the Molecular Signatures Database (MSigDB Version 3.0) provided in the GSEA software package. The GSEA results can be accessed at www1.lf1.cuni.cz/∼vkulv/gsea.
Results
Elevated levels of miR-155 and MYB in B-CLL
The levels of miR-155 become elevated on lymphocyte activation and on EBV infection by a mechanisms involving transcriptional regulation of MIR155HG. Elevated miR-155 levels were reported in several lymphoproliferative disorders, among them B-CLL. The mature form of miR-155 as well as its pri-miRNA are consistently up-regulated more than 5-fold in B-CLL compared with CD19+ B cells.11 Using quantitative PCR, we have observed markedly increased levels of miR-155 (Figure 1A) and of MIR155HG pri-miRNA (2- to 5-fold, using semiquantitative data from microarray profiling; supplemental Figure 1) in primary, peripheral blood-derived B-CLL cells compared with normal B cells (Figure 1A; supplemental Table of Clinical Patient Data). The up-regulation of miR-155 is variable, up to approximately 32-fold (Figure 1A). Whereas the levels of miR-155 expression in B-CLL are increased, the mRNA levels of its established target transcription factor PU.1 are significantly decreased33-35 (Figure 1A). A previous study indicated that miR-155 and its pre-miRNA are increasingly localized within proliferation centers of B-CLL/small lymphocytic lymphoma, in a pattern exclusive with the expression pattern of miR-150,36 a microRNA inhibiting the E-box transcription factor MYB in B cells.37 However, in a significant proportion of B-CLL patients, the miR-150 levels were increased (Figure 1A), an observation also supported by others.11 Unexpectedly, the MYB mRNA level measurements in B-CLL (MYB is a predicted target of both miR-150 and miR-155; Figure 1A) revealed its significant up-regulation relative to B cells, unlike its homologs MYC and MYBL1 (data not shown). Under physiologic conditions, the transcription factor MYB is required for the B-lymphoid development and becomes reduced during the process. However, when aberrantly expressed, it causes lymphoproliferative disorders and autostimulates its own expression.38-40 In normal B-cell development, the highest expression of MYB is observed in pro-B, lower levels in pre-B cells, whereas resting B (and also mature T) cells do not express MYB.37 Increased mRNA levels of MYB in B-CLL were reflected by the elevated MYB protein as determined by immunoblotting of the patient B-CLL and normal B-cell lysates. Interestingly, both primary transcript-derived MYB protein (migrating at ∼ 75 kDa) as well as additional, known oncogenic transcriptional variants that vary in the C-terminal end were detected (Figure 1B).41 The immunoblotting also revealed a marked down-regulation of the PU.1 protein (Figure 1B) in the B-CLL samples.
MicroRNA and mRNA expression in B-CLL. (A) Results of TaqMan PCR in B-CLL and control CD19+ cells (Ctrl). The y-axis represents relative abundance of mature miR-155 (number of B-CLL patients, N(B-CLL) = 118, number of controls, N(Ctrl) = 7) and of miR-150 (N(B-CLL) = 75, N(Ctrl) = 4) relative to RNU44; and of MYB mRNA (N(B-CLL) = 119, N(Ctrl) = 10) and PU.1 mRNA (N(B-CLL) = 119, N(Ctrl) = 5) relative to GAPDH. Data (shown as a fold change [FC]) are baseline normalized to RNA levels obtained in normal controls (control measurements were set equal to 1) and shown in log2 scale. Error bars represent SEM. P values (t test) are shown in parentheses. (B) Immunoblotting analysis of MYB and PU.1 in B-CLL (N = 5) and control (C87, N = 1) B-cell samples. The NB4 cell line was used as a positive control. The optical density (below each lane) of each protein band was measured by Scion Image software for Windows Beta 4.0.2, the values below the Western blots represent a fold change of densities of the specific bands (or their expected positions) in the patient, and control lanes normalized to the levels of the housekeeping protein β-actin. (C) The MYB mRNA levels in primary B-CLL cells positively associate with Rai staging. Three patient subgroups are shown. White bars represent currently investigated patients (OS 0) either treated by first-line therapy or untreated (Th 0–1); gray bars, currently investigated patients (OS 0) who received second or more lines of therapy (Th > 1); and black bars, previously investigated, deceased patients (OS 1) who had received at least one line of therapy (Th ≥ 1). The patient numbers in each expression assay and patient group are indicated in parentheses. Error bars represent SEM. P values (t test) are shown in parentheses on top of the brackets indicating the groups being compared. (D) RNA levels of miR-155 (white bars), PU.1 (light gray bars), MYB (dark gray bars), and miR-150 (black bars) categorized by the B-CLL prognostic parameters. Favorable outcome group = IgVH mutated, CD38 negative, ZAP70 negative. Unfavorable outcome group = IgVH unmutated, CD38+, ZAP70+. The patient numbers in each expression assay are indicated in parentheses. Error bars represent SEM. P values (t test) are shown in parentheses on top of the lines indicating the groups being compared. (E) Expression levels of miR-155 (white bars) and miR-150 (gray bars) in MYB underexpressors (< 1.5-fold decrease, left) and MYB overexpressors (> 1.5 increase; right) relative to control B cells. Patient numbers in each expression assay are indicated in parentheses. Error bars represent SEM. P values (t test) are shown in parentheses on top of the brackets indicating the groups being compared.
MicroRNA and mRNA expression in B-CLL. (A) Results of TaqMan PCR in B-CLL and control CD19+ cells (Ctrl). The y-axis represents relative abundance of mature miR-155 (number of B-CLL patients, N(B-CLL) = 118, number of controls, N(Ctrl) = 7) and of miR-150 (N(B-CLL) = 75, N(Ctrl) = 4) relative to RNU44; and of MYB mRNA (N(B-CLL) = 119, N(Ctrl) = 10) and PU.1 mRNA (N(B-CLL) = 119, N(Ctrl) = 5) relative to GAPDH. Data (shown as a fold change [FC]) are baseline normalized to RNA levels obtained in normal controls (control measurements were set equal to 1) and shown in log2 scale. Error bars represent SEM. P values (t test) are shown in parentheses. (B) Immunoblotting analysis of MYB and PU.1 in B-CLL (N = 5) and control (C87, N = 1) B-cell samples. The NB4 cell line was used as a positive control. The optical density (below each lane) of each protein band was measured by Scion Image software for Windows Beta 4.0.2, the values below the Western blots represent a fold change of densities of the specific bands (or their expected positions) in the patient, and control lanes normalized to the levels of the housekeeping protein β-actin. (C) The MYB mRNA levels in primary B-CLL cells positively associate with Rai staging. Three patient subgroups are shown. White bars represent currently investigated patients (OS 0) either treated by first-line therapy or untreated (Th 0–1); gray bars, currently investigated patients (OS 0) who received second or more lines of therapy (Th > 1); and black bars, previously investigated, deceased patients (OS 1) who had received at least one line of therapy (Th ≥ 1). The patient numbers in each expression assay and patient group are indicated in parentheses. Error bars represent SEM. P values (t test) are shown in parentheses on top of the brackets indicating the groups being compared. (D) RNA levels of miR-155 (white bars), PU.1 (light gray bars), MYB (dark gray bars), and miR-150 (black bars) categorized by the B-CLL prognostic parameters. Favorable outcome group = IgVH mutated, CD38 negative, ZAP70 negative. Unfavorable outcome group = IgVH unmutated, CD38+, ZAP70+. The patient numbers in each expression assay are indicated in parentheses. Error bars represent SEM. P values (t test) are shown in parentheses on top of the lines indicating the groups being compared. (E) Expression levels of miR-155 (white bars) and miR-150 (gray bars) in MYB underexpressors (< 1.5-fold decrease, left) and MYB overexpressors (> 1.5 increase; right) relative to control B cells. Patient numbers in each expression assay are indicated in parentheses. Error bars represent SEM. P values (t test) are shown in parentheses on top of the brackets indicating the groups being compared.
Next we have analyzed the significance of miR-155, MYB, miR-150, and PU.1 RNA expression in relation to staging and prognostic groups. First, we observed that patients with more advanced disease stages (Rai 2-4) display significantly elevated levels of MYB (but not of miR-155 or miR-150) compared with the group with Rai 0 or 1 stages (Figure 1C; supplemental Figure 1C). This is demonstrated in 3 different patient subgroups differing by the number of therapy regimens or the survival status. To understand why more advanced stages are marked by increased MYB expression, we compared patients with favorable prognoses (IgVH mutated, CD38−, ZAP70−) with those displaying all 3 unfavorable parameters (unmutated IgVH, CD38+, ZAP70+). In the unfavorable outcome group, the MYB mRNA is expressed at significantly higher levels compared with the favorable outcome group (Figure 1D). When the patients were divided according to 2 prognostic parameters, the expression of MYB was again significantly higher in the unfavorable groups (supplemental Figure 1D). To determine whether there exists a relationship between elevated MYB levels and miR-155, we have divided the B-CLL patients into 2 groups: either overexpressing MYB (> 1.5-fold, a set that represents approximately half of all patients) or with low expression levels of MYB. As demonstrated in Figure 1E, the levels of miR-155 are significantly higher in the MYB-overexpressing B-CLL patient cells. This observation suggested that MYB might regulate the levels of miR-155 in B-CLL. In addition, a B-CLL patient undergoing B cell–specific treatment by rituximab (anti-CD20 antibody) displayed markedly decreased expression pattern of miR-155 and MYB at 24 hours in the peripheral blood mononuclear cells depleted of lymphocytes (supplemental Figure 1B). These data collectively indicate that the MYB levels in B-CLL are increased in the unfavorable outcome group and also in advanced stages of B-CLL and that this pattern is reflected by the increased miR-155 levels in the patients overexpressing MYB.
MYB binds and stimulates MIR155HG promoter in B-CLL
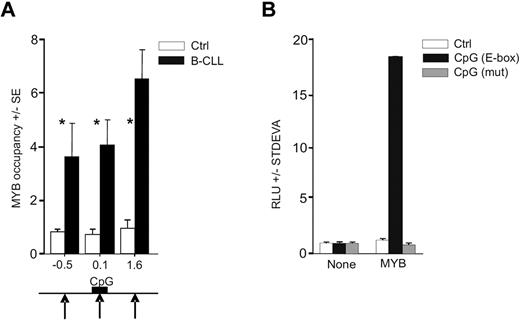
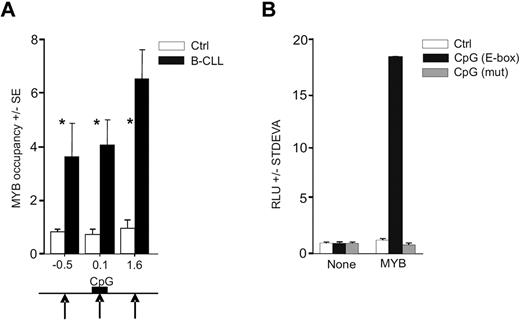
As demonstrated in a previous study, the E-box elements are enriched within the proximal regulatory region of the avian MIR155HG.42 By transcription factor binding site analyses43 on human miR-155 gene sequence (available from www.genome.ucsc.edu), we identified a highly conserved CpG island upstream the MIR155HG (MIR155HG promoter) that ranges from −33 to 349 bp (relative to the TSS; supplemental Data A and B). We found that the human DNA regulatory region upstream of MIR155HG also contains putative MYB binding sites. MYB thus represents a candidate regulator of high miR-155 levels as it may directly associate with MIR155HG via the E-box element binding, in an analogy shown previously in the avian orthologous context.42 In addition, the activation of MIR155HG by a strong transactivator is further suggested by genomic analyses, indicating that the region encoding miR-155 in B-CLL cells is not consistently amplified11 and the putative MYB binding site within the CpG island is not mutated (supplemental Data D). To determine whether MYB is recruited on the MIR155HG promoter, we used chromatin immunoprecipitation (ChIP) using anti-MYB antibody followed by quantitative PCR analyses within the MIR155HG locus using a set of PCR amplicons covering a large proportion of MIR155HG (−3.2 kb to 4.4 kb, relative to TSS). The ChIP was done in primary B-CLL cells and in normal CD19+ B cells obtained from healthy donors. Figure 2A demonstrates that MYB is consistently recruited to MIR155HG in B-CLL exclusively at 3 loci (at −0.5, 0.1, and 1.6 kb relative to TSS in primary B-CLL and nowhere else upstream or downstream within the region studied). The occupancy by MYB is distributed within a relatively extended region of approximately 2 kb, which includes the TSS and the E-box elements within the CpG Island. Other portions of MIR155HG were not occupied by MYB, indicating that the MYB targeting of MIR155HG is TSS-specific. Negligible, background-level occupancy of MYB at MIR155HG in normal B cells indicates that, under normal conditions, MIR155HG may be controlled by other transcription regulators.22
MYB binds and stimulates MIR155HG. (A) MYB occupancy within MIR155HG locus determined by ChIP, using anti-Myb (EP769Y) or control antibodies and carried out on cross-linked chromatin isolated from B-CLL cells (N = 6, average value indicated by black bars, patients included P40, P39, P47, P250, P254, and P255) and normal B cells (N = 6, white bars; “Patients and cells”). The y-axis indicates the relative occupancy of MYB. The x-axis marks the positions (in kilobases) of PCR amplicons relative to TSS (supplemental Data A). Black arrows indicate the positions of MYB DNA binding motifs; and black box, position of the CpG island (supplemental Data A). Error bars represent SEM of 3 independent experiments. *P < .05. (B) HeLa cells transiently transfected with pGL4.17 reporter vector containing the MYB binding site (−399 to −394 bp; black bars), its deletion mutant (gray bars), or a control reporter plasmid (Ctrl, white bars). Reporter vectors were transfected either alone (None) or cotransfected with MYB cDNA expression vector (MYB), and the luciferase activity at 48 hours (RLU) is shown relative to a control vector. The background activity was subtracted, and data were normalized to protein content. Average values and SD of at least 2 independent experiments are plotted.
MYB binds and stimulates MIR155HG. (A) MYB occupancy within MIR155HG locus determined by ChIP, using anti-Myb (EP769Y) or control antibodies and carried out on cross-linked chromatin isolated from B-CLL cells (N = 6, average value indicated by black bars, patients included P40, P39, P47, P250, P254, and P255) and normal B cells (N = 6, white bars; “Patients and cells”). The y-axis indicates the relative occupancy of MYB. The x-axis marks the positions (in kilobases) of PCR amplicons relative to TSS (supplemental Data A). Black arrows indicate the positions of MYB DNA binding motifs; and black box, position of the CpG island (supplemental Data A). Error bars represent SEM of 3 independent experiments. *P < .05. (B) HeLa cells transiently transfected with pGL4.17 reporter vector containing the MYB binding site (−399 to −394 bp; black bars), its deletion mutant (gray bars), or a control reporter plasmid (Ctrl, white bars). Reporter vectors were transfected either alone (None) or cotransfected with MYB cDNA expression vector (MYB), and the luciferase activity at 48 hours (RLU) is shown relative to a control vector. The background activity was subtracted, and data were normalized to protein content. Average values and SD of at least 2 independent experiments are plotted.
To address whether MYB stimulates MIR155HG functionally via the putative MYB binding site (located at position −407 nt adjacent to the CpG island), we generated luciferase reporter constructs carrying either mutated or wild-type MYB binding sites, subcloned as part of a region selected based on high-scoring prediction of binding sites and the ChIP-based MYB location data. The reporter assays were performed in human HeLa cells. Figure 2B demonstrates that the predicted binding site that is occupied by MYB in B-CLL is indeed specifically stimulated by MYB, whereas its mutated version disabled MYB-directed luciferase activity. Interestingly, transfection of MYC did not significantly elevate the reporter activity, indicating that a segment of the MIR155HG promoter is specifically stimulated by MYB (data not shown). These findings collectively indicate that the MIR155HG is a direct transcriptional target of MYB.
Altered chromatin structure at MIR155HG promoter in B-CLL
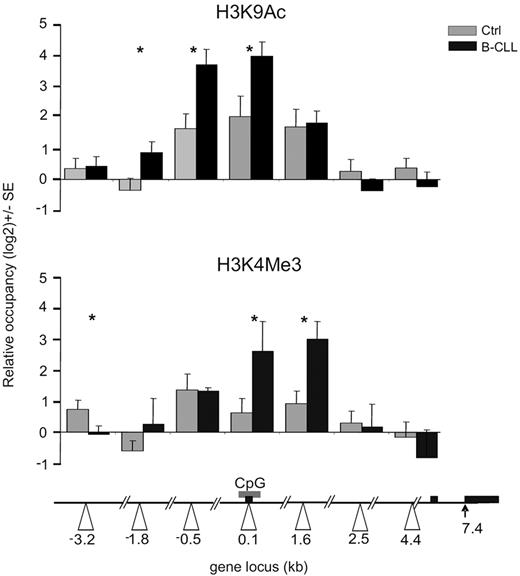
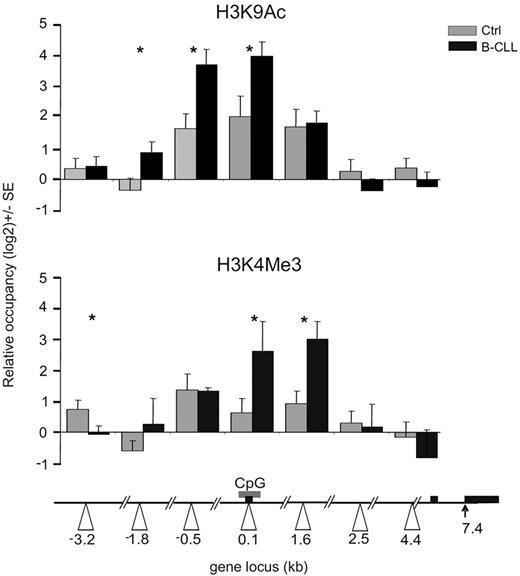
The recruitment of MYB on the MIR155HG promoter associated with highly elevated miR-155 levels suggests its direct involvement in transcriptional activation of the MIR155HG in B-CLL. Furthermore, the lack of genetic changes within the MIR155HG locus11 (supplemental Data D) implicates that the putative mechanism of its transcriptional activation involves epigenetic changes. We first tested the acetylation status of lysine 9 of histone H3 (H3K9), a mark associated with actively transcribed genes.44 As expected, normal CD19+ cells display an acetylation peak of H3K9 within the MIR155HG promoter (region 0.1 kb), in concordance with the basal expressions of miR-155 in B cells (Figure 1A; supplemental Figure 1A). However, the ChIP analysis of H3K9Ac levels in B-CLL displayed significant elevation of this mark at the MIR155HG promoter compared with normal B cells (Figure 3 top panel). In addition, we observed B-CLL-specific, prominent spreading of the H3K9Ac mark both upstream (−3.2 kb) and downstream (1.6 kb) from the center of occupancy observed in normal B cells (Figure 3). Importantly, the H3K9Ac pattern overlaps with the pattern of MYB occupancy (Figure 2A). Furthermore, regression curves indicate a correlation between MYB mRNA levels and the enrichment of H3K9Ac at the MIR155HG promoter (r = 0.65; supplemental Figure 2A), indicating that MYB may be directly responsible for activating MIR155HG expression. B-CLL-specific changes at the levels of both expression and DNA-specific localization of MYB might promote the miR-155 expression by facilitating transcription of MIR155HG. These regulatory effects of histone H3K9 acetylation on miR-155 levels are supported by our next observation demonstrating that, after inhibition of histone deacetylases (HDACs) by trichostatin in hematopoietic murine pro-B progenitors, the levels of miR-155 correspondingly increased (supplemental Figure 2B).
Histone modifications at the MIR155HG promoter in B-CLL. ChIP was performed on crosslinked chromatin from primary B-CLL (N = 9: P250, P254, P255, P143, P130, P161, P162, P164, P179, and P135, black bars) and control B cells (N = 8, gray bars) using the anti-H3K9Ac, H3K4Me3, and control antirabbit IgG antibody, as described in “ChIP.” The levels of H3K9 acetylation and H3K4 trimethylation (y-axis) are expressed relative to control IgG antibody and relative to upstream −4.7 kb locus with consistent low H3K9 acetylation and H3K4 methylation patterns. The x-axes indicate positions of amplicons (in kilobases) relative to TSS (white arrowheads). Gray box represents the position of the CpG; and black box, the exons of human MIR155HG. Error bars represent SEM. *P < .05.
Histone modifications at the MIR155HG promoter in B-CLL. ChIP was performed on crosslinked chromatin from primary B-CLL (N = 9: P250, P254, P255, P143, P130, P161, P162, P164, P179, and P135, black bars) and control B cells (N = 8, gray bars) using the anti-H3K9Ac, H3K4Me3, and control antirabbit IgG antibody, as described in “ChIP.” The levels of H3K9 acetylation and H3K4 trimethylation (y-axis) are expressed relative to control IgG antibody and relative to upstream −4.7 kb locus with consistent low H3K9 acetylation and H3K4 methylation patterns. The x-axes indicate positions of amplicons (in kilobases) relative to TSS (white arrowheads). Gray box represents the position of the CpG; and black box, the exons of human MIR155HG. Error bars represent SEM. *P < .05.
To further study the activation status of MIR155HG promoter, we have analyzed another chromatin mark, trimethylation of H3K4, which associates with gene activation and with the recruitment of DNA-dependent RNA polymerase II to gene promoters.45 Our data in Figure 3 (bottom panel) indicate that H3K4Me3 is indeed significantly enriched in primary B-CLL chromatin near the MIR155HG promoter (regions 0.1, 1.6 kb) compared with normal B cells. These data collectively indicate the importance of the histone H3K9 acetylation and H3K4 methylation status of the MIR155HG locus in regulating miR-155 levels, in a process directed in B-CLL leukemic cells by oncogenic MYB and potentially by other transcription cofactors.
Global gene expression profiling in B-CLL reveals deregulation of MYB and of miR-155 targets
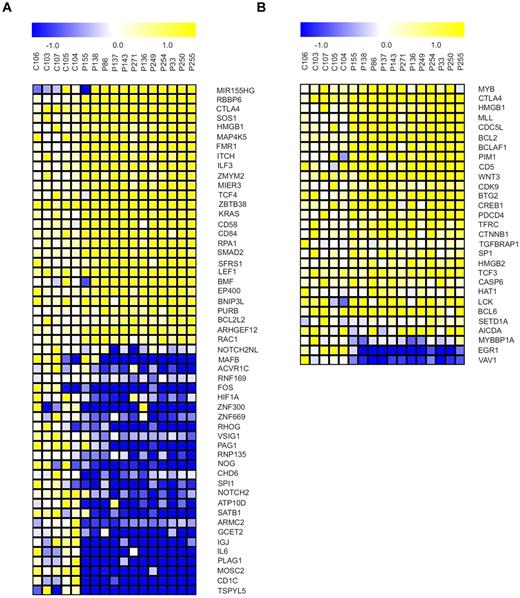
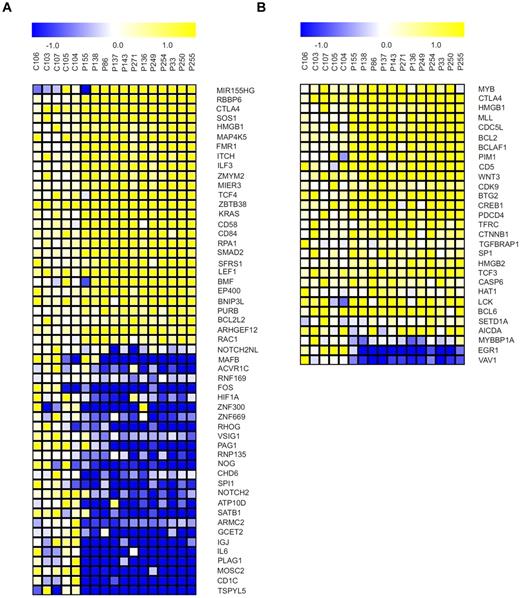
We assessed the behavior of MYB and miR-155 and their downstream targets at the transcriptional level using microarrays (“Microarray mRNA profiling”). We used GSEA to identify genes among the downstream targets of the MYB and miR-155 that are differentially regulated between B-CLL–derived B cells and healthy CD19+ B cells and to detect any significant enrichment (based on expression level) of these target genes in B-CLL samples. The miR-155 targets from the lists produced by a set of prediction tools (“GSEA”) are significantly dysregulated (based on the FDR q value) in the B-CLL phenotype as determined by GSEA. We observed significant down-regulation of target mRNAs, including important transcription factors (such as PU.1) and chromatin remodeling genes (SATB1, CHD6; Figure 4A). We rather surprisingly observed significant changes in up-regulated mRNA targets indicating complex regulatory relationships between microRNAs and their targets and a possibility that these targets are selectively regulated by other mechanisms, related or unrelated to elevated miR-155 levels. Next, the mRNAs in predicted MYB target genes lists produced by prediction tools (“GSEA”) are up-regulated in the B-CLL phenotype according to GSEA (FDR q values < 0.111). Within this list, there are numerous high scoring genes with known functions in B-CLL (BCL-2, CD5; Figure 4B). This paragraph overall indicates that significant expression pattern changes of the miR-155 and MYB target mRNAs are observed in B-CLL.
Targets of miR-155 and MYB become dysregulated in B-CLL. (A) Selected miR-155 target genes exhibiting differential expression between B-CLL (N = 12, patient codes are indicated on top of the heat maps) and normal B-cell samples (P < .05, t test). Genes were selected from the union of 6 lists of either predicted or experimentally validated target genes of miR-155 (“GSEA”). (B) Selected MYB target genes exhibiting differential expression between B-CLL (N = 12, patient codes are indicated on top of the heat maps) and normal B-cell samples (P < .05, t test). Genes were selected from 3 independent sources cataloguing the MYB target genes (“GSEA”).
Targets of miR-155 and MYB become dysregulated in B-CLL. (A) Selected miR-155 target genes exhibiting differential expression between B-CLL (N = 12, patient codes are indicated on top of the heat maps) and normal B-cell samples (P < .05, t test). Genes were selected from the union of 6 lists of either predicted or experimentally validated target genes of miR-155 (“GSEA”). (B) Selected MYB target genes exhibiting differential expression between B-CLL (N = 12, patient codes are indicated on top of the heat maps) and normal B-cell samples (P < .05, t test). Genes were selected from 3 independent sources cataloguing the MYB target genes (“GSEA”).
Manipulations of MYB and miR-155 document their relationship in B-CLL
As demonstrated in the previous paragraph, the target genes of miR-155 and MYB are dysregulated in B-CLL. The relationship between miR-155 targets and MYB levels manifests at the levels of transcriptional regulation of MIR155HG. To test whether a manipulation of MYB levels can influence the miR-155 expression and whether the manipulation of miR-155 directly regulates levels of its targets in B-CLL, we devised and performed the following experiments.
First, the levels of MYB were down-regulated by siRNA-mediated inhibition in B-CLL sample and in the Raji Burkitt lymphoma-derived cell line. As indicated in Figure 5A, significant down-regulation of MYB resulted in significant down-regulation of miR-155 in B-CLL. Analogous results were produced in the Raji cells. Next, the MYB levels were increased by transfecting the expression plasmid encoding murine c-Myb cDNA. This plasmid efficiently induced (5-fold) the expression of MYB protein in HeLa cells (Figure 5B). After the transfection of c-Myb into Raji cells, the levels of miR-155 were dose-dependently and significantly up-regulated in both 0.2 and 2 μg of transfected mixtures (Figure 5B). These experiments demonstrated that ectopic manipulation of MYB levels in vitro leads to corresponding regulatory effects on miR-155 levels, thus providing functional support for the positive regulatory effects of MYB levels on miR-155 levels in B-CLL.
Manipulations of MYB and miR-155 levels in B-CLL. (A) Transfection of primary B-CLL cells (P88) and Raji cell line with siRNA oligos inhibiting MYB expression (final concentrations of 30nM, gray bars; and 100nM, black bars) or with negative control oligos. The y-axis indicates quantitative PCR-determined relative levels of mRNA of indicated genes related to housekeeping gene GAPDH (for MYB, PU.1) or to RNU44 (for miR-155) 48 hours after transfection (“Transfections”). Data (shown as a fold change [FC]) are normalized to mRNA levels obtained in negative control transfection experiment (control measurements were set as baseline = 1). Error bars represent SEM. *P < .05. **P < .01. (B) Raji cells were transfected with MYB cDNA encoding expression plasmid (in 2 final concentrations: 0.2 μg/mL, gray bars; and 2.0 μg/mL, black bars) or with a negative control (pcDNA 3.1) plasmid. The y-axis: quantitative PCR-determined relative levels of mRNA miR-155 compared with the RNU44 internal control (control measurements were set as baseline = 1) 48 hours after transfection (“Transfections”). Error bars represent SEM. *P < .05. **P < .01. Immunoblotting of MYB (and β-actin) in HeLa cells at 48 hours after transfection indicates expression product of the MYB cDNA expressing plasmid. The relative optical density of each protein is indicated below the blots. (C) B-CLL cells (P161) and Raji cell line was transfected with anti–miR-155 oligos (at final concentration of 40nM, gray bars; and 100nM, black bars) and with negative control oligos. After 96 hours, the total RNA was purified, reverse transcribed, and measured by quantitative PCR as described in “RNA expression.” The y-axis indicates relative expression of mRNA of indicated genes relative to the housekeeping gene GAPDH (for PU.1) or to RNU44 (for miR-155). Data are normalized to the mRNA levels obtained in negative control transfection experiment (control measurements were set as baseline = 1). Error bars represent SEM. *P < .05. **P < .01. (D) Raji cell line was transfected with miR-155 oligos (at final concentration 40nM, gray bars; and 100nM, black bars) or with negative control oligos. After 24 hours, total RNA was purified, reverse transcribed, and measured by quantitative PCR as described in “RNA expression.” The y-axis indicates relative expression of mRNA of indicated genes relative to the housekeeping gene GAPDH (for PU.1) or to RNU44 (for miR-155). Data are normalized to the mRNA levels obtained in negative control transfection experiment (control measurements were set as baseline = 1). Error bars represent SEM. *P < .05. **P < .01.
Manipulations of MYB and miR-155 levels in B-CLL. (A) Transfection of primary B-CLL cells (P88) and Raji cell line with siRNA oligos inhibiting MYB expression (final concentrations of 30nM, gray bars; and 100nM, black bars) or with negative control oligos. The y-axis indicates quantitative PCR-determined relative levels of mRNA of indicated genes related to housekeeping gene GAPDH (for MYB, PU.1) or to RNU44 (for miR-155) 48 hours after transfection (“Transfections”). Data (shown as a fold change [FC]) are normalized to mRNA levels obtained in negative control transfection experiment (control measurements were set as baseline = 1). Error bars represent SEM. *P < .05. **P < .01. (B) Raji cells were transfected with MYB cDNA encoding expression plasmid (in 2 final concentrations: 0.2 μg/mL, gray bars; and 2.0 μg/mL, black bars) or with a negative control (pcDNA 3.1) plasmid. The y-axis: quantitative PCR-determined relative levels of mRNA miR-155 compared with the RNU44 internal control (control measurements were set as baseline = 1) 48 hours after transfection (“Transfections”). Error bars represent SEM. *P < .05. **P < .01. Immunoblotting of MYB (and β-actin) in HeLa cells at 48 hours after transfection indicates expression product of the MYB cDNA expressing plasmid. The relative optical density of each protein is indicated below the blots. (C) B-CLL cells (P161) and Raji cell line was transfected with anti–miR-155 oligos (at final concentration of 40nM, gray bars; and 100nM, black bars) and with negative control oligos. After 96 hours, the total RNA was purified, reverse transcribed, and measured by quantitative PCR as described in “RNA expression.” The y-axis indicates relative expression of mRNA of indicated genes relative to the housekeeping gene GAPDH (for PU.1) or to RNU44 (for miR-155). Data are normalized to the mRNA levels obtained in negative control transfection experiment (control measurements were set as baseline = 1). Error bars represent SEM. *P < .05. **P < .01. (D) Raji cell line was transfected with miR-155 oligos (at final concentration 40nM, gray bars; and 100nM, black bars) or with negative control oligos. After 24 hours, total RNA was purified, reverse transcribed, and measured by quantitative PCR as described in “RNA expression.” The y-axis indicates relative expression of mRNA of indicated genes relative to the housekeeping gene GAPDH (for PU.1) or to RNU44 (for miR-155). Data are normalized to the mRNA levels obtained in negative control transfection experiment (control measurements were set as baseline = 1). Error bars represent SEM. *P < .05. **P < .01.
The effects of miR-155 on its targets were tested using overexpression of miR-155 and inhibitory anti–miR-155 in B-CLL samples and in Raji Burkitt lymphoma-derived cell line. As documented by Figure 5C, anti-mir–mediated down-regulation of miR-155 levels leads to efficient and rapid up-regulation of its target, the mRNA encoding transcription factor PU.1. Conversely, a transient increase in miR-155 levels leads to significant down-regulation of PU.1 (Figure 5D). PU.1 represents an important mediator that dose-dependently directs gene expression of all blood lineages, and its very low levels are associated with leukemogenesis. Our data thus provide important evidence that the regulatory pathway MYB → miR-155 ⊣ PU.1 may be of critical importance for the transcriptome reprogramming and consequently for the pathogenic cell phenotype in B-CLL.
Discussion
B-CLL is the most frequent leukemia in the Western world, and it represents an incurable disease despite novel therapeutic modalities, with the underlying mechanisms still not fully known. Our study focuses on identification of molecular mechanisms associated with consistently increased levels of the novel putative pathogenic factor in B-CLL: the microRNA miR-155.10-12 MiR-155 is elevated both at the levels of the mature 22-nt miRNA but also at the levels of the primary transcript generated from the MIR155HG (Figure 1A; supplemental Figure 1A), thus suggesting its own aberrant transcriptional activation. Based on a study in the avian cell context42 and our detailed analyses of the MIR155HG promoter, we hypothesized that the E-box-binding proteins are important players in conveying the transcription regulation of MIR155HG in the humans. Second, we tested levels of miR-150 that were previously associated with B-CLL/small lymphocytic lymphoma and those of its oncogenic target MYB. Indeed, we detected significantly elevated levels of the oncogenic MYB mRNA and protein in the B-CLL tumor cells compared with normal B cells (Figure 1A-B), suggesting that MYB may act as the transactivator of MIR155HG. Interestingly, the MYB-inhibiting hematopoietic transcription factor PU.1,46-48 a target of miR-155,23,24 is reduced in B-CLL cells at both the mRNA (Figure 1A) and protein (Figure 1B) levels. The role of decreased PU.1 levels in lymphoid malignancies was documented by data showing that down-regulation of PU.1 promotes multiple myeloma and T-cell lymphoma cell growth,26,27 whereas restoration of PU.1 levels induces cell cycle arrest and cell death.27 These and the aforementioned observations of up-regulated miR-155, MYB, and down-regulated levels of PU.1 in B-CLL cells are supported using genome-wide arrays, demonstrating significant deregulation of their downstream transcriptomic programs in the set of B-CLL patients (Figure 4).
It has been recently demonstrated that miR-155 levels associate with (1) the increased ZAP70 expression in B-CLL,49 (2) a short duration to the onset of therapy,12 and (3) activated B-cell like phenotype of diffuse large B-cell lymphoma marking unfavorable prognosis.10 In our set of patients, miR-155 was further increased approximately 2-fold in B-CLL samples concomitantly overexpressing MYB (Figure 1E). MYB B-CLL overexpressors were those displaying advanced Rai stage (Figure 1C) and unfavorable prognostic markers (Figure 1D; supplemental Figure 1D). We have not observed significant changes in the expression of MYB and other genes related to cytogenetic aberrations, probably because of the small number of samples with each cytogenetic aberration. These data are currently being expanded in a larger patient group and at least in part indicate that oncogenic transcription factor MYB may represent important step in regulating the progression of B-CLL.
The presence of E-boxes in the CpG Island of the MIR155HG promoter together with elevated MYB levels in B-CLL prompted us to analyze both the transcriptional regulation and the chromatin structure of this region in the B-CLL tumor cells. Other studies previously described a proximal promoter region (AP-1, −40 nt) involved in normal B-cell receptor activation and additional relatively distant regulatory regions (NF-κB, −1150 and −1697 nt) involved in EBV-mediated transformation.22 We add to these findings another tumor and B-cell specific regulatory region based on our following observations: the up-regulated MYB in B-CLL cells is recruited onto MIR155HG promoter near the TSS in relatively broad domain containing multiple putative MYB binding sites (Figure 2A). E-box sequence cloned from the MIR155HG promoter readily responds to MYB in reporter assays (Figure 2B). In normal B cells, the MYB is expressed but not recruited onto the MIR155HG promoter. Thus, MIR155HG represents a novel direct target of MYB in B-CLL cells.
Next, a correlation exists between the MYB mRNA levels and hyperacetylation of H3K9 upstream MIR155HG. Indeed, our ChIP analyses revealed a domain of enhanced H3K9 hyperacetylation compared with normal B-cell chromatin (Figure 3). In addition, H3K4 hypermethylation was also observed in B-CLL, documenting overall transcriptional activation of MIR155HG. Furthermore, the distribution of H3K9Ac and H3K4Me, 2 well-known marks of actively transcribed genes, overlapped with the MYB occupancy in B-CLL cells (Figure 2A). These data are supported by gene expression arrays revealing the down-regulation of several HDACs, including HDAC1, HDAC4, and HDAC9, and up-regulation of histone acetyltransferase HAT1 compared with normal B cells (Figure 4; supplemental Figure/GSEA results). This down-regulation (especially of HDAC1) might represent a very important step in the regulation of miR-155 levels, a notion supported by our observation based on trichostatin treatment of a cell line model that significantly enhanced the miR-155 in vitro (supplemental Figure 2).
The ability of MYB to regulate MIR155HG was further tested using a transient gene manipulation approach to modulate MYB and miR-155 in B-CLL and in a Burkitt lymphoma-derived cell line. In these experiments, MYB is again demonstrated to positively regulate miR-155 levels. The mechanism of activation of miR-155 host gene by MYB is proposed to include acetylation of histone H3K9, a notion supported by others.50 This is further supported by positive correlation between the MYB levels and the histone H3K9 levels upstream of MIR155HG (supplemental Figure 2A). Moreover, there is a significant elevation of the miR-155 levels in the B-CLL patients overexpressing MYB (Figure 1E). However, a direct comparison of MYB levels and miR-155 did not show a strong correlation (r = 0.12), which may be explained by the work of Eis et al,10 showing that the molar ratios of miR-155 to BIC RNA varied from 4 to 35 and do not correlate with the absolute copy numbers of either molecule, and are therefore possibly directed by a combination of factors related to miRNA biogenesis and RNA processing.4,5 By manipulating the miR-155 levels, we demonstrated that it negatively regulates its target, PU.1 (Figure 5C-D). It should be noted that the MYB elevation in B-CLL is possibly important not only for the up-regulation of miR-155 but also for the regulation of additional target genes, including BCL2 and others known to be deregulated in B-CLL41 (Figure 4). The altered levels of MYB and PU.1 in B-CLL, on the other hand, may be of critical importance for differentiation and cell type specification; these mechanisms are tightly regulated by the levels of these transcription factors.39,51,52 Lastly, deregulated programs of MYB, miR-155, and PU.1 in B-CLL potentially harbor important signatures of advanced disease that can be used to delineate in more detail the pathogenesis and treatment options and to improve the clinical outcome in a B-CLL patient subset.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Genome Technology Center at the New York University Langone Medical Center, New York, NY, for expert assistance with microarray gene expression profiling; Drs J. Salkova, K. Benesova, P. Klener, J. Molinsky, M. Klanova, E. Konirova, M. Hnatkova, and J. Koren for useful discussions; M. Hamouzova, K. Plevova, and J. Malcikova for their help with organizing the patient data; Dr M. Dvorak for providing valuable reagents; and Dr A. I. Skoultchi for reading the manuscript.
This work was supported by the Czech Ministries of Health (NS10310-3/2009, NT11218-6/2010), Education (NPVII 2B06077, MSM0021620806, LC06044, MSM0021620808), Industry/Trade (FR-TI2/509), and institutional funding (SVV-2010-254260507).
Authorship
Contribution: T.S. designed the experiments; K.V., M.M., and P. Basova performed RNA expression and transfection; P. Burda performed ChIP; N.C. performed expression/luciferase assays; V.P. performed trichostatin experiments; E.N. and F.S. performed FACS; J.Z., J. Kokavec, and V.K. analyzed profiling data; M.T., J. Karban, P.O., A.B., S.P., and J.M. performed patient and clinical data processing; and T.S. and J.Z. wrote and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tomas Stopka, Pathological Physiology and Center of Experimental Hematology, First Faculty of Medicine, Charles University in Prague, U Nemocnice 5, Prague 2, 12853, Czech Republic; e-mail: tstopka@lf1.cuni.cz.
References
Author notes
K.V., N.C., and P. Burda contributed equally to this study.
T.S., J.Z., and M.T. are senior coauthors.

![Figure 1. MicroRNA and mRNA expression in B-CLL. (A) Results of TaqMan PCR in B-CLL and control CD19+ cells (Ctrl). The y-axis represents relative abundance of mature miR-155 (number of B-CLL patients, N(B-CLL) = 118, number of controls, N(Ctrl) = 7) and of miR-150 (N(B-CLL) = 75, N(Ctrl) = 4) relative to RNU44; and of MYB mRNA (N(B-CLL) = 119, N(Ctrl) = 10) and PU.1 mRNA (N(B-CLL) = 119, N(Ctrl) = 5) relative to GAPDH. Data (shown as a fold change [FC]) are baseline normalized to RNA levels obtained in normal controls (control measurements were set equal to 1) and shown in log2 scale. Error bars represent SEM. P values (t test) are shown in parentheses. (B) Immunoblotting analysis of MYB and PU.1 in B-CLL (N = 5) and control (C87, N = 1) B-cell samples. The NB4 cell line was used as a positive control. The optical density (below each lane) of each protein band was measured by Scion Image software for Windows Beta 4.0.2, the values below the Western blots represent a fold change of densities of the specific bands (or their expected positions) in the patient, and control lanes normalized to the levels of the housekeeping protein β-actin. (C) The MYB mRNA levels in primary B-CLL cells positively associate with Rai staging. Three patient subgroups are shown. White bars represent currently investigated patients (OS 0) either treated by first-line therapy or untreated (Th 0–1); gray bars, currently investigated patients (OS 0) who received second or more lines of therapy (Th > 1); and black bars, previously investigated, deceased patients (OS 1) who had received at least one line of therapy (Th ≥ 1). The patient numbers in each expression assay and patient group are indicated in parentheses. Error bars represent SEM. P values (t test) are shown in parentheses on top of the brackets indicating the groups being compared. (D) RNA levels of miR-155 (white bars), PU.1 (light gray bars), MYB (dark gray bars), and miR-150 (black bars) categorized by the B-CLL prognostic parameters. Favorable outcome group = IgVH mutated, CD38 negative, ZAP70 negative. Unfavorable outcome group = IgVH unmutated, CD38+, ZAP70+. The patient numbers in each expression assay are indicated in parentheses. Error bars represent SEM. P values (t test) are shown in parentheses on top of the lines indicating the groups being compared. (E) Expression levels of miR-155 (white bars) and miR-150 (gray bars) in MYB underexpressors (< 1.5-fold decrease, left) and MYB overexpressors (> 1.5 increase; right) relative to control B cells. Patient numbers in each expression assay are indicated in parentheses. Error bars represent SEM. P values (t test) are shown in parentheses on top of the brackets indicating the groups being compared.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/14/10.1182_blood-2010-05-285064/4/m_zh89991168700001.jpeg?Expires=1763730917&Signature=dwAQsmvdMePQeEbEKXWA9Kbpa~uTDkzmbK86GMPZWJQAXoTd8ddwsVIrRU79NgXwFk2a9xdl1o5ECcDHNJ8a1fkHf2WXDGaiuJMPaT2nNztb5TqSRDd0iKckaoN2uiLUkruLrEd4iGKqJHit4xPR9V-Cu5Q7bobw6oENawxFILjZ1TItlzytSxC4G6MahUP6jjcC2wXSGEHl2ZwWZd3cDGFyXeUtmh1UoamCphh16xGp92EZPTHWnVS7CSHi4JdB7m5U8j~deS6i0~oRBbNlyQ22KRzIOzLkjBIkmHXwohmtxWs87ZP0dN5tJUbleXKzUoncudqkdMTqK3AXbVwdfA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Manipulations of MYB and miR-155 levels in B-CLL. (A) Transfection of primary B-CLL cells (P88) and Raji cell line with siRNA oligos inhibiting MYB expression (final concentrations of 30nM, gray bars; and 100nM, black bars) or with negative control oligos. The y-axis indicates quantitative PCR-determined relative levels of mRNA of indicated genes related to housekeeping gene GAPDH (for MYB, PU.1) or to RNU44 (for miR-155) 48 hours after transfection (“Transfections”). Data (shown as a fold change [FC]) are normalized to mRNA levels obtained in negative control transfection experiment (control measurements were set as baseline = 1). Error bars represent SEM. *P < .05. **P < .01. (B) Raji cells were transfected with MYB cDNA encoding expression plasmid (in 2 final concentrations: 0.2 μg/mL, gray bars; and 2.0 μg/mL, black bars) or with a negative control (pcDNA 3.1) plasmid. The y-axis: quantitative PCR-determined relative levels of mRNA miR-155 compared with the RNU44 internal control (control measurements were set as baseline = 1) 48 hours after transfection (“Transfections”). Error bars represent SEM. *P < .05. **P < .01. Immunoblotting of MYB (and β-actin) in HeLa cells at 48 hours after transfection indicates expression product of the MYB cDNA expressing plasmid. The relative optical density of each protein is indicated below the blots. (C) B-CLL cells (P161) and Raji cell line was transfected with anti–miR-155 oligos (at final concentration of 40nM, gray bars; and 100nM, black bars) and with negative control oligos. After 96 hours, the total RNA was purified, reverse transcribed, and measured by quantitative PCR as described in “RNA expression.” The y-axis indicates relative expression of mRNA of indicated genes relative to the housekeeping gene GAPDH (for PU.1) or to RNU44 (for miR-155). Data are normalized to the mRNA levels obtained in negative control transfection experiment (control measurements were set as baseline = 1). Error bars represent SEM. *P < .05. **P < .01. (D) Raji cell line was transfected with miR-155 oligos (at final concentration 40nM, gray bars; and 100nM, black bars) or with negative control oligos. After 24 hours, total RNA was purified, reverse transcribed, and measured by quantitative PCR as described in “RNA expression.” The y-axis indicates relative expression of mRNA of indicated genes relative to the housekeeping gene GAPDH (for PU.1) or to RNU44 (for miR-155). Data are normalized to the mRNA levels obtained in negative control transfection experiment (control measurements were set as baseline = 1). Error bars represent SEM. *P < .05. **P < .01.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/14/10.1182_blood-2010-05-285064/4/m_zh89991168700005.jpeg?Expires=1763730917&Signature=pePO28I2o0RY9TX5PBg--eNCrChfu3rOPCQDhV3Tf8kbKG4LyCeY~uEJJVGbcrrAU~7Yi50TXTiYfdrOak2GYZ8X7ImK4UJh-XSyuYH8n6kp4zcPAna4L4OLjmUkvvNcKdEPxXKUMdgs5dFpeqwsiVsZaCfcSq7JHBr3jqf4j7rjzr3y4xNJbT4O~RS1jYrJcJjyqup6z2OkacXvvTTdTQ~9t1cDzS0l4pDRueMvi2mWASteuFaZvFeBZ~OMsxV3C-AUJF3sv1mr-m9Odi-Pi6DzkQyG8YUWLXOpK-pwotom5pomaZPvxcLR4q-gvAT53a9TEMSojatAno9w8WLqIA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 1. MicroRNA and mRNA expression in B-CLL. (A) Results of TaqMan PCR in B-CLL and control CD19+ cells (Ctrl). The y-axis represents relative abundance of mature miR-155 (number of B-CLL patients, N(B-CLL) = 118, number of controls, N(Ctrl) = 7) and of miR-150 (N(B-CLL) = 75, N(Ctrl) = 4) relative to RNU44; and of MYB mRNA (N(B-CLL) = 119, N(Ctrl) = 10) and PU.1 mRNA (N(B-CLL) = 119, N(Ctrl) = 5) relative to GAPDH. Data (shown as a fold change [FC]) are baseline normalized to RNA levels obtained in normal controls (control measurements were set equal to 1) and shown in log2 scale. Error bars represent SEM. P values (t test) are shown in parentheses. (B) Immunoblotting analysis of MYB and PU.1 in B-CLL (N = 5) and control (C87, N = 1) B-cell samples. The NB4 cell line was used as a positive control. The optical density (below each lane) of each protein band was measured by Scion Image software for Windows Beta 4.0.2, the values below the Western blots represent a fold change of densities of the specific bands (or their expected positions) in the patient, and control lanes normalized to the levels of the housekeeping protein β-actin. (C) The MYB mRNA levels in primary B-CLL cells positively associate with Rai staging. Three patient subgroups are shown. White bars represent currently investigated patients (OS 0) either treated by first-line therapy or untreated (Th 0–1); gray bars, currently investigated patients (OS 0) who received second or more lines of therapy (Th > 1); and black bars, previously investigated, deceased patients (OS 1) who had received at least one line of therapy (Th ≥ 1). The patient numbers in each expression assay and patient group are indicated in parentheses. Error bars represent SEM. P values (t test) are shown in parentheses on top of the brackets indicating the groups being compared. (D) RNA levels of miR-155 (white bars), PU.1 (light gray bars), MYB (dark gray bars), and miR-150 (black bars) categorized by the B-CLL prognostic parameters. Favorable outcome group = IgVH mutated, CD38 negative, ZAP70 negative. Unfavorable outcome group = IgVH unmutated, CD38+, ZAP70+. The patient numbers in each expression assay are indicated in parentheses. Error bars represent SEM. P values (t test) are shown in parentheses on top of the lines indicating the groups being compared. (E) Expression levels of miR-155 (white bars) and miR-150 (gray bars) in MYB underexpressors (< 1.5-fold decrease, left) and MYB overexpressors (> 1.5 increase; right) relative to control B cells. Patient numbers in each expression assay are indicated in parentheses. Error bars represent SEM. P values (t test) are shown in parentheses on top of the brackets indicating the groups being compared.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/14/10.1182_blood-2010-05-285064/4/m_zh89991168700001.jpeg?Expires=1763730918&Signature=q7jH~elNPT-XcKiQWRKqXQYftEw0GFc1snSYprMh3f-8bf~oxRM-KpVZzOTJQhKs4oqvqf2bduly-RLrXIf4fHuLvcuyQBJbVx46xo4kgNGStQ3FXzmgbdyMskKOXMQlv6PblgEW8O62knqwBkLBT0UP6VTLVQN8gSMxg6HyfHpRSv~h3RgeJYTvGtoekQZOsawi8Pj8zCBaPIrIYQ6SXIklmYqgbdX~31n0exUURocsCrhlp20uA0W3rFhkCQK9nLhPlfUCZZ1PbqFQn6mkea1OlWBOyLRnTCVpy0Qf4Gg-xKDK3qsiJcK66xtCypbs1fmmRpp0dOsXv12XKVMzjQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Manipulations of MYB and miR-155 levels in B-CLL. (A) Transfection of primary B-CLL cells (P88) and Raji cell line with siRNA oligos inhibiting MYB expression (final concentrations of 30nM, gray bars; and 100nM, black bars) or with negative control oligos. The y-axis indicates quantitative PCR-determined relative levels of mRNA of indicated genes related to housekeeping gene GAPDH (for MYB, PU.1) or to RNU44 (for miR-155) 48 hours after transfection (“Transfections”). Data (shown as a fold change [FC]) are normalized to mRNA levels obtained in negative control transfection experiment (control measurements were set as baseline = 1). Error bars represent SEM. *P < .05. **P < .01. (B) Raji cells were transfected with MYB cDNA encoding expression plasmid (in 2 final concentrations: 0.2 μg/mL, gray bars; and 2.0 μg/mL, black bars) or with a negative control (pcDNA 3.1) plasmid. The y-axis: quantitative PCR-determined relative levels of mRNA miR-155 compared with the RNU44 internal control (control measurements were set as baseline = 1) 48 hours after transfection (“Transfections”). Error bars represent SEM. *P < .05. **P < .01. Immunoblotting of MYB (and β-actin) in HeLa cells at 48 hours after transfection indicates expression product of the MYB cDNA expressing plasmid. The relative optical density of each protein is indicated below the blots. (C) B-CLL cells (P161) and Raji cell line was transfected with anti–miR-155 oligos (at final concentration of 40nM, gray bars; and 100nM, black bars) and with negative control oligos. After 96 hours, the total RNA was purified, reverse transcribed, and measured by quantitative PCR as described in “RNA expression.” The y-axis indicates relative expression of mRNA of indicated genes relative to the housekeeping gene GAPDH (for PU.1) or to RNU44 (for miR-155). Data are normalized to the mRNA levels obtained in negative control transfection experiment (control measurements were set as baseline = 1). Error bars represent SEM. *P < .05. **P < .01. (D) Raji cell line was transfected with miR-155 oligos (at final concentration 40nM, gray bars; and 100nM, black bars) or with negative control oligos. After 24 hours, total RNA was purified, reverse transcribed, and measured by quantitative PCR as described in “RNA expression.” The y-axis indicates relative expression of mRNA of indicated genes relative to the housekeeping gene GAPDH (for PU.1) or to RNU44 (for miR-155). Data are normalized to the mRNA levels obtained in negative control transfection experiment (control measurements were set as baseline = 1). Error bars represent SEM. *P < .05. **P < .01.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/14/10.1182_blood-2010-05-285064/4/m_zh89991168700005.jpeg?Expires=1763730918&Signature=u4QwwH0ec6Gd6gwD12taB0bv~VkC8gMGT1KvjjOGKesRPa5Um7rzSKCBiW4q6gfrDLhkA0IsOw0asuD30y6EKyWuZu30ta3iPGI-GPAQLwBSgbNwapF333-fMmKiWv25z5ofSJCYQtKNHnP1gBCPo2zPLQXldmao6N0CNmUQUgfliFtQ4DfsRsh-zuc1fCyz1MfXffLQsxEr4vsHwmDIR19aJ81o1LnoyvMxmseE1AYj1lORguInVEyemsb61z6svTkLfzsmYc3Kv4VXiSK-1otGgG3EuM9PDCNUnvqohsPaMuLlqQcPH6gDCnxA4FwaaS6Mi--LyNZVb9wE~mgMow__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)