Abstract
Here we report that T-cell lymphoma cells carrying the NPM-ALK fusion protein (ALK+ TCL) frequently express the cell-stimulatory receptor ICOS. ICOS expression in ALK+ TCL is moderate and strictly dependent on the expression and enzymatic activity of NPM-ALK. NPM-ALK induces ICOS expression via STAT3, which triggers the transcriptional activity of the ICOS gene promoter. In addition, STAT3 suppresses the expression of miR-219 that, in turn, selectively inhibits ICOS expression. ALK+ TCL cell lines display extensive DNA methylation of the CpG island located within intron 1, the putative enhancer region, of the ICOS gene, whereas cutaneous T-cell lymphoma cell lines, which strongly express ICOS, show no methylation of the island. Treatment of the ALK+ TCL cell lines with DNA methyltransferase inhibitor reversed the CpG island methylation and augmented the expression of ICOS mRNA and protein. Stimulation of the ICOS receptor with anti-ICOS antibody or ICOS ligand-expressing B cells markedly enhanced proliferation of the ALK+ TCL cells. These results demonstrate that NPM-ALK, acting through STAT3 as the gene transcriptional activator, induces the expression of ICOS, a cell growth promoting receptor. These data also show that the DNA methylation status of the intronic CpG island affects transcriptional activity of the ICOS gene and, consequently, modulates the concentration of the expressed ICOS protein.
Introduction
T-cell lymphomas (TCLs) are clinically and biologically diverse with the majority derived from helper/inducer CD4+ T lymphocytes. Among TCLs, those aberrantly expressing anaplastic lymphoma kinase (ALK+ TCL) have been recognized as a distinct entity and designated by the World Health Organization classification as anaplastic large cell lymphomas, ALK-positive (ALK+). The ectopic expression of ALK is the result of chromosomal translocations fusing the ALK gene and nucleophosmin (NPM) gene or, rarely, other partner gene.1 The NPM-ALK fusion protein is not only constitutively expressed but also chronically activated through autophosphorylation. NPM-ALK is highly oncogenic and acts by activating several key cell signal transduction pathways, foremost the STAT3 pathway.2,3 Another form of TCL, cutaneous T-cell lymphoma (CTCL), originally defined as mycosis fungoides, is not only by far the most frequent type of the primary lymphoma of the skin but also the most frequent type of TCL in general.4 The early lesions of CTCL typically present as limited skin patches or plaques involving epidermis and the superficial, papillary dermis. The more advanced lesions form intradermal tumors and can undergo a large cell transformation, which almost invariably results in a highly aggressive clinical course.
ICOS (CD278) is a member of the CD28-costimulatory receptor superfamily involved in generating cell-activating signals together with the T-cell receptor (TCR)–CD3 complex.5 ICOS is mainly expressed by activated, functionally diverse CD4+ T-cell subsets and promotes their expansion. As shown in both mice and humans, expression of ICOS is critical for the proper functioning of the CD4+ T lymphocytes, including the whole spectrum of Th1, Th2, Th17, T follicular helper (Tfh), and regulatory T-cell (Treg) phenotypes.5,6
Transcriptional gene silencing resulting from DNA methylation of gene regulatory regions is a common epigenetic event in functional maturation and diversification of normal T lymphocytes as well as development of lymphoid and other types of malignancies.7,8 Silencing is mediated by members of the DNA methyltransferase family (DNMT), which induce and maintain methylation of CpG dinucleotides typically clustered within the promoters and enhancers of the affected genes forming the so-called CpG islands. The gene silencing can be reversed by DNMT inhibitors, such as 5′-aza-2′-deoxycytidine (ADC), and is typically associated in malignant cells with the re-expression of tumor suppressor protein(s) and, consequently, impaired cell growth and survival.
Here we report that ALK+ TCL/anaplastic large cell lymphoma ALK+ cells express ICOS at an intermediate concentration compared with mitogen-activated peripheral blood mononuclear cells/phytohemagglutinin (PBMCs/PHA) and CTCL cells. ICOS expression is induced by NPM-ALK that acts through STAT3, which directly activates ICOS gene transcription and indirectly enhances ICOS protein expression by inhibiting expression of miR-219, the negative regulator of ICOS translation. In contrast to CTCL cells, which strongly express ICOS, ALK+ TCL cells display methylation of the CpG island located within first intron of the ICOS gene, and ADC-induced demethylation of the island is associated with enhanced ICOS expression by ALK+ TCL cells. Finally, cross-linking of ICOS markedly increased proliferative rate of ALK+ TCL cells.
Methods
Cells and tissue samples
NPM-ALK–expressing SUDHL-1, JB6, SUP-M2, Karpas 299, SR-786, and L-82 cell lines were derived from ALK+ TCL patients.9,10 IL-2–dependent T-cell lines Sez-4 and SeAx and IL-2–independent Myla2059 and MyLa3675 were derived from CTCL, and PB-1, 2A, and 2B were derived from a patient with a primary skin CD30+ lymphoproliferative disorder.11,12 Jurkat cell line was developed from lymphoblastic T-cell lymphoma. The Epstein-Barr virus (EBV)–negative cell lines, Ly18 and Val, were derived from diffuse large B-cell lymphoma and Ramos from Burkitt lymphoma.13,14 The EBV-positive, latency type III lymphoblastoid B-cell lines (LCLs) MM and HH were established by the EBV-mediated immortalization of peripheral blood B lymphocytes.13 BCBL-1, BC-1, and JSC-1 were derived from primary effusion lymphoma and are human herpesvirus 8-positive.14 PBMCs harvested from healthy adults were isolated by Ficoll/Paque centrifugation and stimulated in vitro for 72 hours with a mitogen (PHA; Sigma-Aldrich). The cell lines were cultured at 37°C and 5% CO2 in RPMI 1640 medium, supplemented with 2mM l-glutamine, 10% heat-inactivated FBS, 1% penicillin/streptomycin mixture and, for the Sez-4 and SeAx cell lines, IL-2 (200 U/mL). ALK+ TCL tissues were from lymph nodes or extranodal tumors as excisional biopsies obtained for diagnostic purposes and used in this study under the protocol approved by the Institutional Review Board (protocol #706169). Diagnosis was established by standard morphologic and immunohistochemical criteria, including expression of CD30 and ALK proteins. For DNA isolation, snap-frozen, glass slide-deposited tissue sections were enriched for lymphoma cells by collecting the parts highly enriched in malignant cells as determined by microscopic evaluation of the hematoxylin and eosin-stained control slides.
ALK kinase and STAT3 inhibitors
ALK inhibitors CEP-26939 designated also as compound15 and CEP-14513 and its structurally related ALK noninhibitory counterpart CEP-1198816 were kindly provided by Cephalon. STAT3 inhibitor (Stattic; Santa Cruz Biotechnology) is a small-molecule selective inhibitor of STAT3 phosphorylation and dimerization.
Treatment of ALK+ TCL cells with DNMT inhibitor
The cells were treated with 0.75μM 5-ADC (Sigma-Aldrich) for up to 4 days with replenishing the culture medium with freshly prepared drug after 2 days as described.17 The cells were harvested typically at 24-hour intervals and subjected to DNA, RNA, and protein extraction.
DNA oligonucleotide array
Analysis was performed as described previously.10,11,17 In brief, total RNA from ALK+ TCL SUDHL-1 cells either untreated or treated in triplicate cultures with Stat3 siRNA or control nonspecific siRNA was reverse-transcribed, biotin-labeled, and hybridized to a U133 Plus Version 2.0 array (Affymetrix) containing the 54 000 DNA oligonucleotide probe set. The results were normalized using GeneSpring (Agilent Technologies) and further analyzed using Partek GS 6.3 Spotfire software, and GeneSpring programs.
RNA isolation and quantitative RT-PCR
Total RNA was isolated using the RNeasy Protect Mini kit (QIAGEN), according to the manufacturer's instructions. cDNA was prepared using High Capacity RNA-to-cDNA Master Mix (Applied Biosystems). Quantitative (real-time) PCR was performed using ABI PRISM 7000 Sequence Detection System (Applied Biosystems). PCR reaction components and conditions were as recommended by Applied Biosystems. TaqMan Gene Expression Assay was used for ICOS mRNA detection (Hs00359999_m1). All amplifications were performed in duplicate. The nontemplate controls were included in every run.
Western blot
DNA methylation analysis
DNA isolated using the DNeasy Tissue kit (QIAGEN) was bisulfite-modified using the CpGenome DNA Modification kit (Intergen) and amplified by PCR in 40 cycles under standard conditions using ICOS gene promoter-specific biotinylated primers (5′-TGGAATTTAGGAATTTTTTTAGGATAT-3′ and 5′-ATTAAAAACTTTCACCCAAT CTT-3′ and Platinum Taq DNA polymerase (Invitrogen) as described before.9,17 The PCR amplification products were purified and rendered single-stranded on a Pyrosequencing workstation (Pyrosequencing AB) and annealed to the sequencing primers (S1: 5′-TTATTAATTTTTTTTTTGAGG-3′; S2: 5′-AGGTTGGAGTGTAGTGG-3′; S3: 5′-AGTAGGTGAGATTATAGGTATT-3′; 5′-TTATAGTGTTAGTTAGGATGGTT-3′; S4: 5′-TTTTAGTTTTAGTTTTTTAAAGTG-3′). Quantitative DNA methylation analysis was performed on a PSQ 96MA system with the PyroGold SQA reagent kit (Pyrosequencing), and results were analyzed using the Q-CpG software (Version 1.0.9; Pyrosequencing).
Luciferase reporter assay
The ICOS promoter DNA sequence was amplified by PCR with primers 5′-TATAGGTACCCAGGCTAAGGGAAGTCCAG-3′ (sense) and 5′-ATACTCGAGGGAGGTAAAGTAGACAATAACAACAA-3′ (antisense; the italicized sequences represent nucleotides added to the complementary sequences to generate KpnI- and XhoI-specific restriction digest sites). The 528-bp PCR product was gel-purified and cloned into the pGL3-basic luciferase reporter construct (Promega) to generate the ICOS-promoter-pGL3 construct. SUDHL-1 cells were transiently transfected in duplicate with the construct using SuperFect Transfection Reagent (QIAGEN) according to the manufacturers' directions with a combination of luciferase construct and phRL (Renilla luciferase) TK plasmid (Promega), the latter serving as a measure of transfection efficiency. Twenty-four hours after transfection, the cells were washed, lysed, and sequentially evaluated for luciferase activity using the Dual-Luciferase Reporter Assay system (Promega). Luciferase activity was measured for 10 seconds after a 2-second delay using a BD Monolight 3010 luminometer (BD Biosciences). Variation in transfection efficiency was normalized by dividing the construct luciferase activity by the corresponding Renilla luciferase activity. Promoter activity is reported as the mean ± SD.
Short interfering RNA assay
We introduced a mixture of 4 ALK, STAT3, and ICOS-specific or nontargeting siRNAs (all from Dharmacon RNA Technologies) or mimic-miR-219 or miR-48 (both from ABI) into cells using Lipofectamine 2000 as described before.9
Electrophoretic mobility shift assay
Nuclear proteins were incubated with biotin-labeled DNA probe 5′-ACCTGCTTCGGTTAAGAGTGA-3′ corresponding to the ICOS gene promoter region that contains STAT3 binding sites. The proteins were separated by gel electrophoresis and transferred to membranes as described.9 Protein bands were visualized using HPR system (Thermo Scientific).
Chromatin immunoprecipitation assays
Lysates isolated from formaldehyde-fixed and sonicated cells were incubated with antibodies against STAT3 and IgG (Santa Cruz Biotechnology). DNA-protein immunocomplexes were precipitated with protein A-agarose beads and treated with RNase A and proteinase K. DNA was extracted with phenol-chloroform, precipitated with ethanol, and quantitatively PCR-amplified using primers specific for the ICOS gene promoter (5′-TGGATCTAGCATCTTGGAAGC-3′ and 5′-GAACCTGGCCCAAGTCATATAG-3′).
Flow cytometry
The cells were incubated with the phycoerythrin-conjugated anti-ICOS or control IgG antibody (both from R&D Systems) or anti-CD4 or control IgG (both from BD Biosciences) for 30 minutes at room temperature. After washing in PBS, flow cytometric analysis (FACSCalibur; BD Biosciences) was performed using the CellQuest Pro Version 6.0.3 software.
MTT enzymatic conversion assay
The cells were cultured at 37°C in microtiter plates at 2 × 104/well for up to 3 days, labeled with 10 μL 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide (MTT; Promega) at 5 mg/mL for 4 hours, and solubilized overnight with 10% SDS in 0.01M HCl. The absorbance was determined at 570 nm with a Titertek Multiskan reader.
In vitro cell proliferation (BrdU incorporation) assay
After 48 hours of culture with 100pM of siRNA or ICOS beads, cell proliferation was evaluated by detection of 5′-bromo-2′-deoxyuridine (BrdU) incorporation using the commercially available Cell Proliferation ELISA kit (Calbiochem) according to the manufacturer's protocol. In brief, the treated cells were seeded in 96-well plates (Corning) at a concentration of 2 × 104 cells/well in RPMI medium supplemented with 10% FBS and labeled with BrdU overnight. After the culture plate centrifugation (10 minutes at 300g), supernatant removal, and the plate drying, the cells were fixed and DNA denatured by adding 200 mL FixDenat reagent. The amount of incorporated BrdU was determined by incubation with a specific antibody conjugated to peroxidase followed by colorimetric conversion of the substrate and optical density evaluation using an ELISA plate reader.
Cell proliferative rate evaluation assay
CFSE-labeled ALK+ TCL cell line SUDHL-1 was cocultured for 2 days in duplicate with the unlabeled ICOSL+ HH or ICOSL− JSC B cells at the cell ratio of 1:1 and analyzed by FACS for the CFSE labeling pattern of the responder cells. In some experiments, the cocultures were performed in the presence of the anti-ICOS blocking antibody.
Human IL-10 enzyme immunoassay
The concentration of IL-10 in culture supernatants was evaluated using a Quantikine Human IL-10 kit (R&D Systems). In brief, 200 μL/well of sample or standard was added to an IL-10 antibody-precoated plate followed by replacement with 200 μL of the IL-10 antibody peroxidase conjugate followed by optical density determination.
Results
ALK+ TCL cells frequently express ICOS
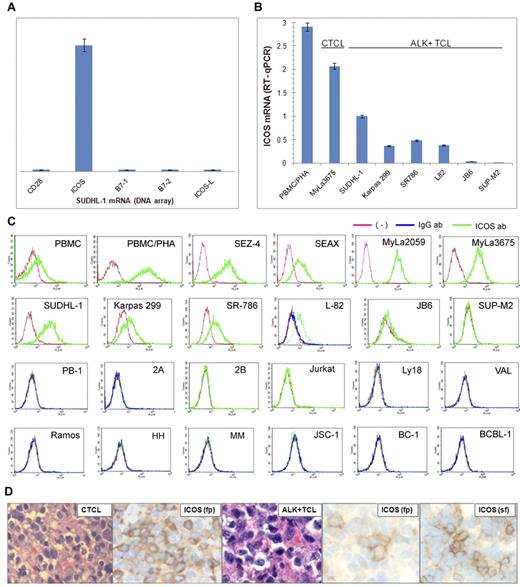
To identify gene protein products that may play a role in the pathogenesis of ALK+ TCL, we examined the ALK+ TCL-derived cell line SUDHL-1 for gene expression using DNA oligonucleotide array-based genome-scale gene expression profiling. The important receptor expressed by these cells was ICOS. ICOS expression was selective within the TCR costimulatory receptor and their ligand families because neither CD28 nor B7.1, nor B7.2, nor ICOS ligand (ICOS-L) was expressed (Figure 1A). To determine ICOS expression status in a larger pool of ALK+ TCL, we examined 6 different ALK+ TCL-derived cell lines. Quantitative RT-PCR analysis revealed that 5 of the cell lines expressed the ICOS-encoding mRNA to a variable degree (Figure 1B). This expression was weaker than in the control, mitogen-stimulated PBMCs/PHA and the CTCL-derived cell line MyLa3675. This overall moderate level of ICOS expression by ALK+ TCL cells was also observed at the protein level as determined by flow cytometry (Figure 1C). In contrast, the CTCL-derived cell lines expressed ICOS at a higher concentration that almost matched the ICOS concentration in PBMC/PHA and 3 of the 4 cell lines examined. Other cell lines, including 3 cell lines (PB-1/Mac-1, 2A/Mac-2A, and 2B/Mac-2B) derived from the progressive primary cutaneous CD30+ lymphoproliferative disorder that does not express ALK (1 line from T-cell lymphoblastic lymphoma (Jurkat) and 8 cell lines from various types of B-cell lymphoma), did not express the ICOS protein.
ICOS expression in ALK+ TCL cells. (A) The relative expression of ICOS, CD28, and their ligands by SUDH-L1 cells detected by genome-scale DNA oligonucleotide array. (B) Expression of ICOS mRNA determined by quantitative RT-PCR in the depicted ALK+ TCL cell lines compared with a CTCL cell line (MyLa3675) and mitogen-stimulated PBMCs (PBMC/PHA). (C) Expression of the ICOS protein determined by flow cytometry on ALK+ TCL- and other TCL-derived cell lines and control cell populations. (Topmost panel) Normal PBMC, PBMC/PHA, and CTCL-derived cell lines. (Top middle panel) ALK+ TCL cell lines. (Bottom middle panel) Cell lines from a progressive primary cutaneous CD30+ lymphoproliferative disorder (PB-1, 2A, and 2B), T-cell lymphoblastic lymphoma (Jurkat), and germinal center-derived diffuse large B-cell lymphoma (Ly18 and VAL). (Bottom panel) Cell lines from Burkitt lymphoma (Ramos), EBV-mediated B-cell transformation (HH and MM), and Kaposi sarcoma virus-associated body cavity lymphoma (JSC-1, BC-1, and BCBL-1). These results are representative of at least 2 independent experiments. (D) Expression of ICOS in CTCL (left panels) and ALK+ TCL (middle and right panels) tissues (original magnification ×400). Microscope: Olympus BX 40; objective: Plan Apox/0.95; camera: Leica DFC 420; software: Leica DFC Twain 6.9.0 and Microsoft Office PowerPoint 2003. Biopsy tissue sections were either formalin-fixed, paraffin-embedded (fp) or snap-frozen (sf) and stained with hematoxylin and eosin or anti-ICOS antibody.
ICOS expression in ALK+ TCL cells. (A) The relative expression of ICOS, CD28, and their ligands by SUDH-L1 cells detected by genome-scale DNA oligonucleotide array. (B) Expression of ICOS mRNA determined by quantitative RT-PCR in the depicted ALK+ TCL cell lines compared with a CTCL cell line (MyLa3675) and mitogen-stimulated PBMCs (PBMC/PHA). (C) Expression of the ICOS protein determined by flow cytometry on ALK+ TCL- and other TCL-derived cell lines and control cell populations. (Topmost panel) Normal PBMC, PBMC/PHA, and CTCL-derived cell lines. (Top middle panel) ALK+ TCL cell lines. (Bottom middle panel) Cell lines from a progressive primary cutaneous CD30+ lymphoproliferative disorder (PB-1, 2A, and 2B), T-cell lymphoblastic lymphoma (Jurkat), and germinal center-derived diffuse large B-cell lymphoma (Ly18 and VAL). (Bottom panel) Cell lines from Burkitt lymphoma (Ramos), EBV-mediated B-cell transformation (HH and MM), and Kaposi sarcoma virus-associated body cavity lymphoma (JSC-1, BC-1, and BCBL-1). These results are representative of at least 2 independent experiments. (D) Expression of ICOS in CTCL (left panels) and ALK+ TCL (middle and right panels) tissues (original magnification ×400). Microscope: Olympus BX 40; objective: Plan Apox/0.95; camera: Leica DFC 420; software: Leica DFC Twain 6.9.0 and Microsoft Office PowerPoint 2003. Biopsy tissue sections were either formalin-fixed, paraffin-embedded (fp) or snap-frozen (sf) and stained with hematoxylin and eosin or anti-ICOS antibody.
To evaluate ICOS expression in TCL tissues, we examined by immunohistochemistry formalin-fixed, paraffin-embedded tissue samples from 17 cases of CTCL representing various histologic stages of the disease and from 39 cases of ALK+ TCL. We detected ICOS expression in 9 CTCL samples (Figure 1D left panels: representative images of a positive case; supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), which contains a negative case as well as staining of control reactive lymphoid tissue. The ICOS expression was present at all stages of CTCL from patch to large cell transformation. Although 21 ALK+ TCL samples displayed ICOS staining, it was rather weak and focal (Figure 1D middle panels: representative images). Based on the cell line data showing a markedly lower ICOS expression in the ALK+ TCL cell lines compared with the CTCL cell lines (Figure 1B), we reasoned that the former may express ICOS at a concentration that is below the level of detection in formalin-fixed tissues. Indeed, analysis of snap-frozen tissues permitted detection of ICOS in 13 of the 14 ALK+ TCL cases (Figure 1D right panels: representative images).
NPM-ALK induces expression of ICOS
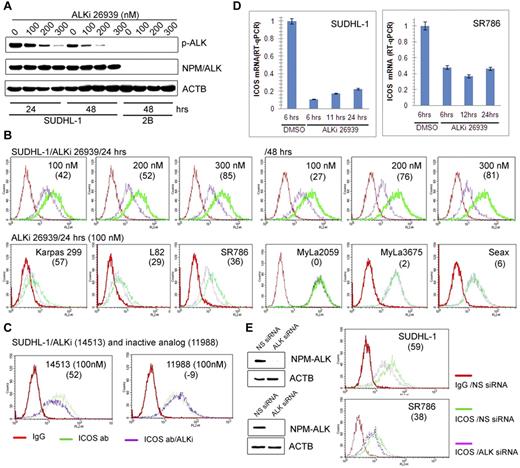
Because NPM-ALK–mediated cell transformation involves modulation of gene expression,2,3 we examined whether NPM-ALK plays a role in promoting ICOS expression. Exposure of SUDHL-1 cells to different doses of a highly specific ALK inhibitor CEP-2693915 resulted in a decrease in ALK autophosphorylation (Figure 2A) as well as ICOS protein expression (Figure 2B top panel), both of which corresponded to the inhibitor concentration. The ALK inhibitor also suppressed ICOS expression in 3 other ALK+ TCL cell lines but not in the 3 control CTCL-derived cell lines (Figure 2B bottom panel). The ICOS inhibition was specific because the expression of CD4 T-cell receptor present in one of the ALK+ TCL cell lines SR-786 remained unaffected, whereas ICOS expression decreased in a time-dependent manner (supplemental Figure 2).
ICOS expression is induced by NPM-ALK. (A) Dose- and time-dependent inhibition of NPM-ALK enzymatic activity by the small-molecule ALK inhibitor (ALKi) CEP-26939 as identified in Western blot using phospho-ALK-specific antibody in the ALK+ TCL cell line SUDHL-1 with the ALK- TCL cell line 2B serving as a negative control. (B-D) Effect of ALK inhibition on ICOS expression. The depicted ALK+ TCL cell lines were examined by flow cytometry for ICOS expression after treatment with 2 different ALK inhibitors (ALKi), CEP-2693915 (B) and CEP-14513,18 (C) at the indicated concentrations. The ALK-inactive, CEP-14153-related CEP-11988 compound22 served as a negative control. The effect of 100nM CEP-26939 on expression of ICOS mRNA was also determined (D). (E) Effect of siRNA-mediated NPM-ALK depletion on expression of ICOS in the 2 depicted ALK+ TCL cell lines. (B-E) The numbers in parentheses indicate the percentage of ICOS expression inhibition relative to controls.
ICOS expression is induced by NPM-ALK. (A) Dose- and time-dependent inhibition of NPM-ALK enzymatic activity by the small-molecule ALK inhibitor (ALKi) CEP-26939 as identified in Western blot using phospho-ALK-specific antibody in the ALK+ TCL cell line SUDHL-1 with the ALK- TCL cell line 2B serving as a negative control. (B-D) Effect of ALK inhibition on ICOS expression. The depicted ALK+ TCL cell lines were examined by flow cytometry for ICOS expression after treatment with 2 different ALK inhibitors (ALKi), CEP-2693915 (B) and CEP-14513,18 (C) at the indicated concentrations. The ALK-inactive, CEP-14153-related CEP-11988 compound22 served as a negative control. The effect of 100nM CEP-26939 on expression of ICOS mRNA was also determined (D). (E) Effect of siRNA-mediated NPM-ALK depletion on expression of ICOS in the 2 depicted ALK+ TCL cell lines. (B-E) The numbers in parentheses indicate the percentage of ICOS expression inhibition relative to controls.
As even the most specific kinase domain inhibitors display cross-reactivity with other kinases, we exposed SUDHL-1 cells to another, structurally different ALK inhibitor as well as its ALK-inactive derivative, both of which have been described in detail previously.11,16 As can be seen (Figure 2C), the active inhibitor CEP-14153 suppressed ICOS expression, but the inactive variant CEP-11988 did not. To document that NPM-ALK inhibition affects ICOS mRNA, we treated 2 ALK+ TCL cell lines SUDHL-1 and SR-786 with the selective ALK inhibitor CEP-26939 for up to 24 hours and examined the mRNA expression by quantitative RT-PCR. As shown in Figure 2D, NPM-ALK inhibition markedly decreased ICOS mRNA at all evaluated time points. Finally, to provide additional evidence that NPM-ALK induces ICOS expression, we depleted NPM-ALK in ALK+ TCL cell lines using siRNA. As shown in Figure 2E, the depletion diminished expression of both NPM-ALK protein (left panels) and ICOS protein (right panels) in both cell lines. In a similar experiment, the siRNA-mediated NPM-ALK depletion suppressed expression of not only of NPM-ALK and ICOS proteins but also of ICOS mRNA (supplemental Figure 3).
NPM-ALK induces ICOS expression through STAT3
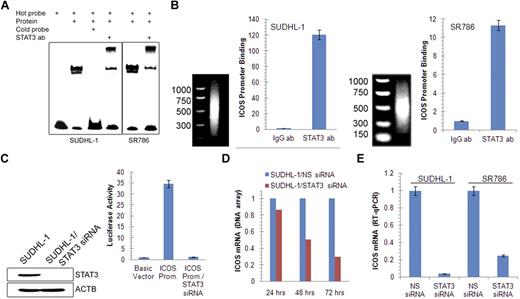
Accumulating evidence indicates that STAT3 is the main effector of NPM-ALK–mediated oncogenesis through its capacity to modulate gene expression.3 We therefore examined whether STAT3 is involved in the induction of ICOS expression. Sequence analysis of the ICOS promoter identified a potential STAT3 binding site and an electromobility shift assay with a 23-mer DNA oligonucleotide corresponding to the identified site showed binding of the probe to proteins isolated from 2 ALK+ TCL cell lines (Figure 3A). The protein binding was specific as demonstrated by abrogation of binding on addition of an unlabeled (cold) probe. The band was also specific for STAT3 as established in the electromobility shift assay super-shift assay in which preincubation of the ALK+ TCL cell line protein extract with the STAT3-specific antibody resulted in formation of the “shifted,” slower-migrating band.
STAT3-mediated induction of ICOS gene expression. (A) Binding of STAT3 to the ICOS gene promoter in vitro detected by electromobility shift assay in the 2 depicted ALK+ TCL cell lines. (B) Binding of STAT3 to the endogenous ICOS gene promoter in vivo detected by chromatin immunoprecipitation. (C) Impact of siRNA-mediated depletion of STAT3 on ICOS gene promoter activity detected by the luciferase gene reporter assay. (D) Kinetics of STAT3 siRNA-mediated inhibition of ICOS mRNA expression detected by a genome-scale DNA oligonucleotide array. The results are depicted as fold increase in miR-21 mRNA expression in the STAT3 siRNA-treated compared with the control, non-sense siRNA (NS siRNA)–treated cells. (E) STAT3 siRNA depletion-induced decrease in ICOS mRNA expression detected by quantitative RT-PCR.
STAT3-mediated induction of ICOS gene expression. (A) Binding of STAT3 to the ICOS gene promoter in vitro detected by electromobility shift assay in the 2 depicted ALK+ TCL cell lines. (B) Binding of STAT3 to the endogenous ICOS gene promoter in vivo detected by chromatin immunoprecipitation. (C) Impact of siRNA-mediated depletion of STAT3 on ICOS gene promoter activity detected by the luciferase gene reporter assay. (D) Kinetics of STAT3 siRNA-mediated inhibition of ICOS mRNA expression detected by a genome-scale DNA oligonucleotide array. The results are depicted as fold increase in miR-21 mRNA expression in the STAT3 siRNA-treated compared with the control, non-sense siRNA (NS siRNA)–treated cells. (E) STAT3 siRNA depletion-induced decrease in ICOS mRNA expression detected by quantitative RT-PCR.
To demonstrate that STAT3 binds to the ICOS promoter also in live, intact ALK+ TCL cells, we performed chromatin precipitation assay using STAT3 antibody and PCR primers specific for the ICOS promoter. Binding of the STAT3 antibody to the promoter was at least 10-fold higher than binding of control IgG as determined by quantitative PCR in both ALK+ TCL cell lines examined (Figure 3B).
To provide direct functional evidence that STAT3 activates the ICOS promoter, we performed a luciferase reporter assay (Figure 3C). On transfection into SUDHL-1 cells that constitutively express activated STAT3, the ICOS promoter construct displayed 35-fold higher activity than the control construct containing only the luciferase gene. Concomitant depletion of STAT3 by siRNA caused reduction in promoter activity to the control baseline level. Furthermore, siRNA-mediated depletion of STAT3 resulted in a gradual decrease in ICOS mRNA as determined by DNA oligonucleotide array-based genome-scale gene expression profiling on one ALK+ TCL cell line (Figure 3D) and confirmed by quantitative RT-PCR in 2 other cell lines (Figure 3E). Similar results were obtained using a direct STAT3 inhibitor (supplemental Figure 4). Of note, the inhibitor suppressed ICOS mRNA expression also in a CTCL cell line MyLa2059, suggesting that STAT3 role in ICOS regulation is not limited to the ALK+ TCL cells.
STAT3 enhances ICOS expression by inhibiting expression of miR-219
Analysis of ICOS mRNA identified a putative binding site for miR-219 in its 3′ untranslated region (Figure 4A). Furthermore, several potential STAT3 binding sites are present in the miR-219 gene region (Figure 4B). Analysis of miR-219 expression in the large set of TCL cell lines and mitogen-activated normal PBMCs (Figure 4C) showed an inverse correlation with ICOS expression (Figure 1B-C), strongly suggesting that miR-219 indeed negatively regulates ICOS. To determine whether STAT3 is capable of regulating miR-219 expression, we performed miR-219-specific quantitative RT-PCR after treating SUDHL-1 cells with either STAT3-specific or control siRNA. STAT3 depletion markedly increased miR-219 expression in 2 ALK+ TCL cell lines (Figure 4D). In turn, transduction of SUDHL-1 cells with a miR-219 mimic but not the control miR-48 mimic diminished ICOS expression (Figure 4E), indicating that miR-219 does target ICOS.
STAT3 inhibits miR-219 expression and miR-219 inhibits ICOS expression. (A) Schematic representation of the miR-219 binding site in 3′-untranslated region of ICOS mRNA and the sequence complementarity of miR-219 and the ICOS 3′-untranslated region target sequence. (B) Schematic depiction of miR-219 gene and molecular structure. The potential STAT3 binding sites in the miR-219 gene 3′ region, including the promoter, are indicated by arrows. (C) miR-219 expression in the depicted T-cell lymphoma lines and PHA-stimulated PBMCs detected by quantitative RT-PCR. (D) STAT3 siRNA-induced miR-219 expression detected by quantitative RT-PCR. (E) Inhibitory effect of miR-219 mimic on ICOS expression by SUDHL-1 cells detected by quantitative RT-PCR. Cell treatment with miR-48 mimic was used as control. The number in parentheses indicates the percentage of ICOS expression inhibition by miR-219 mimic relative to the miR-48 mimic control.
STAT3 inhibits miR-219 expression and miR-219 inhibits ICOS expression. (A) Schematic representation of the miR-219 binding site in 3′-untranslated region of ICOS mRNA and the sequence complementarity of miR-219 and the ICOS 3′-untranslated region target sequence. (B) Schematic depiction of miR-219 gene and molecular structure. The potential STAT3 binding sites in the miR-219 gene 3′ region, including the promoter, are indicated by arrows. (C) miR-219 expression in the depicted T-cell lymphoma lines and PHA-stimulated PBMCs detected by quantitative RT-PCR. (D) STAT3 siRNA-induced miR-219 expression detected by quantitative RT-PCR. (E) Inhibitory effect of miR-219 mimic on ICOS expression by SUDHL-1 cells detected by quantitative RT-PCR. Cell treatment with miR-48 mimic was used as control. The number in parentheses indicates the percentage of ICOS expression inhibition by miR-219 mimic relative to the miR-48 mimic control.
Methylation of the ICOS gene CpG island in ALK+ TCL cells
Evaluation of the ICOS gene structure identified the presence of a CpG island within intron 1 (Figure 5A), the region that frequently contains gene enhancer. The island showed a distinct methylation pattern that was highly TCL-type specific and inversely correlated with the degree of ICOS expression (Figure 5B). All 6 ALK+ TCL cell lines that express ICOS in the moderate to nondetectable manner (Figure 1B top panel) showed overall a high degree of CpG island methylation (Figure 5B top middle panel). Similarly, 2 cell lines (2A and 2B) derived from cutaneous ALK-CD30+ lymphoproliferative disorder that do not express ICOS (Figure 5B bottom panel) also displayed a high degree of CpG island methylation (Figure 5B bottom middle panel). In contrast, 4 CTCL-derived cell lines that express ICOS at a strong to moderate level (Figure 1B top panel) displayed no or little methylation of the island (Figure 5B bottom panel).
Impact of CpG island DNA methylation on ICOS expression. (A) Schematic diagram of the ICOS gene structure depicting the position of CpG island within intron 1 of the gene. (B) Patterns of CpG island methylation in various TCL-derived cell lines. (Top panel) ALK+ TCL-derived cell lines. (Bottom panel) Cell lines from cutaneous CD30+ lymphoproliferative disorder (2A and 2B) and CTCL (Sez-4, SeAx, MyLa2059, and MyLa3675). (C) Changes in ICOS gene CpG island methylation induced in SUDHL-1 cells by treatment with ADC for 0, 48, and 88 hours. (D) Increase in ICOS mRNA expression in ALK+ TCL cell lines treated with ADC for 96 hours detected by quantitative RT-PCR. (E) Flow cytometry–detected increase in ICOS protein expression in the depicted ALK+ TCL cell lines treated with ADC for 96 hours.
Impact of CpG island DNA methylation on ICOS expression. (A) Schematic diagram of the ICOS gene structure depicting the position of CpG island within intron 1 of the gene. (B) Patterns of CpG island methylation in various TCL-derived cell lines. (Top panel) ALK+ TCL-derived cell lines. (Bottom panel) Cell lines from cutaneous CD30+ lymphoproliferative disorder (2A and 2B) and CTCL (Sez-4, SeAx, MyLa2059, and MyLa3675). (C) Changes in ICOS gene CpG island methylation induced in SUDHL-1 cells by treatment with ADC for 0, 48, and 88 hours. (D) Increase in ICOS mRNA expression in ALK+ TCL cell lines treated with ADC for 96 hours detected by quantitative RT-PCR. (E) Flow cytometry–detected increase in ICOS protein expression in the depicted ALK+ TCL cell lines treated with ADC for 96 hours.
DNA methylation partially suppresses transcription of the ICOS gene
To assess the impact of CpG island methylation on the transcriptional activity of the ICOS gene, we treated ALK+ TCL cell lines with the DNMT inhibitor ADC. As shown in Figure 5C depicting the SUDHL-1 cell line, the treatment diminished CpG island methylation. More importantly, it led to a profound increase in expression of ICOS mRNA, as determined by quantitative RT-PCR (Figure 5D) and ICOS protein, as detected by flow cytometry (Figure 5E).
Engagement of ICOS enhances proliferation of ALK+ TCL cells
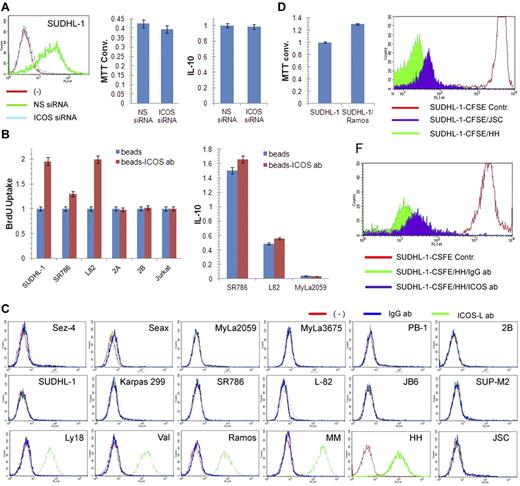
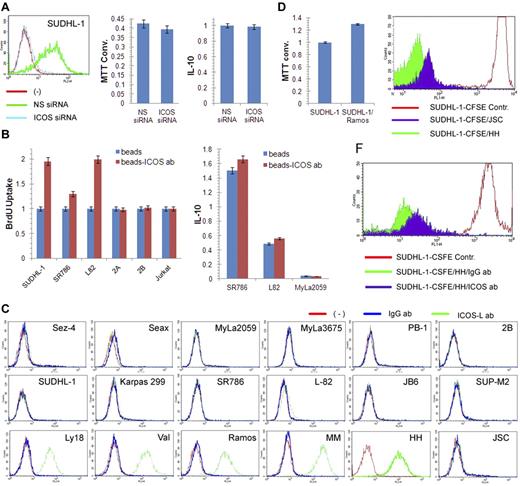
Given the known stimulatory nature of ICOS, we examined its role in the growth of ALK+ TCL cells. However, siRNA-mediated depletion of ICOS did not affect cell growth or synthesis of IL-10 (Figure 6A), the cytokine reported to be associated with ICOS expression in some experimental systems.5 In contrast, cross-linking of ICOS with bead-immobilized anti-ICOS antibody5,19 markedly enhanced proliferation of 3 ICOS-expressing ALK+ TCL cell lines but not the 3 control, ICOS-negative TCL cell lines. The cross-linking had minimal, if any, augmenting effect on IL-10 synthesis (Figure 6B).
ICOS cross-linking induced increase in proliferation of ALK+ TCL cells. (A) Lack of impact of siRNA-mediated ICOS depletion (left panel) on growth of SUDHL-1 cells as detected by MTT enzymatic conversion assay (middle panel) and IL-10 synthesis as determined by enzyme immunoassay (right panel). (B) Effect of beads-coupled ICOS antibody on BrDU uptake-detected proliferative rate of ALK+ TCL cells (left panel) and enzyme immunoassay–detected IL-10 synthesis (right panel) with uncoated beads serving as negative control in both assays. (C) Expression of ICOS-L protein as determined by flow cytometry in the transformed T- and B-cell lines and control cell populations. (Top panel) Cell lines derived from CTCL (Sez-4, SeAx, MyLa2059, and MyLa3675) and cutaneous CD30+ lymphoproliferative disorder (PB-1 and 2B). (Middle panel) ALK+ TCL cell lines. (Bottom panel) B-cell lines from the germinal center–derived diffuse large B-cell lymphoma (Ly18 and VAL). (Bottom panel) Cell lines from the Burkitt lymphoma (Ramos), EBV-mediated transformation (MM and HH), and Kaposi sarcoma virus-associated body cavity lymphoma (JSC-1). (D) Effect of coculture of ALK+ TCL SUDHL-1 cells with irradiated ICOS-L+ Ramos B cells on growth of SUDHL-1 cells as determined by the MTT enzymatic conversion assay. (E) Effect of coculture of CFSE-labeled SUDHL-1 cells with ICOS-L+ HH or ICOS-L− JSC B cells on proliferation of SUDHL-1 cells as determined by FACS for the cell CSFE labeling pattern. (F) Impact of the blocking anti-ICOS antibody on proliferation of the CFSE-labeled SUDHL-1 cells cocultured with ICOS-L+ HH.
ICOS cross-linking induced increase in proliferation of ALK+ TCL cells. (A) Lack of impact of siRNA-mediated ICOS depletion (left panel) on growth of SUDHL-1 cells as detected by MTT enzymatic conversion assay (middle panel) and IL-10 synthesis as determined by enzyme immunoassay (right panel). (B) Effect of beads-coupled ICOS antibody on BrDU uptake-detected proliferative rate of ALK+ TCL cells (left panel) and enzyme immunoassay–detected IL-10 synthesis (right panel) with uncoated beads serving as negative control in both assays. (C) Expression of ICOS-L protein as determined by flow cytometry in the transformed T- and B-cell lines and control cell populations. (Top panel) Cell lines derived from CTCL (Sez-4, SeAx, MyLa2059, and MyLa3675) and cutaneous CD30+ lymphoproliferative disorder (PB-1 and 2B). (Middle panel) ALK+ TCL cell lines. (Bottom panel) B-cell lines from the germinal center–derived diffuse large B-cell lymphoma (Ly18 and VAL). (Bottom panel) Cell lines from the Burkitt lymphoma (Ramos), EBV-mediated transformation (MM and HH), and Kaposi sarcoma virus-associated body cavity lymphoma (JSC-1). (D) Effect of coculture of ALK+ TCL SUDHL-1 cells with irradiated ICOS-L+ Ramos B cells on growth of SUDHL-1 cells as determined by the MTT enzymatic conversion assay. (E) Effect of coculture of CFSE-labeled SUDHL-1 cells with ICOS-L+ HH or ICOS-L− JSC B cells on proliferation of SUDHL-1 cells as determined by FACS for the cell CSFE labeling pattern. (F) Impact of the blocking anti-ICOS antibody on proliferation of the CFSE-labeled SUDHL-1 cells cocultured with ICOS-L+ HH.
To confirm the pro-proliferative activity of ICOS, we cultured ALK+ TCL cells with cells expressing ICOS-L. As shown in Figure 6C, none of the TCL cell lines expressed ICOS-L regardless of the lymphoma type. In contrast, all but one B-cell line strongly expressed the protein. Coculture of the ALK+ TCL cell line SUDH-L1 with an irradiated, ICOS-L–expressing B-cell line Ramos enhanced growth of the former (Figure 6D). Similarly, coculture of the labeled ALK+ TCL cells with ICOS-L–expressing HH B cells enhanced their proliferative rate to much higher degree than coculture with ICOS-L-negative JSC B cells (Figure 6E). Of note, the stimulatory capacity of the ICOS-L–positive HH cells was markedly inhibited by the soluble, blocking anti-ICOS antibody (Figure 6F).
Discussion
Here we report that ALK+ TCL cells frequently express ICOS at a moderate level compared with CTCL cells. Furthermore, expression of ICOS is induced by NPM-ALK thanks to its ability to activate STAT3; STAT3 transcriptionally activates the ICOS gene and suppresses expression of the ICOS inhibitor miR-219. Furthermore, the degree of ICOS expression strictly correlates with DNA methylation of the putative enhancer region within intron 1 of the ICOS gene. Finally, although ICOS depletion has no effect on IL-10 synthesis and cell growth, cross-linking of the receptor with bead-immobilized ICOS-specific antibody substantially enhanced cell proliferation but did not modulate IL-10 production.
Since its discovery more than a decade ago,20 ICOS has been studied extensively in normal immune cells, foremost in murine CD4+ T lymphocytes.5 It is clear that this coactivating receptor plays a key role in expansion of essentially all functional subsets of CD4+ T cells. Whereas original studies implicate ICOS in Th2 differentiation, later reports document its important role in Th1, Th17, Tfh, and Treg cells.5,6,19 Although it is not easy to fit ALK+ TCL into a specific functional CD4+ T-cell subset considering the sometime overlapping features of the subsets and malignant, aberrant phenotype of the ALK+ TCL cells, it can be argued that the lymphoma cells resemble type 1 subcategory of Treg cells because of the production of large amounts of IL-10 and lack of expression of FoxP3.12,21 Of note, ALK+ TCL also express an immunosuppressive cell-surface protein PD-L110 ; but in contrast to IL-10 and FoxP3 expression, PD-L1 has not so far been associated with any given category of normal Treg cells.
Our previous study identified strong ICOS expression in lymphomas derived from Tfh cells,22 but not other types of T- and B-cell lymphomas using formalin-fixed tissues. In this study, we show that CTCL, a lymphoma type not evaluated previously, expresses ICOS in abundance expanding the spectrum of T-cell lymphomas strongly expressing this costimulatory cell-surface receptor. Furthermore, we show that ICOS expression, albeit at a much lower concentration, is frequently detected in ALK+ TCL, particularly when frozen rather than formalin-fixed tissues are studied. Therefore, it is possible that relatively low, yet functionally relevant (Figure 6) ICOS expression may be present also in other types of lymphoma. Of note, although ICOS acts as a costimulatory receptor that amplifies the signal generated by the antigen-specific TCR in normal T cells, ALK+ TCL do not express the TCR complex18 and yet increase their growth rate in response to ICOS stimulation (Figure 6). The previously reported capacity of NPM-ALK to substitute for TCR signals23 is the most likely explanation for this phenomenon.
Several transcription factors have been implicated so far in regulating expression of ICOS in murine T lymphocytes, including NFATc2 and ERK, and have been shown to bind independently to the ICOS promoter and to up-regulate its transcription.24,25 Strikingly, the combination of the ICOS transcriptional activators appears to be strictly dependent on the T-cell phenotype. Whereas in Th1 cells, NFATc2 cooperates with the Th1-defining factor T-bet, in Th2 it interacts with GATA-3.25 In turn, the Th2-associated IRF4 transcription factor binds to ICOS gene promoter in Treg cells targeting Th2 cells.26 Given the critical role of STAT3 in the Th17 cells as well as Th17-suppressing Treg cells, it is probable that STAT3, identified by us as the transcriptional activator of the ICOS gene in ALK+ TCL (Figure 3), controls ICOS in these and possibly other functional types of normal CD4+ T lymphocytes.
We also found that miR-219 partially inhibits ICOS expression and that its gene is targeted by STAT3 (Figure 4). This latter observation indicates that STAT3 fosters ICOS expression on 2 different levels by simultaneously acting as transcriptional inhibitor of the miR-219 gene in addition to directly activating transcription of the ICOS gene (Figure 3). A recent study has demonstrated that another member of the miR family, miR-155, is expressed in normal Treg cells in a manner dependent on activity of the FoxP3 transcription factor.27 miR-155 targets the negative regulator of IL-2 signaling, SOCS1 in the Treg cells, and, consequently, augments their response to the cytokine. Even more recently, it was found that another miR, miR-146a, impacts Th1-directed suppression of Treg cells, probably by inhibiting expression of STAT1.28 In light of these reports, our finding implicating miR-219 in modulating expression of ICOS strongly suggests that miRs play an important role in immune responses by regulating expression of diverse and probably many key immunomodulatory proteins.
We found that CTCL cells strongly express ICOS and ALK+ TCL comparatively weakly (Figure 1). This differential expression is reminiscent of the “high” versus “low” ICOS expression dichotomy described for subsets of normal CD4+ T lymphocytes, with the former being enriched in cells with a Treg phenotype, at least in the evaluated anti-CTL4 antibody-induced experimental colitis model.29 It would be very interesting to determine whether the association between the degree of ICOS expression and the methylation status of the CpG island in the intron 1 of the ICOS gene (Figure 5) also occurs in normal immune CD4+ T lymphocytes. The strict correlation between lymphoma type and methylation as well as the tendency of the CTCL cells to express the Treg-associated FoxP3 transcription factor12 (Zhang et al, manuscript in preparation), strongly suggest that this indeed may be the case.
Our findings identify ICOS as a potential therapeutic target in ALK+ TCL and other T-cell lymphomas (Figure 1).19 In principle, administration of a blocking, nonagonistic anti-ICOS antibody may prove beneficial in treatment of lymphoma patients. Although concomitant inhibition of normal immune response against the lymphoma cells may be an undesired side effect of ICOS blockade, the functionally differentiated immune cells may be able to circumvent ICOS inhibition using CD28 or other costimulatory receptors. Furthermore, administration of the antibody may be combined with infusion of effector T cells expanded in vitro,19 given that ICOS is required for expansion of the effector T cells, rather than execution of their function. Alternatively, signaling pathways controlling ICOS gene expression may be targeted. Indeed, NPM-ALK and the oncogenic forms of ALK emerge as a very attractive therapeutic target for the direct small-molecule inhibitors in ALK+ TCL and other ALK-driven malignancies. Treatment with ALK inhibitor used as a single agent was effective in 90% of patients with ALK+ lung carcinoma30 ; a similar clinical trial is ongoing in ALK+ TCL. However, the observed responses were typically either partial tumor regressions or lack of the disease progression, and only 1 of the 82 treated patients reached the complete remission.30 Furthermore, the development of resistance to ALK inhibitor have already been noted.31 Therefore, combination therapies targeting not only ALK but also directly ICOS and other growth-promoting molecules may prove the most effective and potentially curative for ALK+ TCL and other ALK+ malignancies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by National Cancer Institute grants R01-CA89194 and R01-CA96856.
National Institutes of Health
Authorship
Contribution: Q.Z. designed and performed research and analyzed data; H.Y.W. analyzed genomic data and performed research; K.K., J.C.P., and X.L. performed research; A.S. and C.P. contributed vital new reagents; M.C.M., S.T., and T.M. designed research; N.O. contributed vital new reagents and designed research; and M.A.W. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mariusz A. Wasik, University of Pennsylvania Medical Center, Department of Pathology and Laboratory Medicine, 413 Stellar-Chance Bldg, 422 Curie Blvd, Philadelphia, PA 19104; e-mail: wasik@mail.med.upenn.edu; and Qian Zhang, University of Pennsylvania Medical Center, Department of Pathology and Laboratory Medicine, 413 Stellar-Chance Bldg, 422 Curie Blvd, Philadelphia, PA 19104; e-mail: qian2@mail.med.upenn.edu.