Abstract
Cytogenetically normal acute myeloid leukemia (CN-AML) compose between 40% and 50% of all adult acute myeloid leukemia (AML) cases. In this clinically diverse group, molecular aberrations, such as FLT3-ITD, NPM1, and CEBPA mutations, recently have added to the prognostic accuracy. Aberrant DNA methylation is a hallmark of cancer, including AML. We investigated in total 118 CN-AML samples in a test and a validation cohort for genome-wide promoter DNA methylation with Illumina Methylation Bead arrays and compared them with normal myeloid precursors and global gene expression. IDH and NPM1 mutations were associated with different methylation patterns (P = .0004 and .04, respectively). Genome-wide methylation levels were elevated in IDH-mutated samples (P = .006). We observed a negative impact of DNA methylation on transcription. Genes targeted by Polycomb group (PcG) proteins and genes associated with bivalent histone marks in stem cells showed increased aberrant methylation in AML (P < .0001). Furthermore, high methylation levels of PcG target genes were independently associated with better progression-free survival (odds ratio = 0.47, P = .01) and overall survival (odds ratio = 0.36, P = .001). In summary, genome-wide methylation patterns show preferential methylation of PcG targets with prognostic impact in CN-AML.
Introduction
Malignant transformation is a complex process, partly driven by genetic lesions, facilitated by environmental, inheritable, and most likely also immunologic factors.1 In addition, it is now widely recognized that epigenetic changes are an integral part of the process of neoplastic transformation and clonal evolution, causing aberrant gene expression.2 Epigenetic regulation include mRNA modulation by noncoding RNAs, various chromatin modifications such as histone tail acetylation, methylation, phosphorylation, ubiquitination, and sumoylation as well as promoter DNA methylation and global demethylation.3,4
Acute myeloid leukemia (AML) is the most prevalent type of acute leukemia in adults. The mainstay of treatment ever since the 1970s is a combination of cytarabine and anthracyclines. The major reason for the improved treatment results in AML seen over the past 40 years is the refined use of allogeneic transplantation by both better selection of candidate patients and better treatment protocols, including reduced intensity conditioning regimens and improved supportive care.5 Risk stratification of AML patients as a selection tool for intensified treatment, including allogeneic stem cell transplantation, is thus crucial. Current stratification protocols include chromosomal lesions validated in large cohorts of patients,6 and recently, the emergence of molecular diagnostics, such as NPM1, FLT3, and CEBPA mutational analysis.7 Molecular diagnostics are especially important in the large group of AML with normal karyotype composed of approximately 45% of all adult AMLs with a very heterogeneous prognosis. Furthermore, understanding the molecular mechanisms behind hematologic malignancies facilitates the development of new classes of targeted drugs exemplified by FLT3 inhibitors in leukemia and JAK2 inhibitors in myeloproliferative diseases.
Epigenetic changes in AML, although extensively studied on individual gene level and to a much lesser extent on global level, have not yet found their place in risk stratification and prognostication. In part, this may be explained by the lack of standardized methodology in the epigenetic field. Even so, clinical studies with presumably epigenetically acting drugs, such as DNA methyltransferase inhibitors and histone deacetylase inhibitors, are ongoing.
Thus, the need for increased knowledge of epigenetic mechanisms in AML is substantial, especially in cytogenetically normal (CN) AML. In this study, we used a state-of-the-art platform to evaluate a test cohort of 58 well-characterized CN-AML patients for DNA methylation, correlating our findings with molecular parameters, clinical outcomes, gene expression, and genome-wide studies of chromatin marks. Furthermore, we validated our findings in a separate cohort of 60 CN-AMLs. We show that aberrant promoter methylation is of prognostic relevance and, importantly, correlate this to genes associated with Polycomb group proteins (PcG) and bivalent chromatin marks in stem cells.
Methods
Patients, samples, and molecular analyses
Primary bone marrow samples from 58 de novo CN-AML patients were obtained at diagnosis from patients at the Karolinska University Hospital, Huddinge, Uppsala University Hospital and Örebro University Hospital. Informed consent was obtained in accordance with the Declaration of Helsinki, and the study was approved by the regional institutional review boards. All patients were treated with standard induction regimens containing cytarabine and anthracycline, according to the national AML guidelines. All patients were eligible for consolidation treatment, including allogeneic stem cell transplantation. Clinical characteristics of all patients are shown in Table 1. A validation cohort of 60 de novo CN-AML patients was set up. Thirty-one of these patients were diagnosed and analyzed in Angers, France and the remaining 29 were diagnosed Ulm, Germany and in the Swedish centers mentioned previously. The clinical characteristics are shown in supplemental Table 1. DNA was extracted after mononuclear cell isolation (Lymphoprep; Axis-Shield PoC). Chromosome banding was done using standard laboratory techniques. Mutation analyses of FLT3 (internal tandem duplications [FLT3-ITD] and tyrosine kinase domain [FLT3-TKD] mutations at codon D835 and I836), NPM1, CEBPA, IDH1, and IDH2 (exon 4) were performed as previously described.7,8 Standard diagnostic and remission criteria were used.9 BM from healthy donors (n = 9) was separated for mononuclear cells by Lymphoprep (Axis-Shield). BM CD34+ cells were further separated by MACS, indirect CD34 microbead kit (Miltenyi Biotec), according to the manufacturer's instructions. In 5 cases, the common myeloid progenitor (CMP) and granolucyte-macrophage progenitor (GMP) cells were purified from the CD34+ pool with FACS, using antibodies specified in supplemental Table 2 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article) as previously described,10 and CD34+ cells were used in 4.
Methylation analysis
Genome-wide DNA methylation profiling was performed using the Illumina Infinium HumanMethylation27 BeadChip (Illumina) in the whole test cohort and in 31 of 60 cases in the validation cohort. Twenty-nine cases were analyzed with the Illumina HumanMethylation 450K array where only the 25 978 CpG sites overlapping with the 27k array were considered. To exclude sources of technical bias, we excluded all CpG sites with detection P values > .01 in > 5 samples; n = 126. Furthermore, a systematic bias dependent on sex was found: why only probes on autosomes were used for further analyses except for validation purposes. The analyses were performed at the BEA core facility at Karolinska Institute and the genomics core facility in Angers, France. The EZ DNA methylation kit (Zymo Research) was used for bisulfite conversion of 500 ng of DNA, and the remaining assay steps were performed as previously published, using Illumina-supplied reagents and conditions.11 The readout from the array is a β-value, which is defined as the ratio between the fluorescent signal from the methylated allele to the sum of both methylated and unmethylated allele and thus correlates to the level of DNA methylation. A β-value of 1.0 corresponds to complete methylation and 0 is equal to no DNA methylation. The array data in this publication have been deposited in NCBI's Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE32252.
Bisulfite pyrosequencing was performed for the CDKN2A, CDH1, HIC1, and CDKN2B promoter. After bisulfite conversion pyrosequencing was performed on the PyroMark12 platform (QIAGEN) as previously described.12 Primer sequences are available at: www.techsupport.pyrosequencing.com. Samples were considered methylated at mean levels of more than 15% methylation.
Global DNA methylation was quantified by luminometric assay (LUMA) as previously described.13 A total of 500 ng genomic DNA was cleaved with HpaII plus EcoRI or MspI plus EcoRI in separate reactions. After the digestion step, the extent of cleavage was quantified by pyrosequencing. HpaII and MspI are methylation-sensitive isoschizomers. DNA methylation was defined as 1 − (HpaII/MspI ratio); fully methylated DNA gives a ratio that approaches 1, whereas if methylation is completely absent, the ratio approaches 0. The assay was carried out in duplicate.
Gene expression
Preparation of mRNA was performed using the RNeasy kit (QIAGEN) according to instructions, and quality was controlled using an Agilent Bioanalyzer (Agilent Technologies). Whole genome expression arrays were performed by the BEA core facility using the Illumina HumanHT-12 Version 4 Expression BeadChip using the Direct Hybridization Assay and iScan system (Illumina).
Statistics
Statistical analysis of the methylation data was carried out in the statistical computing language R (www.r-project.org). To search for the differentially methylated genes between the different prognostic subgroups, the data were arcsin transformed and an empirical Bayes moderated t test was then applied,14 using the “limma” package.15 The P values were adjusted using the method of Benjamini and Hochberg,16 and a level of P < .05 was used as a cut-off. An additional filter for the average β-geometric difference was also applied to assure only genes with absolute differences of > 0.10 β between the groups remained. Integration of gene expression array data with methylation array data and integration of previously published ChIP-chip results were made with the BeadStudio Version 3.2 software (Illumina). Unsupervised hierarchical clustering analysis and principal components analysis were performed with the Genesis 1.7.6 software package.17 Hierarchical clustering analysis distance was measured using Pearson correlation with a complete clustering algorithm. Samples were normalized using a global mean centering method. Methylation levels of different molecular subgroups were measured as continuous variables. Hypermethylation of individual CpG sites was defined as β-values > 0.7, unmethylated as β < 0.3, and values in between were called “methylated.”
Survival data are presented using the Kaplan-Meier method and compared with the log-rank test. Logistic regression was used for univariate and multivariate analysis of complete remission (CR) rates and Cox regression for survival. For multivariate analyses, both the test and validation cohorts were pooled. Odds ratios (ORs) are shown with 95% CIs. Correlations were calculated using Spearman ρ or Pearson correlation as indicated. Comparisons between 2 groups of continuous parameters were made using the Student t test or Mann-Whitney as appropriate and categorical data compared with χ2 or Fisher exact test. ANOVA was used for multiple group comparisons with the Tukey post-hoc test if not otherwise indicated. Statistical calculations were performed with the PASW Version 18.0 software (SPSS). Gene ontologic analyses were performed using the DAVID functional annotation tool (www.david.abcc.ncifcrf.gov). All P values are 2-sided.
Results
Validation of the Illumina methylation array
Bisulfite pyrosequencing of the HIC1, CDH1, CDKN2B, and CDKN2A promoter was performed in 14 to 17 samples. The β-values were higher for methylated (> 15%) than unmethylated genes in all samples (P = .01, .06, and .03 for HIC1, CDH1, and CDKN2B, respectively; Figure 1A). For CDKN2A, all AML samples were unmethylated by pyrosequencing with matching low methylation levels on the array.
Validation of the Illumina methylation assay. (A) Average β-levels for 15 to 17 samples (y-axis) according to bisulfite pyrosequencing results. Bisulfite pyrosequencing was considered methylated at a cut-off of 15%. P = .03, .01, and .06 for CDKN2B, HIC1, and CDH1, respectively. There were no methylated samples of CDKN2A according to bisulfite pyrosequencing. (B) Technical replicates; 2 samples were replicated on the 27k array (top panel) and 2 replicates on both the 27k and 450k array (bottom panel). All were highly correlated (P < .0001) with Pearson correlation coefficients of r = 0.99, 0.94 and r = 0.98, 0.98, clockwise from top left. (C) There is an inverse relationship between promoter methylation assessed by the Illumina HumanMethylation27 array and global CpG methylation as measured by LUMA (Pearson r = −0.45, P = .02, n = 29).
Validation of the Illumina methylation assay. (A) Average β-levels for 15 to 17 samples (y-axis) according to bisulfite pyrosequencing results. Bisulfite pyrosequencing was considered methylated at a cut-off of 15%. P = .03, .01, and .06 for CDKN2B, HIC1, and CDH1, respectively. There were no methylated samples of CDKN2A according to bisulfite pyrosequencing. (B) Technical replicates; 2 samples were replicated on the 27k array (top panel) and 2 replicates on both the 27k and 450k array (bottom panel). All were highly correlated (P < .0001) with Pearson correlation coefficients of r = 0.99, 0.94 and r = 0.98, 0.98, clockwise from top left. (C) There is an inverse relationship between promoter methylation assessed by the Illumina HumanMethylation27 array and global CpG methylation as measured by LUMA (Pearson r = −0.45, P = .02, n = 29).
Two technical replicates were run on the 27k array and 2 replicates on both the 27k and 450k arrays. They were compared before normalization. All were highly correlated (P < .0001) with Pearson correlation coefficients of r = 0.99, 0.94 and r = 0.98, 0.98, respectively (Figure 1B). Even though the Illumina HumanMethylation27 array covers 27 578 CpG sites in 14 495 genes, the global CpG content is not necessarily reflected as the array is biased toward promoter CpG island methylation. Indeed, there is often an inverse relationship between promoter methylation and global CpG methylation in tumors.12,18 To investigate this in the current cohort, we measured a surrogate of the global 5-methylcytosine content with LUMA, a nonbisulfite, unbiased method using the methylation-sensitive isoschizomeric restriction enzymes MspI and HpaII. These enzymes target CCGG sites, which are distributed throughout the genome with approximately 20% in CpG islands and the rest reflecting intergenomic and intragenomic DNA as well as repetitive sequences.19,20 Twenty-nine samples were investigated, and we found an inverse relationship between promoter methylation assessed by the Illumina HumanMethylation27 array and global CpG methylation as measured by LUMA (Pearson r = −0.45, P = .02, Figure 1C), indicating that promoter methylation and nonpromoter methylation are inversely correlated in AML as in many solid tumors. Because one of the X-chromosomes is epigenetically silenced and hypermethylated in females, we further validated the arrays by calculating average β-values for the 1084 X-chromosomal CpG sites common in the 27k and 450k arrays. There was a significant increase in methylation of female samples compared with male (average β = 0.51 vs 0.38, P = 2 × 10−16; supplemental Figure 1). In a logistic regression model, the X-chromosome methylation level was able to predict sex. The hazard ratio for male sex was 0.01 when methylation levels were high (β > 0.42, P = 2.2 × 10−9).
Methylation in relation to molecular subtypes, age, and morphology
Differences in average β-values were investigated for DNMT3A, NPM1, FLT3-ITD, IDH1, IDH2, FLT3-TKD, and CEBPA mutated samples compared with their wild-type counterparts. The associations between age and methylation as well as differences in CpG-island versus non-CpG-island methylation in the different subgroups were investigated. Results are shown in supplemental Table 3. In short, a mutation of any IDH gene was associated with increased β-values (P = .006). Interestingly, in IDH1 mutated samples, only CpG-island annotated sites showed increased methylation (P = .02), whereas in IDH2 mutated samples, only non-CpG island sites were hypermethylated (P = .0002). Average methylation was lower in FLT3-TKD mutated samples compared with unmutated (P = .01); however, only 12 samples had an FLT3-TKD mutation. Age and morphologic French-American-British class were not significantly associated with differences in β-values, nor were DNMT3A, FLT3-ITD, NPM1, or CEBPA mutational status.
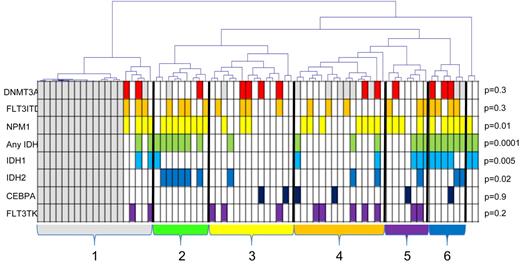
Unsupervised hierarchical clustering of the samples (Pearson complete clustering) was performed (Figure 2). Six clusters were identified and 2 outliers. The normal progenitor samples clustered together within cluster 1. Differences in frequencies of common molecular aberrations between clusters were assessed with the Fisher-Freeman-Halton test. There was an unequal distribution of NPM1 and IDH mutations (P = .01 and P = .0001, respectively), with concordantly high frequencies in cluster 2 and 6 and low in clusters 3 and 4.
Unsupervised complete linkage hierarchical clustering of samples according to methylation of the 2764 differentially methylated CpG sites. There are 2 outliers and 6 major clusters of samples. The normal samples clustered together within cluster one (gray) and are marked by gray boxes below the dendrogram. Mutational status is indicated below; white boxes represent wild-type; and colored boxes, mutations. P values are given for unequal distribution between clusters for each mutation using the Fisher-Freeman-Halton test (outliers were disregarded). The frequency of NPM1 mutations is increased in cluster 2 and 6, IDH1 in cluster 6, and IDH2 in cluster 2.
Unsupervised complete linkage hierarchical clustering of samples according to methylation of the 2764 differentially methylated CpG sites. There are 2 outliers and 6 major clusters of samples. The normal samples clustered together within cluster one (gray) and are marked by gray boxes below the dendrogram. Mutational status is indicated below; white boxes represent wild-type; and colored boxes, mutations. P values are given for unequal distribution between clusters for each mutation using the Fisher-Freeman-Halton test (outliers were disregarded). The frequency of NPM1 mutations is increased in cluster 2 and 6, IDH1 in cluster 6, and IDH2 in cluster 2.
Promoter methylation is inversely correlated to gene expression
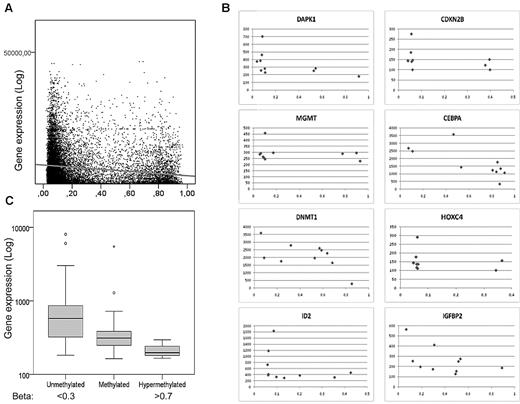
It is well known that promoter DNA methylation does not always correlate with gene expression.21 To examine this in our cohort, we performed gene expression arrays for 10 of the samples. There was a moderate, but highly significant, inverse correlation between average β-values and average gene expression (Spearman rho = −0.17, P = 2 × 10−88), verifying an association between promoter methylation and transcription (Figure 3A). Plots for individual genes are also shown in Figure 3B. Methylation of CpG islands versus non-CpG islands affected gene expression to a similar extent (Spearman rho = −0.14, P < .0001 and Spearman rho = −0.14, P < .0001, respectively). To further investigate this, Spearman correlation coefficients were calculated for every CpG site annotated to valid expression data for the quartile of genes with the highest SD between samples (n = 1979 genes annotated to 3851 CpGs). Overall, 29.2% of genes had a moderate to strong inverse correlation (ie, < −0.3 rho) between methylation and expression, whereas only 11.9% had a moderate to strong positive correlation (ie, > 0.3 Rho), corroborating our findings.
Inverse correlation of methylation and gene expression. (A) Log average gene expression is plotted on the y-axis and corresponding average β-values on the x-axis for 7 samples. Data for 6541 CpG sites corresponding to 3395 genes had significant signals on both expression and methylation arrays in more than 6 of the samples. Spearman correlation Rho = −0.17, P = 2 × 10−88. (B) Examples of correlation between expression (y-axis) and β-values (x-axis) for 12 genes. (C) Average gene expression of Polycomb-associated genes according to average methylation levels in 10 samples, showing an inverse relation in this subgroup.
Inverse correlation of methylation and gene expression. (A) Log average gene expression is plotted on the y-axis and corresponding average β-values on the x-axis for 7 samples. Data for 6541 CpG sites corresponding to 3395 genes had significant signals on both expression and methylation arrays in more than 6 of the samples. Spearman correlation Rho = −0.17, P = 2 × 10−88. (B) Examples of correlation between expression (y-axis) and β-values (x-axis) for 12 genes. (C) Average gene expression of Polycomb-associated genes according to average methylation levels in 10 samples, showing an inverse relation in this subgroup.
Different methylation in normal myeloid progenitors and AML
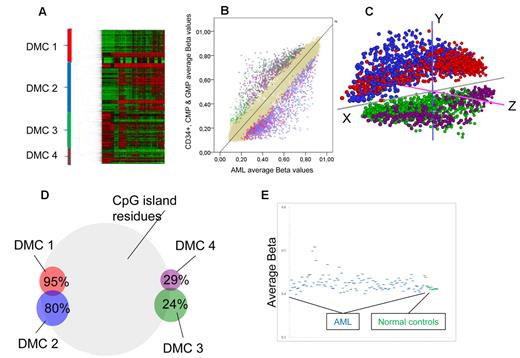
Myeloid progenitor cells from the bone marrow CD34+ pool from 9 healthy donors were FACS separated and cells from the CMP and GMP-stages were selected in 5 because their gene expression signatures are most similar to AML gene expression signatures (Bo Porse, BRIC/University of Copenhagen, personal verbal communication, November 2010). Differences in average methylation levels between the CD34+, CMP, and GMP stages were negligible (Figure 4E), and they were thus pooled together for further comparisons. To identify groups of genes that were differentially methylated between progenitors and CN-AML while keeping false positives to a minimum, we applied a Bayes modified, Benjamini-Hochberg adjusted t test for difference set to < 0.05 and a minimum geometric average distance between groups of 0.10 β. With these criteria, 2764 CpG residues corresponding to 2304 genes were found to be differentially methylated. Using unsupervised hierarchical clustering, 4 clusters of differentially methylated CpGs (DMCs) were defined (Figure 4A). DMC 1 and DMC 2 were less methylated in normal progenitors than in AML samples. DMC 3 and DMC 4 were more methylated in progenitors than in AML (Figure 4B). A principal components analysis of the same CpGs clearly separated clusters 1 and 2 from 3 and 4, confirming their relevance. However, DMC 1 and DMC 2 separated only on the z-axis, as did DMC 3 and 4 (Figure 4C). On the whole array, 72.5% of all CpG residues are located in CpG islands. In comparison, DMC 1 consisted overwhelmingly of CpG residues within CpG islands, 95% (P = 1 × 10−40), whereas in DMC 3 and DMC 4, only 24% and 29% of the CpG residues were located in CpG islands, respectively (P < 1 × 10−65 for both). DMC 2 had a slight increase in the proportion of CpG island-annotated sites, 80% (Figure 4D). Gene ontologic analyses showed that DMC 3 and DMC 4 were enriched for genes involved in defense responses, DMC 1 and DMC 2 for genes related to cell-cell signaling, neuron differentiation, embryonic morphogenesis, and cell fate commitment, mainly homeobox (HOX) genes (supplemental Table 4). The average β-values of the differentially methylated CpG sites were higher in most AML samples compared with normal progenitors (P = .0003; Figure 4E). However, the individual AML samples showed a great variability from close to normal to extreme hypermethylation. To confirm our findings, we repeated the hierarchical clustering and gene ontologic analyses in the validation AML cohort and normal controls with similar results as in the test cohort: 4 major clusters with similar gene ontologic enrichment as in the test cohort (data not shown).
Different methylation in normal progenitors and AML. (A) Two-way unsupervised hierarchical analysis (Pearson correlation, complete clustering) of differentially methylated CpG residues (n = 2764) between CN-AML and myeloid progenitor cells. Each column represents a sample and each row a CpG site. CpG sites are divided into 4 major DMCs, which are color-coded: red represents DMC 1; blue, DMC 2; green, DMC 3; and purple, DMC 4. (B) An average plot showing differentially methylated CpGs. Each autosomal CpG residue is represented with its average β-value for myeloid progenitors on the y-axis, and the corresponding average β-value of all CN-AML samples on the x-axis. Color codes mark the clustered CpGs defined in Figure 3A, and beige dots represent nondifferentially methylated CpG residues. (C) Principal components analysis of the differentially methylated CpG sites shows separation of the DMC 1 + DMC 2 from DMC 3 + DMC 4, whereas only z-axis differences separate DMC 1 from DMC 2 and DMC 3 from DMC 4. (D) The DMCs have significantly different percentages of CpG sites within CpG islands shown here in a Venn diagram. On the whole array, 72.5% of CpG sites are within CpG islands. (E) Scatterplot showing the average β-values of the differentially methylated genes on the y-axis for all CN-AML samples (blue) and the myeloid progenitors from normal bone marrow (green), highlighting the variability among AML samples who have increased average β-values compared with the normal controls (P = .0003).
Different methylation in normal progenitors and AML. (A) Two-way unsupervised hierarchical analysis (Pearson correlation, complete clustering) of differentially methylated CpG residues (n = 2764) between CN-AML and myeloid progenitor cells. Each column represents a sample and each row a CpG site. CpG sites are divided into 4 major DMCs, which are color-coded: red represents DMC 1; blue, DMC 2; green, DMC 3; and purple, DMC 4. (B) An average plot showing differentially methylated CpGs. Each autosomal CpG residue is represented with its average β-value for myeloid progenitors on the y-axis, and the corresponding average β-value of all CN-AML samples on the x-axis. Color codes mark the clustered CpGs defined in Figure 3A, and beige dots represent nondifferentially methylated CpG residues. (C) Principal components analysis of the differentially methylated CpG sites shows separation of the DMC 1 + DMC 2 from DMC 3 + DMC 4, whereas only z-axis differences separate DMC 1 from DMC 2 and DMC 3 from DMC 4. (D) The DMCs have significantly different percentages of CpG sites within CpG islands shown here in a Venn diagram. On the whole array, 72.5% of CpG sites are within CpG islands. (E) Scatterplot showing the average β-values of the differentially methylated genes on the y-axis for all CN-AML samples (blue) and the myeloid progenitors from normal bone marrow (green), highlighting the variability among AML samples who have increased average β-values compared with the normal controls (P = .0003).
Polycomb target genes and genes with bivalent histone marks in stem cells are preferably methylated in CN-AML
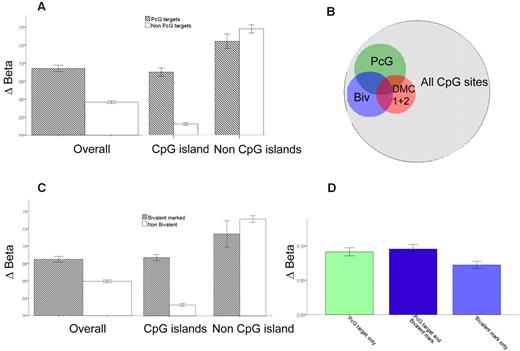
It has been reported that genes in stem cells associated with PcG transcriptional repressor proteins become hypermethylated in various solid tumors.22,23 To investigate this on a genome-wide scale in AML, we integrated chromatin immunoprecipitation sequencing (ChIP-seq) data of Polycomb repressor complex 2 (PRC2) proteins in human embryonic fibroblasts by Bracken and Helin23 with our own methylation data. We recorded the difference in β-values between AML samples in the test cohort (n = 58) and normal progenitor cells (Δβ) for every CpG residue present on the Illumina array and compared Δβ for all CpGs annotated to the genes implicated as PcG targets in the ChIP-seq dataset with those that were not. Indeed, PcG target genes in the Bracken and Helin study23 had a greater increase in methylation (Δβ, 0.09 vs 0.05, P < .0001). The difference was restricted to CpG islands (Δβ, 0.08 vs 0.02, P = 1.9 × 10−144), whereas in non-CpG island residues, there was actually a slight but significant decrease of methylation in PcG-associated genes (Δβ 0.12 vs 0.13, P = .005; Figure 5A). Furthermore, PcG target genes in human embryonic fibroblasts were 2- to 4-fold more common in DMC 1 and DMC 2 (OR = 3.5; 95% CI, 3.0-4.1; and OR = 2.1; 95% CI, 1.7-2.4, respectively, P < 1 × 10−20 for both, Figure 5B). Interestingly, PcG-targeted genes were significantly more common among hypermethylated genes in IDH1-mutated versus nonmutated samples (33% vs 17%, P = 3 × 10−8), whereas in IDH2-mutated samples they were not (19 vs 17%, P = .10). There was a clear association between DNA methylation and silencing also in PcG-targeted genes (Figure 3C).
Preferential methylation of PcG targets. The difference between average β-values for AML minus the average β-values in normal CD34+, CMP, and GMP progenitors is defined as Δβ. (A) Δβ for PcG-targeted genes; overall and for CpG residues within and outside CpG islands. (B) Venn diagram showing the overlap of PcG targets, bivalently marked genes, and DMC 1 + DMC 2 in relation to all measured CpG sites. The OR for overlap between PcG targets and DMC 1 + DMC 2 is 2.8 (P < 1 × 10−20) and for bivalently marked genes and DMC1 + DMC 2 is 4.4 (P < 1 × 10−20). (C) Δβ for genes with bivalent chromatin marks; overall and for CpG residues within and outside CpG islands. (D) Δβ for genes both bivalently marked and PcG-targeted (middle, dark blue, n = 694), PcG-targeted only (left, light green, n = 1662), and bivalently marked only (right, light blue, n = 1180). Error bars represent 95% CI.
Preferential methylation of PcG targets. The difference between average β-values for AML minus the average β-values in normal CD34+, CMP, and GMP progenitors is defined as Δβ. (A) Δβ for PcG-targeted genes; overall and for CpG residues within and outside CpG islands. (B) Venn diagram showing the overlap of PcG targets, bivalently marked genes, and DMC 1 + DMC 2 in relation to all measured CpG sites. The OR for overlap between PcG targets and DMC 1 + DMC 2 is 2.8 (P < 1 × 10−20) and for bivalently marked genes and DMC1 + DMC 2 is 4.4 (P < 1 × 10−20). (C) Δβ for genes with bivalent chromatin marks; overall and for CpG residues within and outside CpG islands. (D) Δβ for genes both bivalently marked and PcG-targeted (middle, dark blue, n = 694), PcG-targeted only (left, light green, n = 1662), and bivalently marked only (right, light blue, n = 1180). Error bars represent 95% CI.
The PRC2 catalyzes the trimethylation of lysine 27 on histone H3 (H3K27Me3). Concurrent methylation of lysine 4 on histone H3 (H3K4Me) coincides with H3K27Me3 at transcription factors important for developmental control in stem cells.24 The relevance of such “bivalent marks” was investigated by applying ChIP-seq data of bivalently marked genes from a study in hematopoietic stem cells by Cui et al in a similar fashion as described for PcG targets.25 Δβ was greater among genes with bivalent marks (Δβ, 0.08 vs 0.05, P < .0001), again restricted to CpG islands (Δβ, 0.08 vs 0.01, P < .0001, Figure 5C). Among non-CpG island residues, there was a slight decrease of methylation of bivalently marked genes (Δβ, 0.11 vs 0.13, P = .006). As for PcG targets, genes with bivalent marks were enriched in DMC 1 and DMC 2 (OR = 5.3; 95% CI, 4.6-6.2; and OR = 2.3; 95% CI, 2.0-2.6, respectively, P < 1 × 10−20 for both, Figure 5B). Similar results were attained when using gene sets from LIN1- hematopoietic stem cells with bivalent marks26 as well as ChIP for the PRC 2 protein SUZ1227 (data not shown). PcG target genes and genes with bivalent marks were significantly overlapping. Overlapping genes and genes that were PcG targets but not bivalently marked had similar Δβ-values (P = .8), which was higher than for genes with bivalent marks that were not PcG targets (P < .0001, Figure 5D).
Promoter methylation and prognosis
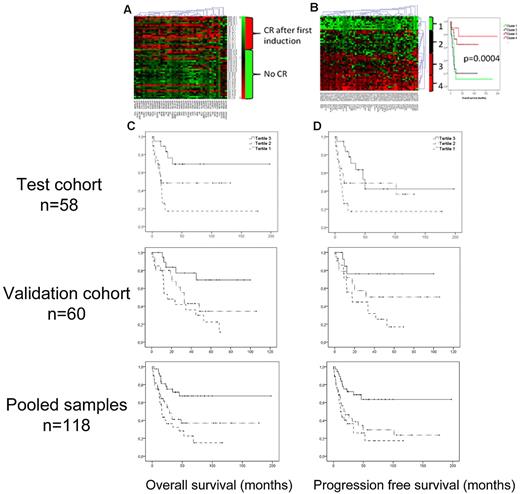
The CpG sites that were most significantly coupled to CR and 2-year survival were selected by a Bayes moderated t test (adjusted P < .05, geometric difference > 0.10 beta). Of the 42 CpG sites most significantly coupled to CR in this supervised analysis, 38% were hypermethylated (ie, β-values > 0.6) in patients achieving CR and 18% among patients who did not (P = .0002; Figure 6A; supplemental Table 5). Similarly, of the 62 CpG sites most significantly correlated to 2-year survival, 64% were hypermethylated in the group of patients that achieved 2-year survival compared with 30% in the group who did not (P = 6 × 10−11; Figure 6B). Interestingly, 33 of the selected 62 CpG residues were located in the PcG target-enriched DMC 2, which is 31 times more than would be expected by chance (P = 1.4 × 10−97).
Methylation and prognosis. Using a Bayes moderated t test, the most significant CpG sites for achieving CR after one induction (n = 42) and for 2-year survival (n = 62) were selected. Red represents more methylation; and green, less methylation. (A) Heat map showing a one-way hierarchical clustering analysis of CpG residues (columns) according to CR after first induction (rows). (B) Left panel: Two-way hierarchical clustering of CpG residues (columns) and samples (rows) according to the most significant residues for 2-year survival. Right panel: Kaplan-Meier diagrams demonstrate the discriminating ability of the selected CpG sites on OS. (C-D) Kaplan-Meier diagram showing the impact of the methylation levels of PcG-marked genes on OS and PFS. The samples were divided in tertiles according to the average methylation levels of PcG-marked genes. The tertile with most methylation (tertile 3), marked with a solid line, had significantly better OS (C) and PFS (D) than the less methylated tertiles; in the test cohort (top panels): P(trend) = .001 and .002, respectively. In the validation cohort (bottom panels): P(trend) = .009 and P(trend) = .035 (middle panels), and for pooled samples: P(trend) = .00009 and .0002, respectively.
Methylation and prognosis. Using a Bayes moderated t test, the most significant CpG sites for achieving CR after one induction (n = 42) and for 2-year survival (n = 62) were selected. Red represents more methylation; and green, less methylation. (A) Heat map showing a one-way hierarchical clustering analysis of CpG residues (columns) according to CR after first induction (rows). (B) Left panel: Two-way hierarchical clustering of CpG residues (columns) and samples (rows) according to the most significant residues for 2-year survival. Right panel: Kaplan-Meier diagrams demonstrate the discriminating ability of the selected CpG sites on OS. (C-D) Kaplan-Meier diagram showing the impact of the methylation levels of PcG-marked genes on OS and PFS. The samples were divided in tertiles according to the average methylation levels of PcG-marked genes. The tertile with most methylation (tertile 3), marked with a solid line, had significantly better OS (C) and PFS (D) than the less methylated tertiles; in the test cohort (top panels): P(trend) = .001 and .002, respectively. In the validation cohort (bottom panels): P(trend) = .009 and P(trend) = .035 (middle panels), and for pooled samples: P(trend) = .00009 and .0002, respectively.
The methylation levels of Polycomb target genes and bivalently marked genes are associated with outcome
Because there was a relative enrichment of PcG-associated genes in DMC 2, where also the most predictive CpG residues for overall survival (OS) and progression-free survival (PFS) were found, we further investigated the potential of the methylation status of all PcG targets to predict prognosis. Indeed, 20 of the 62 CpG sites most predictive of 2-year survival were annotated to genes targeted by PcG, a significant enrichment (P = .001). Average β-values of PcG-annotated CpG residues were calculated for every sample (betaPcG) in the test cohort. Construction of Kaplan-Meier diagrams for the tertiles of betaPcG showed that increasing betaPcG was significantly associated with OS and PFS, P(trend) = .001 and .002, respectively (Figure 6C-D top panels). The analysis was repeated in the validation cohort with similar results: P = .009 for OS and P = .035 for DFS (Figure 6C-D middle panels), and highly significant for pooled samples, P = .00009 and .0002 for OS and DFS, respectively (Figure 6C-D bottom panels).
There was a trend toward increased CR rates for samples with high betaPcG (OR = 0.36; 95% CI, 0.12-1.06, P = .06) in univariate analysis, which however was lost in logistic regression analysis correcting for age, WBC count, FLT3-ITD, and NPM1 status (OR = 0.7, P = .48). A Cox regression analysis of PFS and OS, entering the same variables at baseline, showed an independent association of high betaPcG with better PFS (OR = 0.47; 95% CI, 0.26-0.85, P = .01) and OS (OR = 0.36; 95% CI, 0.19-0.68, P = .001, Table 2). Both cohorts were pooled before multivariate analyses to maximize statistical strength.
Discussion
This study investigates 118 CN-AML samples using the Illumina HumanMethylation Bead arrays. Novel findings are that PcG target genes and genes with bivalent marks are preferentially methylated compared with other genes and that the level of this aberrant methylation is an independent prognostic factor for clinical outcome in CN-AML.
Furthermore, we describe that differentially methylated CpG sites between normal progenitors and AML cluster in groups with distinct ontologic functions, inferring that aberrant DNA methylation and demethylation are not random events in CN-AML. We also show associations between NPM1 and IDH mutations and clustering of samples. The clustering of AML samples according to various genetic and molecular aberrations has been reported previously, in most detail by Figueroa et al who found 16 different clusters in a mixed cohort of AML samples.28 Nine of the 16 clusters primarily consisted of CN-AML samples. Of these, clusters 12, 13, 14, and 16 were characterized by frequent NPM1 mutations as in our cluster 2 and 6. Cluster 4, 5, 9, and 15 by Figueroa et al,28 to the contrary, had a low frequency of NPM1 mutations as in our clusters 3 and 4, highlighting similarities of our findings. A notable difference is that we cannot detect any sample clustering according to CEBPA mutational status as noted in that study. However, there were only 6 CEBPA mutated samples in our cohort, which may explain the absence of clustering. It is also important to stress the difference in methylation analysis techniques used in our studies with substantial differences in sensitivity and specificity for different types and areas of methylation between the methods. The clusters in this study differed from each other, particularly by the different rates of IDH and NPM1 mutations that overlapped. Interestingly, sample cluster 2 is characterized by IDH2 mutations and cluster 6 by IDH1 mutations, indicating that they have different epigenetic profiles, differing not least in the proportion of PcG targets that were preferably methylated in IDH1 mutated compared with IDH2 mutated samples. The mechanistic link behind this is not clear. However, IDH1 is a cytosolic enzyme and IDH2 mitochondrial, making it feasible that their influence on DNA methylation would differ. The link between DNA methylation and IDH mutations has recently been explored by others. Mutant IDH enzymes produce 2-hydroxyglutarate, which in turn is an inhibitor of TET2 activity, affecting the conversion of 5-methylcytosine to 5-hydroxymethylcytosine with implications on DNA methylation homeostasis.29-31 Our findings confirm the presence of generally increased DNA methylation and a specific epigenetic signature in IDH mutated samples, in line with what was recently shown by Figueroa et al.31
In this study, we show that Polycomb target genes implicated in human embryonic fibroblasts are generally more hypermethylated than other genes compared with normal progenitor cells. The PcG of transcriptional repressors is central for stem cell maintenance and differentiation and is also implicated in cancer. Polycomb proteins are also critical regulators of both benign and malignant hematopoiesis, as reviewed recently by Martin-Perez et al.32 It has been suggested that Polycomb-associated genes in stem cells are silenced by aberrant DNA hypermethylation in cancer,33 and this has also been shown for several hematopoietic neoplasms, such as follicular lymphoma, acute lymphatic leukemia, and chronic myeloid leukemia, but not previously in AML.34 The association between genes with bivalent marks in stem cells and hypermethylation in cancer may in part, but not completely, be the result of the overlap with PcG target genes. There are few investigations of this in the literature and, to our knowledge, none in AML; Ohm et al showed that several tumor suppressor genes hypermethylated in various malignancies had bivalent marks in stem cells.35 Rodriguez et al had similar findings in CpG islands at a region of chromosome 5q in colon cancer,36 and McGarvey et al showed it functionally on a global scale using a DNMT1 and 3 double-knockout cell line of colon cancer.21 Our results are in agreement with these findings, strengthening the hypothesis that there is an association of bivalent marks in stem cells and DNA hypermethylation in multiple malignancies.
Furthermore, we show an association between clinical outcome and the degree of promoter methylation among the PcG target genes. Increased methylation of PcG targets was independently associated with disease-free survival and OS in multivariate analyses. One plausible explanation of this finding could be the central role of Homeobox (HOX) gene methylation in AML pathogenesis. HOX genes encode DNA binding proteins central in embryonic development and hematopoiesis, and their expression is epigenetically regulated by PcG and Trithorax proteins.37 They are highly enriched in our cohort in DMC 1 and DMC 2 (supplemental Table 4). HOX genes are overexpressed in AML with mixed lineage leukemia translocations, which is a marker of poor prognosis.38 Several authors have found that high expression of HOX genes correlates with poor prognosis and low expression with favorable prognosis,39,40 compatible with our results. Other possible explanations are offered when looking at the most significant genes associated with outcome (supplemental Table 5), many of which are PcG targets. Interestingly, several potassium channel encoding, PcG-targeted, genes were on the list of methylated genes associated with 2-year survival. Potassium channels have been associated with AML prognosis, with low expression being a favorable prognostic factor.41,42
The genome-wide approach applied in a well-defined cohort of CN-AML enabled us to show an association between PcG targets and the AML methylome, with increased aberrant methylation of targeted genes, a finding that has not previously been reported. We also found possible prognostic implications of PcG target methylation that could be reproduced in a separate validation cohort. These findings suggest that there is a “CpG island methylator phenotype” among CN-AMLs with separate clinical characteristics. However, further investigation of the relationship between PcG target silencing, molecular mutations, and outcome would be warranted to define such an entity for use in clinical practice.
In conclusion, we show that NPM1 and IDH mutations associate with specific clusters of samples, that PcG target genes have an increase of aberrant methylation compared with other genes, and that the level of PcG target methylation may be an independent prognostic factor for clinical outcome in CN-AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Keji Zhao for sharing his ChIP-seq data on bivalently marked genes in hematopoietic stem cells, Björn Wahlin for advice on survival analysis, Hanna Göransson Kultima for bioinformatics analysis, and Tor Olofsson for FACS separation of progenitor cells.
This work was supported by the Swedish Cancer Foundation, Swedish Children Cancer Foundation, Swedish Research Council, Göran Gustafssons Foundation for Research in Natural Sciences, Sigurd och Elsa Golje Foundation, Ligue Contre le Cancer (2008), and La Région Pays de la Loire/Appel à Projets du Cancéropôle Grand-Ouest (2009).
Authorship
Contribution: S.D., P.G., A.L., Y.Q., O.B., M.K., and S.B. performed research; S.D., P.G., O.B., H.N., B.U., U.T., C.P., and M.H. collected clinical data and samples from the AML patients; A.L. and K.E. separated and analyzed the myeloid progenitors; V.G. and K.D. characterized the AML samples; S.D., A.L., K.E., C.P., and S.L. designed and interpreted the experiments; and S.D. wrote the manuscript, with contribution from all coauthors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stefan Deneberg, M54, Karolinska University Hospital Huddinge, 14186 Stockholm, Sweden; e-mail: stefan.deneberg@ki.se.