As controversy exists regarding the prognostic significance of genomic rearrangements of CRLF2 in pediatric B-precursor acute lymphoblastic leukemia (ALL) classified as standard/intermediate-risk (SR) or high-risk (HR), we assessed the prognostic significance of CRLF2 mRNA expression, CRLF2 genomic lesions (IGH@-CRLF2, P2RY8-CRLF2, CRLF2 F232C), deletion/mutation in genes frequently associated with high CRLF2 expression (IKZF1, JAK, IL7R), and minimal residual disease (MRD) in 1061 pediatric ALL patients (499 HR and 562 SR) on COG Trials P9905/P9906. Whereas very high CRLF2 expression was found in 17.5% of cases, only 51.4% of high CRLF2 expressors had CRLF2 genomic lesions. The mechanism underlying elevated CRLF2 expression in cases lacking known genomic lesions remains to be determined. All CRLF2 genomic lesions and virtually all JAK mutations were found in high CRLF2 expressors, whereas IKZF1 deletions/mutations were distributed across the full cohort. In multivariate analyses, NCI risk group, MRD, high CRLF2 expression, and IKZF1 lesions were associated with relapse-free survival. Within HR ALL, only MRD and CRLF2 expression predicted a poorer relapse-free survival; no difference was seen between cases with or without CRLF2 genomic lesions. Thus, high CRLF2 expression is associated with a very poor outcome in high-risk, but not standard-risk, ALL. This study is registered at www.clinicaltrials.gov as NCT00005596 and NCT00005603.
Introduction
Since the initial description of deregulated CRLF2 expression in B-cell precursor acute lymphoblastic leukemia (ALL) in 2009,1,2 several studies have described various correlations between elevated CRLF2 mRNA expression, genomic lesions affecting CRLF2, clinical features, and treatment outcome.3,,–6 Two genomic lesions have been identified that result in elevated expression of wild-type CRLF2: a cryptic chromosomal translocation that juxtaposes CRLF2, located in the pseudoautosomal region (PAR1) of chromosome X or Y, to the immunoglobulin heavy chain locus (IGH@), or, an interstitial deletion of some of the PAR1 region centromeric to CRLF2, resulting in CRLF2 expression being driven by the P2RY8 promoter. Depending on the composition of the ALL cohort, deletions involving P2RY8 are usually more common than the IGH@-CRLF2 translocation,3,4,7,8 although comprehensive analyses have not been performed in all series. Another possible mechanism for elevated CRLF2 expression, although not yet well studied, may be related to the presence of additional copies of the CRLF2 locus, presumably through chromosomal gain.1,3,4 A less common alteration of CRLF2 is a point mutation at codon 232 (F232C), which substitutes a phenylalanine with a cysteine.6,9
To date, several studies have examined the prognostic impact of elevated CRLF2 expression and/or CRLF2 genomic lesions with differing conclusions. High CRLF2 expression and genomic lesions are seen nearly exclusively in ALL cases classified as standard/intermediate risk (SR) or high risk (HR), being virtually absent in low- and very high-risk disease.1,,,,–6 In our initial studies of a highly selected subgroup of National Cancer Institute (NCI) HR patients from the Children's Oncology Group (COG) P9906 trial, we found that essentially all patients with high levels of CRLF2 mRNA also had CRLF2 genomic lesions and that patients with these lesions had a very poor outcome.5 In contrast, Mullighan et al studied a cohort of pediatric ALL cases that included NCI low and standard-risk cases and Down syndrome (DS-ALL) patients and reported that, although there was a high frequency of CRLF2 genomic lesions in DS-ALL, these lesions did not convey a poorer outcome.2 Studying a cohort of high- and standard-risk pediatric ALL patients, Cario et al from the Berlin-Frankfurt-Münster group reported that CRLF2 genomic lesions were present in only a subset of cases with high CRLF2 expression; P2RY8-CRLF2 lesions, but not high CRLF2 expression, had prognostic significance in multivariate analyses.3 Ensor et al from the United Kingdom Medical Research Council trials found that CRLF2 genomic lesions in standard-risk ALL patients were associated with an adverse prognosis in univariate, but not multivariate, analyses.4
There are several possible explanations for these discordant conclusions, including a different composition of the patient cohorts, different treatment regimens, and the frequency of other prognostic covariates that may frequently accompany CRLF2 genomic lesions. The best characterized of these covariates include IKZF1 deletions or mutations, JAK mutations, DS-ALL, and NCI risk group. Although CRLF2 genomic lesions occur in about half of children with DS-ALL,2,8 such patients do not appear to have a poorer outcome than non-DS-ALL patients and they less frequently have IKZF1 alterations.8 Another recently described potential covariate is mutation of IL7R; gain-of-function mutations were reported to be relatively frequent in ALL (6%-10% in B-precursor ALL and T-ALL) and were associated with CRLF2 rearrangements.10,11
To further study the prognostic significance of CRLF2 in ALL, we assessed the status of CRLF2 mRNA expression, genomic lesions in CRLF2 (IGH@-CRLF2, P2RY8-CRLF2, CRLF2 F232C), genomic lesions in IKZF1, JAK, and IL7R, which are frequently associated with aberrant CRLF2, and minimal residual disease (MRD) measured at end-induction using flow cytometric techniques in 1061 pediatric ALL patients classified as SR or HR enrolled in COG clinical trials P9905 and P9906.
Methods
Patient cohort characteristics
The 1061 pediatric and adolescent B-precursor ALL patients reported in this study were selected from COG Trials P990512 (NCT00005596, www.clinicaltrials.gov) and P9906 (NCT00005603, www.clinicaltrials.gov)13 and included all patients with residual pretreatment leukemic specimens available in the COG Repository. The detailed treatment regions may be found at www.clinicaltrials.gov. This cohort included 562 ALL patients classified as NCI SR (with ages ranging from 1 to 9.99 years and initial white blood cell count [WBC] < 50 000/μL) and 499 patients classified as NCI HR (age ≥ 10 years or initial WBC ≥ 50 000/μL; Table 1). The 165 patients derived from P9906 represented a selected subset of NCI HR patients with a particularly poor outcome, defined primarily by older age (mean, 13.6 years) and higher WBC according to the Shuster algorithm.14 Patients with MLL translocations were eligible only for COG P9906, regardless of other clinical factors. The remaining 896 ALL patients were derived from P9905 and included 344 patients with traditional NCI HR features and 552 patients with NCI SR features. NCI SR patients were not eligible for P9905 if they had either t(12;21)/ETV6-RUNX1 or trisomy of chromosomes 4 and 10 and also lacked CNS or testicular leukemia. Similarly, ALL patients with “very high-risk” features (t(9;22)/BCR-ABL1, hypodiploidy (< 44 chromosomes), M3 marrow (> 25% leukemic blasts) on day 29, or M2 (5%-25% leukemic blasts) or M3 marrow on day 43) were not eligible for either P9905 or P9906 and were not included in this study. Trisomies of chromosomes 4 and 10 were detected by FISH and ETV6-RUNX1 fusions by RT-PCR. Intrachromosomal amplification of chromosome 21 (iAMP21) data were not available in these trials. MRD was detected using flow cytometry on day 29 of induction therapy and considered positive if more than or equal to 0.01%.15 Informed consent was obtained from patients and/or their guardians according the Declaration of Helsinki, and the treatment protocol was approved by the NCI and the institutional review boards of the participating institutions.
CRLF2 expression
Quantitative RT-PCR assays for CRLF2 and EEF2 (a control gene) were performed using the Applied Biosystems model 7900HT instrument (Invitrogen). A total of 1 μg of total RNA from pretreatment leukemic cells was converted to cDNA using MMLV-RT and random hexamers as previously described.16 Inventoried assays for CRLF2 (Hs00845692_m1) and EEF2 (Hs00157330_m1) were run using the default conditions recommended by the manufacturer (10 minutes at 95°C, followed by 40 cycles of 15 seconds at 95°C and 1 minute at 60°C). The 165 P9906 cases were run in the 384-well low density array (LDA) format, in which the 1-μL reaction vessels contained specimen cDNA equivalent to approximately 10 ng of starting RNA. The LDA thermal cycling conditions were the defaults recommended by the manufacturer (10 minutes at 94.5°C, followed by 40 cycles of 30 seconds at 97°C and 1 minute at 59.7°C) and duplicates were run for each of the analytes. The 896 P9905 cases were run in 25-μL reactions (96-well format) using specimen cDNA equivalent to approximately 50 ng starting RNA. Each assay was run in duplicate and the average was used for calculations of ΔCt (CRLF2 Ct − EEF2 Ct). A maximum Ct threshold of 30 was set for the control gene EEF2 to include as many specimens as possible while still assuring good sample quality and quantity.
Detection of CRLF2 genomic rearrangements
FISH was performed to detect genomic rearrangements of CRLF2 as previously described.5 In our prior studies,2,5 genomic lesions of CRLF2 were found only in ALL patients with high CRLF2 expression, and JAK mutations almost exclusively in patients with CRLF2 genomic lesions. Thus, to identify patients with CRLF2 rearrangement, FISH was performed on all patients with high CRLF2 RNA expression (defined as ΔCt ≤ 8; n = 186) and/or known JAK mutations (n = 179), and a sampling of 125 additional ALL cases with low CRLF2 expression (ΔCt > 8). A CRLF2 break-apart assay was used composing BAC probes flanking the CRLF2 locus (telomeric: RP11-309M23; centromeric: RP11-261P4).5 Because the centromeric BAC was deleted in cases with the P2RY8-CRLF2 fusion, all cases were evaluated for break-apart of the CRLF2 locus (IGH@-CRLF2) as well as loss of the centromeric BAC. Specimens were scored as P2RY8-CRLF2 if the RP11-261P4 signal was absent or they were positive for the P2RY8-CRLF2 RT-PCR assay. Cases were also evaluated for the presence of extra copies of the CRLF2 locus by scoring the number of copies of the RP11-309M23 BAC in each FISH sample. RT-PCR for the P2RY8-CRLF2 fusion was performed as previously described on all cases studied by FISH that had available RNA.2
Detection of IKZF1 deletions
Leukemic DNA samples from patients enrolled on P9905 were hybridized to a custom designed oligonucleotide array (Agilent Technologies). The high-density, 8 × 15k array format was enhanced and targeted for regions within and flanking the entire IKZF1 gene using the UCSC human genome March 2006/NCBI36/hg18 build. Probes were constructed to allow for genomic tiling with an average probe spacing of 20 bp and a probe length of 50 bp, and included 5000 bp 5′ and 3′ of the IZKF1 gene for a total of 1492 probes within the region. All identified deletions were defined as either focal (within the IKZF1 gene) or broad (extending beyond the IKZF1 gene, including multiple genes, loss of 7p or monosomy 7). Data analyses were performed using Genomic Workbench software (Agilent Technologies).
Sequencing and detection of coding mutations in IKZF1, JAK, CRLF2, and IL7R
Coding mutations in IKZF1 were identified in P9905 by genomic PCR and Sanger sequencing of tumor and matched remission DNA. Mutations and deletions of IKZF1 in P9906 have been previously reported.17,18 Coding mutations in the JAK1, JAK2, and JAK3 kinase and pseudokinase domains were identified by genomic PCR and Sanger sequencing of tumor and matched normal DNA. For POG 9905 patients, the current study included targeted sequence analysis of exons 13, 14, 15, 18, and 19 of JAK1 (Ref Seq: NG_023402.1; accession NM_002227), exons 16, 20, and 21 of JAK2 (Ref Seq: NG_009904.1; accession NM_004972), and exons 18 and 19 of JAK3 (Ref Seq: NG_007273.1; accession NM_000215). For all identified mutations, sequences from tumor and its matched remission DNA were compared with confirm that the observed changes were somatic. Sequencing of CRLF2 exon 6 amplified by PCR was performed on 161 samples identified as high CRLF2 expressors using forward 5′-CATACCCAAGCGACTGGTCA-3′ and reverse 5′-CTCCATAATTTCCATAAAGACAGAA-3′ primers and previously described cycling conditions.6 All PCR products were then subjected to digestion with ApoI (New England BioLabs), which is specific for the wild-type codon. Products from all samples were subjected to agarose gel electrophoresis, and PCR products identified as mutant were purified and submitted for confirmatory Sanger sequencing by Quintara Biosciences. A subset of 335 cases (165 HR and 171 SR) was tested for IL7R mutations. These cases were enriched for high CRLF2 expression (ΔCt ≤ 8; n = 140), CRLF2 lesions (n = 70), and JAK mutations (n = 31). PCR was performed on whole genome amplified patient DNA (QIAGEN) using Phusion HF polymerase and the following M13-tagged primers: forward 5′-tgtaaaacgacggccagtCCTGGTCACCCAAGTCAATGCCTTTT-3′ and reverse 5′-caggaaacagctatgaccTCGTGAAATGCCTTAATCCCCTTTGTG-3′. PCR products were sequenced directly by Beckman Coulter Genomics. Mutation detection was accomplished using SNPdetector and IndelDetector as previously described.19,20
Statistical analysis
Because the COG P9905 and P9906 trials enrolled only patients who had achieved a complete remission (CR) at the end of 4 to 6 weeks of induction therapy, relapse-free survival (RFS) was used as the primary outcome measure, with events defined as relapse, death in CR, or development of a second malignant neoplasm. Patients remaining in continuous CR were censored at the date of last contact. Survival analysis, Cox proportional hazards, Fisher exact test, and stepwise Akaike Information Criterion Cox modeling were performed using R (Version 2.14.0, R Foundation for Statistical Computing).
Results
Clinical, genetic, and biologic features of the ALL cohort
The clinical and biologic features, the frequently recurring ALL-associated cytogenetic abnormalities, and the genetic aberrations detected in CRLF2, JAK, IKZF1, and IL7R in the full cohort of 1061 pediatric and adolescent B-precursor ALL patients and in the NCI HR (n = 499) and NCI SR (n = 562) subsets are provided in Table 1. As patient enrollment in either the COG P9906 or P9905 trials was mutually exclusive and as study entry criteria were based on NCI risk group, age, WBC, and recurring ALL-associated cytogenetic abnormalities (see “Patient cohort characteristics”), it was expected that these variables would differ among the NCI HR and SR subsets. A subset of NCI HR and SR patients with a significantly poorer outcome, as defined by the Shuster criteria,12 were enrolled in P9906; only 10 (6.1%) of the patients accrued to P9906 were classified as NCI SR. In contrast, patients with traditional NCI HR features and patients classified as NCI SR were primarily accrued to P9905. After consideration of these risk assignment and study entry criteria, only MRD at end induction, the frequency of relapse, the frequency of DS, and the frequency of IGH@-CRLF2 translocations were found to be significantly different between the NCI HR and NCI SR patients (Table 1). As expected, the frequency of DS-ALL patients was higher in the NCI SR than the NCI HR group, whereas the rates of end induction MRD and relapse were lower. Interestingly, although P2RY8-CRLF2 fusion was more prevalent in SR than HR ALL (23.8% vs 16.9% of cases tested for CRLF2 genomic lesions), there were nearly 3 times as many IGH@-CRLF2 translocations (15.7% vs 5.6%, P = .004) identified in NCI HR compared with the NCI SR patients tested (Tables 1 and 2). In contrast, the frequency of mutations in any JAK kinase, mutations or deletions in IKZF1, and mutations in IL7R did not differ significantly between the NCI HR and SR cohorts (Table 1). We found IL7R mutations in only 5 of 335 (1.5%) cases tested, a significantly lower frequency than that reported in other studies and only 1 of 5 was among the high CRLF2 expressors.9,10
CRLF2 expression in the ALL cohort
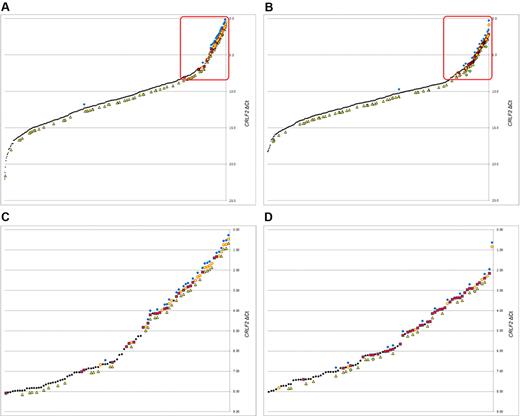
Quantitative RT-PCR was used to determine levels of CRLF2 mRNA transcripts in pretreatment leukemic blasts (Figure 1). Quantitative levels of CRLF2 transcripts were expressed as CRLF2 ΔCt compared with a control gene (EEF2), with lower ΔCt values indicating higher levels of CRLF2 expression (see “CRLF2 expression”). A quantitative threshold was determined that defined “high” levels of CRLF2 mRNA expression by defining the lowest CRLF2 expression level in a patient with a CRLF2 genomic lesion (ΔCt = 8.08, see “Identification of CRLF2 genomic rearrangement to high CRLF2 expression”). Thus, ALL patients with a ΔCt less than or equal to 8.08 were subsequently considered “high” CRLF2 expressors; these patients are highlighted with red boxes in Figure 1. The overall pattern of CRLF2 expression was similar between the NCI HR (Figure 1A) and SR (Figure 1B) cases, although the HR cases appeared to have higher levels of CRLF2 expression among the very highest CRLF2 expressors. This is related to a higher frequency of cases with IGH@-CRLF2 translocations compared with the P2RY8-CRLF2 fusion among the HR cases; the IGH@-CRLF2 translocation yields a higher level of CRLF2 mRNA than the P2RY8-CRLF2 fusion (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The presence of P2RY8-CRLF2, IGH@-CRLF2, CRLF2 F232C, and IKZF1 deletions/mutations was superimposed on the CRLF2 expression plots (Figure 1). As we previously reported in HR ALL,5 virtually all of the JAK mutations and CRLF2 genomic lesions are clustered within the highest CRLF2-expressing ALL cases, whereas mutations and deletions of IKZF1 are also observed in cases lacking elevated expression of CRLF2 (Figure 1; supplemental Figure 1).
Expression profiles of CRLF2 by NCI risk group.CRLF2 expression by NCI risk group is shown along with the corresponding IKZF1 deletions and mutations, CRLF2 lesions and mutations, and JAK mutations. (A-B) All samples for NCI HR and SR, respectively, with the red box indicating the cases with “high” CRLF2 expression. The threshold for high was set at the point where the CRLF2 lesion with the lowest expression (ΔCt = 8.08) was found. (C-D) Enlarged regions within the boxes for NCI HR and NCI SR, respectively, to help illustrate the patterns of lesions and mutations within each. Black dot represents CRLF2 intensity; yellow circle, IGH@-CRLF2; red square, P2RY8-CRLF2; blue diamond, JAK mutations; yellow triangle, IKZF1 deletions or mutations; and green circle, CRLF2 F232C mutation.
Expression profiles of CRLF2 by NCI risk group.CRLF2 expression by NCI risk group is shown along with the corresponding IKZF1 deletions and mutations, CRLF2 lesions and mutations, and JAK mutations. (A-B) All samples for NCI HR and SR, respectively, with the red box indicating the cases with “high” CRLF2 expression. The threshold for high was set at the point where the CRLF2 lesion with the lowest expression (ΔCt = 8.08) was found. (C-D) Enlarged regions within the boxes for NCI HR and NCI SR, respectively, to help illustrate the patterns of lesions and mutations within each. Black dot represents CRLF2 intensity; yellow circle, IGH@-CRLF2; red square, P2RY8-CRLF2; blue diamond, JAK mutations; yellow triangle, IKZF1 deletions or mutations; and green circle, CRLF2 F232C mutation.
These plots reveal that the NCI HR patients have a higher frequency of IGH@-CRLF2 genomic rearrangements than SR patients and tend to have a higher frequency of JAK mutations (Figure 1). In addition, in both NCI risk groups, all JAK mutations except for one in each cohort (a JAK2 mutation in a SR case and a JAK1 mutation in a HR case) occurred in patients with CRLF2 genomic rearrangements and high CRLF2 expression.
Identification of CRLF2 genomic rearrangements relative to high CRLF2 expression
Because this cohort is more heterogeneous than those we have studied previously,21,22 we selected a group of 304 patients to be tested for CRLF2 genomic lesions, including all cases with “high” CRLF2 expression as defined from unpublished data from our prior P9906 studies (a ΔCt ≤ 8.0; n = 179), all patients with identified JAK mutations, and an additional 125 patients spanning the lower expression ranges considered CRLF2 “low” or “negative” (a ΔCt of 8.0-22.04; Figure 1). As shown in Tables 2 and 3, genomic rearrangements of CRLF2 were detected in 93 of these patient samples. In 92 of 93 (98.9%) cases, CRLF2 genomic lesions were found in cases with a high level of CRLF2 expression (ΔCt ≤ 8.0). Only 1 additional case in the remaining 125 tested, with a CRLF2 expression level of ΔCt equal to 8.08, contained a P2RY8-CRLF2 lesion. All remaining cases with low or negative CRLF2 expression lacked CRLF2 genomic lesions. Three patients were identified with CRLF2 F232C gain-of-function mutations; all 3 had high CRLF2 expression (ΔCt ≤ 8.08). Although it is possible that CRLF2 lesions occur among cases with low or negative CRLF2 expression, our data suggest that this is a very uncommon event. Based on these findings, we defined “high” CRLF2 expression as ΔCt less than or equal to 8.08.
Applying the threshold of ΔCt less than or equal to 8.08, 186 of 1061 (17.5%) of the ALL cases in our cohort were defined as having high CRLF2 expression; the clinical features of these cases are provided in Table 3 and supplemental Table 1. ALL cases with high CRLF2 mRNA expression were significantly different from the remaining cases with respect to several features (Table 3; supplemental Table 1). When compared with the remaining ALL cases in the full cohort (n = 875) and consistent with our prior studies,2,5,14 the 186 ALL patients with high CRLF2 mRNA expression had higher rates of end induction MRD (30% vs 21.3%, P = .016), higher rates of relapse (38.2% vs 22.1%, P < .001), and more frequently had DS (15.3% vs 2.5%, P < .001). Patients with high CRLF2 mRNA expression also contained all of the CRLF2 genomic lesions (IGH@-CRLF2, P2RY8-CRLF2, and CRLF2 F232C), virtually all of the JAK mutations (37 of 39), and a higher frequency of IKZF1 deletions and mutations (43.3% vs 18.9%, P < .001; supplemental Table 1; supplemental Figure 1). In contrast, high CRLF2-expressing ALL cases lacked common ALL-associated sentinel cytogenetic lesions. Interestingly, only one of 5 ALL cases with an IL7R mutation had high CRLF2 expression; the remaining 4 cases were low/negative CRLF2 expressors (Table 3). The higher frequency of trisomy of chromosomes 4 and 10 in the high CRLF2-expressing ALL cases is a reflection of the enrollment criteria for P9905 and P9906. Cases with trisomies were downgraded from P9905 to a lower risk study unless they had other HR features, which resulted in an over-representation of trisomies among the P9905 HR patients. All 27 of the trisomies among the high CRLF2-expressing patients (supplemental Table 1) were P9905 NCI HR.
High CRLF2-expressing ALL cases with and without CRLF2 genomic lesions
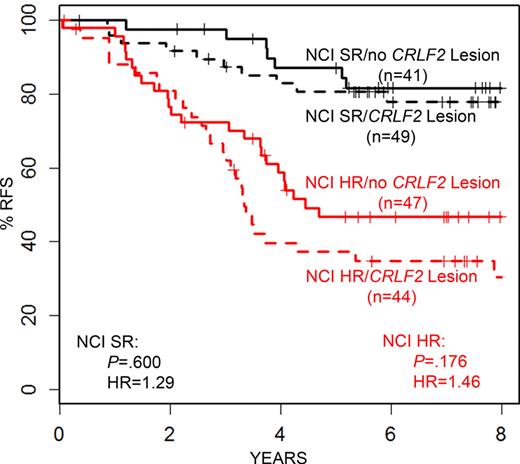
Of the 186 high CRLF2-expressing ALL cases, it was surprising that only 93 of the 181 patient samples with available material for testing (51.4%, Table 3) had genomic rearrangements of CRLF2 (IGH@-CRLF2, P2RY8-CRLF2) known to result in high levels of CRLF2 expression. Three additional high CRLF2 expressors had CRLF2 F232C mutations, and each of these mutations occurred in a patient with either IGH@-CRLF2 or P2RY8-CRLF2 (Table 3). The mechanism that underlies the significant elevation of CRLF2 expression in ALL cases lacking known CRLF2 genomic lesions (IGH@-CRLF2, P2RY8-CRLF2), or, in cases that harbor only a CRLF2 F232C, remains to be determined. Cario et al3 also identified ALL cases with high CRLF2 expression lacking CRLF2 genomic lesions. Of note, we found that 60.7% of high CRLF2-expressing ALL cases in our cohort had a duplication of the full X or Y chromosome, or the X/Y pseudo-autosomal region containing CRLF2, or an increased copy number of the CRLF2 locus alone as determined by FISH (Table 3; and data not shown). Ensor et al also reported gains of X/Y in patients with CRLF2 genomic lesions but did not correlate this with expression.4 However, such chromosomal duplication occurred with relatively equal frequency in high CRLF2 expressors, whether they contained the established CRLF2 genomic lesions or not (Table 5), as well as at a lower frequency (28.5%) in cases with low/negative CRLF2 expression (supplemental Table 1). Within the HR ALL cases, whereas the frequency of relapse in the high CRLF2-expressing ALL cases containing known CRLF2 genomic lesions was very high (63.6%, Table 5), the high CRLF2 expressors lacking CRLF2 genomic lesions also had an unexpectedly high rate of relapse compared with the full ALL cohort (51.1% vs 24.9%, P < .001, Tables 1 and 5). Within the SR ALL cases, there was a similar, but lower, rate of relapse between high CRLF2 expressors with or without genomic lesions (20.4% vs 17.1%, Table 6). In a comparison of the clinical and genetic features between the high CRLF2-expressing cases with genomic lesions and those without, there were clues to the poor outcome in those cases lacking CRLF2 genomic lesions. Among the HR cases (Table 5), high CRLF2 expressors with genomic lesions tended to have a higher rate of end induction MRD (43.9% vs 32.6%) and significantly higher frequencies of JAK mutations and IKZF1 deletions and mutations. Among the SR cases (Table 6), high CRLF2-expressors with genomic lesions had a higher frequency of DS and JAK mutations.
Modeling predictors for RFS
Univariate and multivariable statistical analyses were performed to identify variables associated with variation in RFS (Table 4). Within the full cohort, 6 variables (NCI risk status, end-induction MRD, high CRLF2 expression, the presence of known CRLF2 genomic lesions [IGH@-CRLF2, P2RY8-CRLF2], IKZF1 deletions and/or mutations, and JAK mutations) were each significantly associated with outcome in univariate analysis. However, only 4 variables (NCI risk status, MRD, high CRLF2 expression, and IKZF1 deletions/mutations) retained independent prognostic significance in multivariable analysis (Table 4; Figure 2). As shown in Figure 2A, RFS in NCI HR patients was 67.2% compared with 84.1% in SR patients (P < .001). Measures of end-induction MRD refined outcome prediction in SR (90.1% vs 60.9% RFS) as well as HR ALL cases (76.8% vs 38.9%; P < .001; Figure 2B). Within the HR ALL cases, whereas MRD, high CRLF2 expression, CRLF2 genomic lesions, IKZF1 genomic lesions, and JAK mutations were each associated with a poorer RFS in univariate analysis, only MRD and high CRLF2 expression retained independent predictive power for a poorer RFS in multivariate analysis (Table 4; Figures 2 and 3). High CRLF2 expression predicted a poorer outcome in NCI HR cases (42.6% vs 72.8%, P < .001) but not in SR cases (Figures 2C and 3A). IKZF1 deletions/mutations also predicted a poorer outcome in HR (42.4% vs 70.9%, P < .001) but not in SR cases (Figure 2D). In the NCI SR cases, only end-induction MRD was predictive of outcome in univariate and multivariable analysis (Table 4; Figure 2).
Variables independently predictive of outcome in full cohort. After stepwise multivariate analysis, 4 variables were independently correlated with outcome among the cases tested: (A) NCI risk group, (B) MRD, (C) high CRLF2 expression, and (D) dIKZF1 status. Their correlations with outcome are depicted in Kaplan-Meier survival plots, stratified by NCI risk group. Black lines indicate NCI SR; and red lines, NCI HR. The hazard ratios and log-rank P values were calculated separately for the 2 NCI risk groups and are shown in black (NCI SR) and red (NCI HR) at the bottom of each plot.
Variables independently predictive of outcome in full cohort. After stepwise multivariate analysis, 4 variables were independently correlated with outcome among the cases tested: (A) NCI risk group, (B) MRD, (C) high CRLF2 expression, and (D) dIKZF1 status. Their correlations with outcome are depicted in Kaplan-Meier survival plots, stratified by NCI risk group. Black lines indicate NCI SR; and red lines, NCI HR. The hazard ratios and log-rank P values were calculated separately for the 2 NCI risk groups and are shown in black (NCI SR) and red (NCI HR) at the bottom of each plot.
Variables independently predictive of outcome in NCI HR. After stepwise multivariate analysis, only 2 variables were independently correlated with outcome among the NCI HR cases. Their univariate impacts are shown: (A) MRD and (B) high CRLF2 expression. (C) The performance of the final model of these 2 variables. (D) The model split by its 4 categories to illustrate the relative impact of the variables.
Variables independently predictive of outcome in NCI HR. After stepwise multivariate analysis, only 2 variables were independently correlated with outcome among the NCI HR cases. Their univariate impacts are shown: (A) MRD and (B) high CRLF2 expression. (C) The performance of the final model of these 2 variables. (D) The model split by its 4 categories to illustrate the relative impact of the variables.
Figure 3 presents the association of high CRLF2 expression (found in 19% of the HR ALL cases) with end-induction MRD, which was present in 26.2% of HR ALL cases. The combined modeling of these 2 variables (either high CRLF2 or MRD) identifies 39.3% of the NCI HR cases as higher risk and has a hazard ratio greater than either of the 2 alone (44.3% vs 81.1% RFS, P < .001, Figure 3C). Although the majority of the HR ALL patients in our cohort were positive for either end-induction MRD or high CRLF2 mRNA alone, the 33 (7.3%) cases positive for both high CRLF2 and end-induction MRD had the worst RFS (20.9% RFS, Figure 3D). Interestingly, as shown in Table 5, only 47 of 91 (51.6%) of the HR ALL with high CRLF2 expression had detectable, known CRLF2 genomic lesions. Yet, there was an equivalently poor outcome in the high CRLF2 expressors regardless of the presence or absence of a known CRLF2 genomic lesion (Figure 4).
Impact of CRLF2 lesions on outcome among patients with high CRLF2 expression. The Kaplan-Meier survival curves are shown for NCI HR (red) and SR (black) patients according to their CRLF2 lesion status. Cases without CRLF2 lesions have equivalently poor outcome to those with lesions, regardless of NCI risk group.
Impact of CRLF2 lesions on outcome among patients with high CRLF2 expression. The Kaplan-Meier survival curves are shown for NCI HR (red) and SR (black) patients according to their CRLF2 lesion status. Cases without CRLF2 lesions have equivalently poor outcome to those with lesions, regardless of NCI risk group.
Correlation of quantitative CRLF2 expression with RFS
Our threshold for the definition of high CRLF2 expression (ΔCt ≤ 8.08) was based on identifying the lowest level of CRLF2 expression in a patient with a known CRLF2 genomic lesion. Regardless of the clinical trial on which the patients were enrolled, the survival curves for the NCI HR patients were comparable (supplemental Figure 3B). On finding the strong significance of CRLF2 mRNA expression levels with RFS, we sought to determine whether a different quantitative threshold might be more appropriate for outcome prediction. CRLF2 expression ranges were evaluated in ΔCt bins of 1 unit, with the exception of the highest (ΔCt ≤ 3.0) and the lowest (ΔCt > 9) expressing cases. For the highest expressing, a cutoff of ΔCt ≤ 3.0 was chosen to assure sufficient numbers of cases in each bin. As for the lowest expressing cases, there were no detectable differences seen between any of the bins from ΔCt of 9 to 22 so they were grouped together. As shown in Figure 5, ALL cases with the highest levels of CRLF2 mRNA expression had the worst outcome, whereas those with the lowest had the best (Figure 5). There was not a detectable difference in outcome between any of the bins with ΔCt between 3 and 8. The clear difference in outcome between ΔCt 7-8 and ΔCt 8-9 confirms that a threshold of a ΔCt of 8 is effective not only for prediction of CRLF2 lesions and JAK mutations but also for predicting outcome.
Relationships of CRLF2 expression range and outcome in NCI HR. The expression of CRLF2 was partitioned into ranges, and the survival curves of each were plotted. Given the small number of very high expressing cases, patients with a ΔCt ≤ 3.0 were grouped together. Likewise, patients with a ΔCt > 9 were grouped together because there was no distinguishable difference in outcome between any of the ranges in these low expressing cases.
Relationships of CRLF2 expression range and outcome in NCI HR. The expression of CRLF2 was partitioned into ranges, and the survival curves of each were plotted. Given the small number of very high expressing cases, patients with a ΔCt ≤ 3.0 were grouped together. Likewise, patients with a ΔCt > 9 were grouped together because there was no distinguishable difference in outcome between any of the ranges in these low expressing cases.
Discussion
Herein we report the frequency and prognostic significance of CRLF2 mRNA expression, genomic lesions in CRLF2 (IGH@-CRLF2, P2RY8-CRLF2, CRLF2 F232C), genomic lesions in IKZF1, JAKs, and IL7R frequently associated with elevated CRLF2 expression and/or genomic lesions, and end-induction MRD in 1061 B-precursor pediatric and adolescent ALL patients, the largest cohort reported to date.
Whereas very high levels of CRLF2 expression were found in 186 of 1061 (17.5%) of the ALL cases in the full cohort (in 19% of HR and 16.2% of SR cases), surprisingly only 93 of 181 (51.4%) of the cases with high CRLF2 expression had known CRLF2 genomic lesions (IGH@-CRLF2, P2RY8-CRLF2). An additional 3 patients had the CRLF2 F232C mutation, resulting in 9.0% (96 of 1061) of our patients having CRLF2 genomic lesions. Given that low- and very high-risk patients were excluded from our study (supplemental sections 4 and 5), the overall frequency of CRLF2 genomic lesions in B-precursor ALL cases is probably lower, comparable with the 6% to 7% previously estimated by ourselves and Ensor et al.2,4 Importantly, we did not find any CRLF2 genomic alterations in any patients with low CRLF2 expression, as defined by ΔCt > 8.08. Although we cannot exclude the possibility that such lesions might exist in this subgroup, they are probably exceedingly rare. Although the ratio of P2RY8-CRLF2 to IGH@-CRLF2 in our study reveals P2RY8-CRLF2 to be more common (2.1:1), we find a lower frequency than the 5:1 ratio described by others.2,4 Interestingly, however, there were nearly 3 times as many IGH@-CRLF2 translocations seen in the NCI HR compared with the SR patients. In a related report, Ensor et al4 found patients with IGH@-CRLF2 to be older than those with P2RY8-CRLF2 (median of 8 years vs 4). There was no outcome difference among DS-ALL patients within each NCI risk group (supplemental Figure 2).
The mechanism underlying the significant elevation of CRLF2 expression in ALL cases lacking currently known CRLF2 genomic lesions remains to be determined. Ensor et al speculated that elevated CRLF2 expression might result from extra X/Y chromosomes and/or amplification of the CRLF2 locus.4 We found that 60.7% of high CRLF2-expressing ALL cases had a duplication of the full X or Y or the X/Y pseudo-autosomal region containing CRLF2, or an increased copy number of the CRLF2 locus alone as determined by FISH, whereas these same aberrations were seen in only 28.5% of the ALL cases in our cohort with low CRLF2 expression (P < .001). However, such chromosomal gains occurred with relatively equal frequency in high CRLF2 expressors, whether or not they contained the established CRLF2 genomic lesions. Thus, the role of chromosomal duplication or duplication/amplification of the CRLF2 locus requires more detailed study. Given recent reports of additional genetic events (such as iAmp21) and mutations (IL7R) associated with CRLF2 lesions,10,23,24 it is probable that further comprehensive genomic characterization of these cases will be required to uncover the genetic mechanisms promoting higher CRLF2 expression in cases lacking known CRLF2 genomic lesions. However, we do not think that mutations in IL7R are likely to be an explanation for this phenomenon, as in contrast to a prior report,10 we found that these mutations are rare (0.7%) in B-cell precursor ALL and do not usually occur in patients with high CRLF2 expression and/or CRLF2 genomic lesions.
Consistent with our earlier findings, JAK mutations were nearly exclusively limited to the patients with the highest CRLF2 expression. Of 40 JAK mutations, 38 were among the highest CRLF2 expressing (ΔCt ≤ 8.08) cases (95.0%). Only one case with a JAK1 mutation and one with a JAK2 mutation were found among those with lower expression. Likewise, these mutations are found principally among the cases with CRLF2 lesions (35 of 38, 92.1%). The same was true for the CRLF2 F232C mutations. Although our observed frequency of 2.3% (3 of 131) is lower than in some other reports,6,8 all 3 cases occurred among patients with high CRLF2 and CRLF2 lesions (2 with P2RY8-CRLF2 and 1 IGH@-CRLF2). Notably, there were no JAK mutations among these 3.
When correlating CRLF2 mRNA expression with outcome, we found that CRLF2 predicts for a poorer RFS in HR ALL, but not in cases classified as SR. Within the full cohort, 4 variables (NCI risk group, MRD, high CRLF2 expression, and IKZF1 deletions/mutations retained independent prognostic significance in multivariate analysis. When the cohort was then stratified by NCI risk group, only MRD and high CRLF2 expression retained significance in HR ALL cases. One of our most notable findings was that high CRLF2 expression predicted for a poor outcome regardless of the presence or absence of currently known CRLF2 genomic lesions. More detailed genomic characterization of these cases should provide further insights.
As high CRLF2 mRNA expression is strongly associated with a poor outcome in ALL, particularly in HR ALL cases, prospective identification of high CRLF2 expressors may be useful in future risk stratification and targeting. In our initial studies,5 we found that 50% of ALL cases with known genomic lesions of CRLF2 had JAK mutations. In this report, we find that 21.8% of ALL patients with high CRLF2 expression have JAK mutations. Furthermore, using next-generation sequencing methods and transcriptomic sequencing, we have recently found that many of the ALL cases with high CRLF2 expression or BCR-ABL1–negative cases with a gene expression signature similar to that of BCR-ABL1–positive ALL contain cryptic translocations involving other tyrosine kinases (PDGFRB, ABL1, EPOR, and JAK).25 These studies suggest that initial screening with CRLF2 and other genes derived from such kinase signatures may facilitate the identification of those ALL patients with activated tyrosine kinases, who can then be targeted to unique therapeutic regimens targeting these specific mutations.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Ali Bourguet-Vincent, Richard Vestal, and Bradley Rodgers for performing the LDA experiments with the P9906 cases; Jared Becksfort of Information Sciences at St Jude Children's Research Hospital for performing the IL7R analysis; and Denise Ell at Nationwide Children's Hospital for performing JAK and IKZF1 experiments. FISH analysis was performed in the University of New Mexico & Cancer Center Fluorescence Microscopy Shared Resource.
C.L.W. is the Maurice and Marguerite Liberman Distinguished Chair in Cancer Research. S.P.H. is the Ergen Family Chair in Pediatric Cancer. M.L.L. is a Clinical Scholar of the Leukemia Lymphoma Society. C.G.M. is a Pew Scholar in the Biomedical Sciences and a St Baldrick's Scholar. This work was supported by the National Institutes of Health (grants to the COG): U10 CA98543 (COG Chair's grant), U10 CA98413 (COG Statistical Center), and U24 CA114766 (COG Specimen Banking), as well as the National Cancer Institute (U01 CA114762: Strategic Partnerships to Evaluate Cancer Gene Signatures, C.L.W.; GO 5RC2 CA14852902; Targeted Therapies for Childhood Acute Lymphoblastic Leukemia: Translating Discovery into Practice; R01 CA086011, M.J.B.), Leukemia & Lymphoma Society (SCOR grant 7388-06, C.L.W.), and ALSAC of St Jude Children's Research Hospital (C.G.M).
National Institutes of Health
Authorship
Contribution: I.-M.C. and R.C.H. designed and performed experiments, analyzed data, performed statistical analyses, and prepared the manuscript; H.K. performed statistical analyses; C.G.M., J.G.-F., D.P.-T., S.K.T., C.E.C., and S.R. performed experiments and analyzed and interpreted data; A.J.C. performed cytogenetic analysis; M.J.B. performed flow MRD testing; W.W. reviewed and prepared the manuscript; B.M.C., D.J.P., M.L.L., N.J.W., M.D., G.H.R., W.P.B., W.L.C., and S.P.H. designed COG studies, performed data analysis, and reviewed the manuscript; C.L.W. oversaw all aspects of this project and prepared the manuscript; and all authors critically reviewed the manuscript before submission.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Cheryl L. Willman, UNM Cancer Center, 1201 Camino de Salud NE, Rm 4630, MSC07-4025, 1 University of New Mexico, Alberqueque, NM 87131-001; e-mail: cwillman@salud.unm.edu.