Abstract
LMO2 regulates gene expression by facilitating the formation of multipartite DNA-binding complexes. In B cells, LMO2 is specifically up-regulated in the germinal center (GC) and is expressed in GC-derived non-Hodgkin lymphomas. LMO2 is one of the most powerful prognostic indicators in diffuse large B-cell (DLBCL) patients. However, its function in GC B cells and DLBCL is currently unknown. In this study, we characterized the LMO2 transcriptome and transcriptional complex in DLBCL cells. LMO2 regulates genes implicated in kinetochore function, chromosome assembly, and mitosis. Overexpression of LMO2 in DLBCL cell lines results in centrosome amplification. In DLBCL, the LMO2 complex contains some of the traditional partners, such as LDB1, E2A, HEB, Lyl1, ETO2, and SP1, but not TAL1 or GATA proteins. Furthermore, we identified novel LMO2 interacting partners: ELK1, nuclear factor of activated T-cells (NFATc1), and lymphoid enhancer-binding factor1 (LEF1) proteins. Reporter assays revealed that LMO2 increases transcriptional activity of NFATc1 and decreases transcriptional activity of LEF1 proteins. Overall, our studies identified a novel LMO2 transcriptome and interactome in DLBCL and provides a platform for future elucidation of LMO2 function in GC B cells and DLBCL pathogenesis.
Introduction
Diffuse large B-cell lymphoma (DLBCL), the most common subtype of non-Hodgkin lymphoma (NHL), is a genetically and clinically heterogeneous disease. Marked advances in understanding DLBCL pathobiology have been made by application of gene expression arrays leading to identification of germinal center B (GCB)– and activated B cell (ABC)–like DLBCL subtypes characterized by distinct clinical outcomes. These studies also allowed identification of genes that are preferentially expressed in germinal centers (GCs) and GCB-like DLBCL, such as LMO2.
LMO2 (previously known as RBTN2 or TTG2) is a cysteine-rich protein of 156 amino acids containing 2 zinc-binding LIM domains and a short amino-terminal domain with a potential transcriptional transactivational activity. However, it indirectly mediates gene expression, by mediating protein-protein interactions with other transcriptional factors, facilitating the formation of multipartite DNA-binding complexes.1,2 It is mainly expressed in endothelial and hematopoietic cells, and is implicated in angiogenesis,3 hematopoiesis,4 and hematopoietic stem cell (HSC) maintenance.5 In the erythroid lineage, LMO2 is essential for yolk sac formation and erythropoiesis.6 There are reports supporting LMO2 roles in tumor neovascularization and angiogenesis.7
LMO2 forms distinct multipartite DNA-binding complexes in different cells. In HSCs and endothelial cells it complexes with TAL1, E2A, GATA2, and LDB1 proteins.8 In erythroid cells, LMO2 forms heterodimeric complexes with E proteins, TAL-1, GATA1, and LDB19 recognizing a specific bipartite DNA sequence comprising an E box and a GATA site.9 In erythroid cells LMO2 regulates expression of c-Kit,3 glycophorin A,10 p21CIP11, Gfi-1b,11 and other genes important for erythroid differentiation, and can act as a repressor or activator of transcription, depending on cell stage-specific recruitment of distinct partner proteins.12 A similar complex has also been found in megakaryocytes.13
In contrast, in T cells LMO2 is only expressed in immature CD4/CD8 double-negative thymocytes, and its expression is extinguished early in T-cell development.14 Aberrant expression of LMO2 by chromosomal translocations or aberrations such as t(11;14)(p13;q11), t(7;11)(q35;p13), del(11)(p12-p13) or retroviral integration induces leukemogenesis.15-17 Transgenic mice expressing LMO2 accumulate CD4/CD8 double-negative thymocytes and eventually develop T-cell leukemia.18,19 LMO2 leads to accumulation of early lymphoid precursors and oncogenic transformation by induction of thymocyte self-renewal mediated by Hhex expression, resulting in childhood T-cell acute lymphoblastic leukemia (T-ALL).20 A slightly different multiprotein complex was identified in mouse models of LMO2-induced T-ALL, comprising LMO2, TAL-1, E2A, and LDB1, which binds to bipartite E-box motifs.12 In this system the LMO2 complex inhibits E2A/HEB activity affecting the expression of its target genes such, as pre-T α-chain.21 In addition, the LMO2 complex can bind to GATA3 protein and regulate expression of TALLA1,22 Retinaldehide dehidrogenase 2 (RALDH2),23 and other proteins through the GATA3 binding motif.
In the B-cell lineage, LMO2 has been shown to be expressed at high levels in GC lymphocytes24 and GC-derived non-Hodgkin lymphomas and is a powerful survival predictor in DLBCL patients.25 However, neither its function in B cells or its role in the DLBCL pathogenesis are known.
Herein we characterize LMO2 transcriptome and interactome in DLBCL cells. We demonstrate that in DLBCL cells LMO2 controls expression of a specific set of genes by interacting with LDB1, E2A, HEB, Lyl1, and SP1 proteins. We also identified novel LMO2 interacting partners: ELK1, nuclear factor of activated T-cells (NFATc1), and lymphoid enhancer-binding factor1 (LEF1) proteins. Further, we show that LMO2 expression leads to centrosome amplification.
Methods
Cell culture and transfection
Human chronic myeloid leukemic K562 cell line, human embryonic kidney 293T, Burkitt lymphoma Raji cells, and DLBCL cell lines SU-DHL-6, Rck8, VAL, OCILy19, SU-DHL-4, DOHH2, U2932, WSU-NHL, OCILy3, OCILy7, OCILy10, OCILy1 and Riva were grown as previously reported26 and detailed in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). VAL, Rck8, DOHH2, and Raji lymphoma cells were transiently transfected with Amaxa Nucleofector methodology (Amaxa) following the manufacturer instructions, as previously reported.26
Plasmids and antibodies
pIREShrGFP II LMO2 and pCDNA3.1LMO2 plasmids were constructed using standard molecular biology methods and used for generation of the Rck8 LMO2 stable transfected cell line. OCILy1 cells were transduced with doxycycline-inducible lentivirus containing green fluorescent protein (GFP) and FLAG double-tagged LMO2. pCMV-SPORT6 LEF1(Invitrogen) was subcloned into pCDNA3.1LEF1V5. pCSret-E12, pCSret-E47 were a generous gift of Dr Cornelis Murre (University of California, San Diego, CA); pCAN HACLIM2 was a generous gift of Dr Luc Sabourin (University of Ottawa, Ottawa, ON); pCMV4-SP1/flu was a generous gift of Dr Jonathan Horowitz (North Carolina State University, NC); pMIK-neo-HA-Lyl1 was a generous gift of Dr Serban San-Marina (Ontario Cancer Institute, ON) hHEB MSCV-pgh-EGFP was a generous gift of Dr Trang Hoang (IRCM, Montreal, QC). CMV-Elk1 plasmid and E743-tk80 luciferase plasmid were a generous gift of Dr Ralf Janknecht (University of Oklahoma Health Sciences Center).27 M50 Super8x TOPFlash containing 8 TCF1/LEF binding sites, pREPNFATc1, and pGL3NFAT reporter containing 3xNFAT binding sites were obtained from Addgene.
Mouse monoclonal anti-LMO2 antibody was generated in our laboratory, as previously reported.28 Additional antibodies were from the following sources: TAL1 and SP1 (Abcam); LDB1, GATA1, Lyl1, HEB, E2A, ETO2, GATA2, GATA3, NFATc1, (Santa Cruz Biotechnology), LEF1 (Cell Signaling Technology), and ELK1 (Epitomics).
Whole cell and nuclear extract preparation, Western blot analysis, and inmunoprecipitation
Whole cell and nuclear extract preparation, Western blot analysis and inmunoprecipitation were performed as previously reported,29 and briefly described in supplemental Methods.
RNA isolation, reverse transcription reaction, and real-time PCR
Total cellular RNA was isolated from transfected cells using the Trizol reagent (Invitrogen) according to the manufacturer's instructions. RNA (2 μg) was reverse transcribed using the high-capacity cDNA Archive kit (Applied Biosystems) according to the manufacturer's protocol. Real-time polymerase chain reactions (PCRs) were performed using the ABI PRISMs 7900HT Sequence Detection System Instrument (Applied Biosystems), as previously reported.25 Commercially available Assays-on-Demand gene expression products (Applied Biosystems) were used for measurement of gene expression and normalized to the 18S endogenous control.
Microarray processing and analysis
RNA (100 ng) from 4 distinct Rck8 clones stably transfected with LMO2 and 4 control clones stably transfected with pIREShrGFP II were used for microarray assays using Gene Chip WT 1.0 ST Arrays from Affymetrix as previously reported,26 and briefly described in supplemental Methods. The data from the microarray experiments is available at the GEO database (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE34576.
Centrosome quantification
Centrosome number was assessed by immunofluorescence microscopy in the Rck8 cells transfected with LMO2 or control plasmid at 72 hours after transfection and in induced or noninduced OCILy1-LMO2 cells. The cells were spun down, fixed in 4% formaldehyde for 20 minutes at room temperature, washed with phosphate-buffered saline (PBS), permeabilized in 0.2% Triton X-100 in PBS, washed again in PBS and stained with γ tubulin antibody (Abcam) and Alexa-555 (Invitrogen) as a secondary antibody, and 4′,6′-diamidino-2-phenylindole (DAPI; Molecular Probes). Cells were visualized with a Zeiss Apotome microscope at 63× magnification. Centrosome number was counted in at least 100 cells per experiment and the presented results represent the average from 3 independent experiments.
ChIP
OCILy1 cells were infected with lentivirus expressing GFP and FLAG double-tagged LMO2. The GFP-positive cells were sorted. LMO2 expression was induced by addition of indicated concentrations of doxycycline. The cells were double crosslinked with 2mM ethylene glycol bis(succinidylsuccinate) (Thermo Scientific) and 1% formaldehyde. After sonication, ChIPs were performed with 5 μg anti-FLAG (M2) or IgG (Sigma-Aldrich). DNA fragments enriched by ChIP were quantified by real-time PCR using the Fast SYBR green kit (Applied Biosystems) and an ABI 7900 HT real-time thermal cycler (Applied Biosystems). The GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene was used as a negative control. The fold enrichment was expressed as the percentage of input. The following primers were used for amplification: DLEU2-P Fwd: 5′-ACAGCTAGGGAGAAGGGAGTTT-3′; DLEU2-P Rev: 5′-TAAGGCTTTGAAGGAAAGTTCG-3′; GAPDH Fwd: 5′-CCACCCCCTTCCTTACAAGT-3′; GAPDH Rev: 5′-TCTTCTGGTAGGAGGGCAGA-3′.
DNA constructs and luciferase assays
A 355 bp region from −378 to −733 upstream of the initiation site of the human DLEU2 promoter that contains an SP1 and an E2A binding site was amplified from the SU-DHL-6 cell line by the Phusion High-Fidelity PCR Master Mix (Finnzymes) using the primers DLEU2-FWD 5′-tgcaaactcccttctcccta-3′ and DLEU2-REV 5′-acttccgccctctctcgt-3′. PCR products were digested with BglII and HindIII (New England Biolabs) and ligated into the pGL3-Basic vector to create the 355 DLEU2-Luc construct.
Raji and DOHH2 cells were transfected with 10 μg of the DLEU2 promoter luciferase reporter, 2 μg of indicated expression vector or control pCDNA3.1 plasmid and 10 ng of the internal control plasmid pRLTK (Promega). Cells were cultured for 24 hours after transfection and subsequently harvested into passive lysis buffer for measurement of luciferase and renilla activities with the Dual Luciferase assay kit (Promega).
For assessment of the effect of LMO2 on LEF1 and NFAT luciferase reporter activity at 48 hours after transfection, 3 × 106 Raji or DOHH2 cells were transfected with 3 μg of the M50Super8xTOPFlash plasmid or 10 μg pGL3NFAT luciferase reporter, respectively; 5 μg of LMO2 expression vector or pCDNA3.1 control plasmid, 10 μg of the LEF1V5 expression vector or 5 μg of the NFATc1 expression vector, and 10 ng of the internal control plasmid pRLTK.
Proliferation and cell death studies
Proliferation and cell death studies were performed as previously reported26 and briefly described in supplemental Methods.
Tissue samples and tissue microarray immunohistochemistry
Tissue microarrays were constructed using duplicate cores of 0.6 mm from representative areas of DLBCL, as previously reported28 and described in supplemental Methods.
Primary antibodies directed at LEF1 (N-17; Santa Cruz Biotechnology) and Ki67 (Dako) were used. LEF1 immunohistochemistry was performed at a dilution of 1:1200 after pretreatment with 20mM ethylenediaminetetraacetic acid (EDTA)/50mM Tris, pH 9.0. Ki67 immunohistochemistry was performed at a dilution of 1:1000 after pretreatment with Tris 0.5M, pH 10. The slides were developed using the Dako Envision method (Dako) and cover-slipped with aqueous-based mounting medium.
Statistical analysis
A 2-tailed Student t test was used for reporter assays and the χ2 test was used for centrosome assays; P < .05 was considered statistically significant.
Results
LMO2 transcriptome in DLBCL
LMO2 regulates gene expression by facilitating formation of multipartite DNA-binding complexes with other transcriptional factors. A thorough understanding of the biologic role of LMO2 in DLBCL depends on the identification of the full set of its target genes. To this end, we examined the effect of LMO2 overexpression on global gene expression in DLBCL. The Rck8 DLBCL cell line that expresses low endogenous levels of the LMO2 protein, although expressing potential LMO2 cofactors, was stably transfected with pIREShrGFP II-LMO2 and control pIREShrGFP II plasmids. Four distinct control and LMO2 over-expressing clones were used for global gene expression analysis using Gene Chip WT 1.0 ST Arrays from Affymetrix (supplemental Figure 1A-B). Genes differentially expressed between control and LMO2 stably transfected cell lines were identified using Rank Products, as described in supplemental Methods.
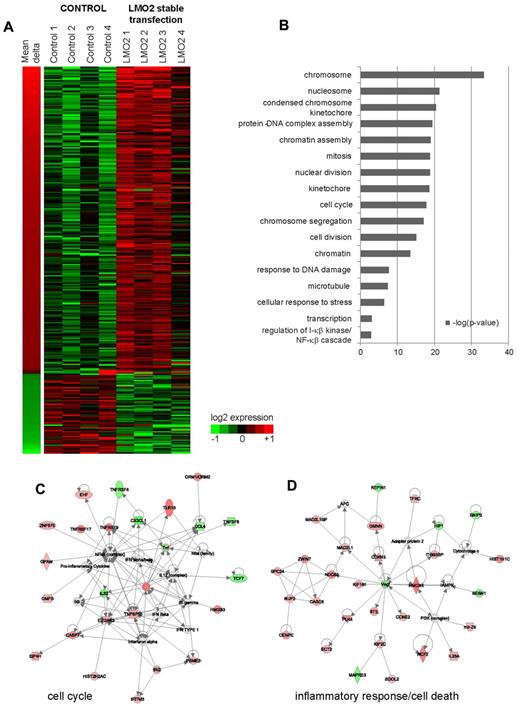
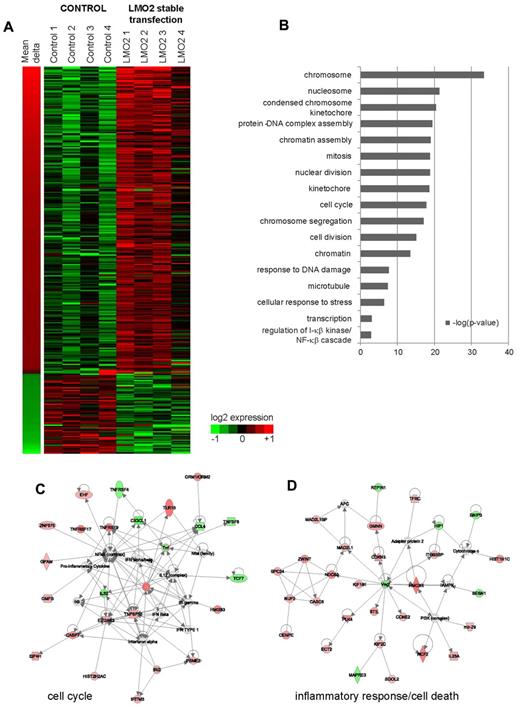
We identified 311 differentially expressed genes (DEGs) between control and stably transfected cells (false discovery rate [FDR] < 5% after correction for multiple hypothesis testing by permutation analysis). Sixty-four genes were down-regulated by LMO2 transfection, whereas 247 were up-regulated (Figure 1A). Prominent among these, were 30 genes encoding proteins from the histone cluster 1 family, that are colocalized on chromosome 6, and were consistently expressed at higher levels in LMO2 expressing cells. At the same time, multiple cell-cycle related genes were also up-regulated by LMO2 transfection, including centromere proteins (CENPE, CENPI, CENPK, CENPN, CENPO, and CENPW) and the kinetochore-associated protein NDC80. Real-time PCR measurement of several selected genes (SPIC, LAX1, DLEU2, DOCK3, CHND2, and TNFRSF9) validated the observed gene expression changes on LMO2 overexpression that ranged between a 2-fold decrease and a 5-fold increase for individual analyzed genes (supplemental Figure 1C).
Genes differentially expressed between control and stably LMO2-transfected cell lines. (A) Three hundred eleven unique genes differentially expressed (5% FDR equivalent) between control cell lines, and cell lines that were stably transfected by LMO2. Shown are the log2 expression values, along with the mean change in expression between control and transfected lines. (B) Significant GO terms (P < .05) associated with genes shown in panel A. (C-D) Network analysis of genes regulated by LMO2 transfection. IPA was used to visualize significant networks of interactions between genes identified as differentially expressed in LMO2 stable transfection experiments. Corresponding to the GO enrichments (B), networks involved in cell cycle/proliferation (C), and inflammatory response/regulation of cell death (D) were significant by IPA analysis.
Genes differentially expressed between control and stably LMO2-transfected cell lines. (A) Three hundred eleven unique genes differentially expressed (5% FDR equivalent) between control cell lines, and cell lines that were stably transfected by LMO2. Shown are the log2 expression values, along with the mean change in expression between control and transfected lines. (B) Significant GO terms (P < .05) associated with genes shown in panel A. (C-D) Network analysis of genes regulated by LMO2 transfection. IPA was used to visualize significant networks of interactions between genes identified as differentially expressed in LMO2 stable transfection experiments. Corresponding to the GO enrichments (B), networks involved in cell cycle/proliferation (C), and inflammatory response/regulation of cell death (D) were significant by IPA analysis.
To determine the potential functional role of these gene expression changes and to visualize known interactions between the genes, we compared the list of DEGs with gene ontology (GO) categories and curated literature using ingenuity pathways analysis (IPA; Figure 1B). This revealed significant (uncorrected P < 10−6) induction of genes involved in chromosome, nucleosome, kinetochore, protein-DNA complex, and chromatin assembly, mitosis and nuclear division, indicating a possible role of LMO2 related to chromosome assembly and segregation in mitosis. At the same time genes related to DNA damage, cellular response to stress and regulation of NFkB cascade were also enriched, although to a lower extent (uncorrected P < 10−4). Furthermore, IPA identified 2 interaction networks enriched for cell-cycle and inflammatory-response/programmed cell death genes, consistent with the GO analysis (Figure 1C-D).
LMO2 induces supernumerary centrosomes
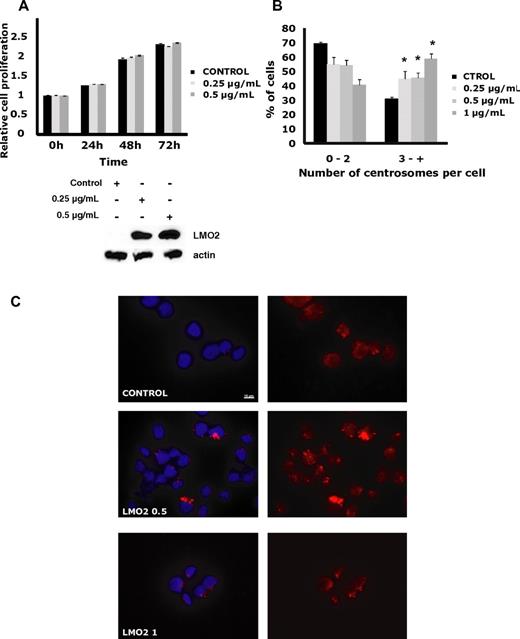
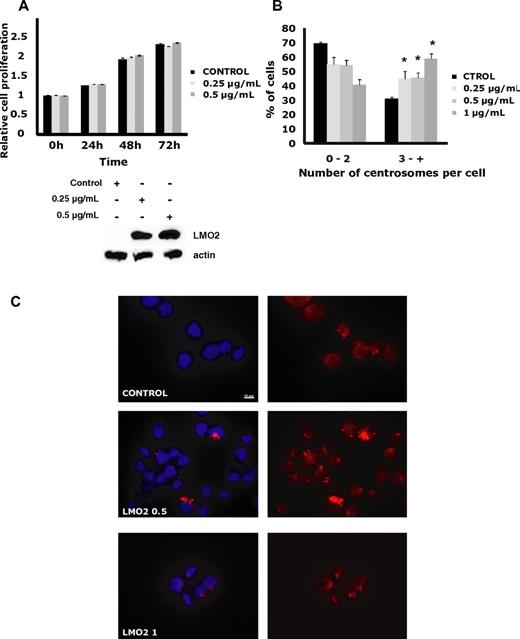
We next analyzed LMO2 effects on several biologic processes affected by the LMO2-regulated genes. Overexpression of LMO2 in the Rck8, Val, Raji, and OCILy1 cell lines did not affect cell proliferation, cell cycle, death, or apoptosis (Figure 2A, supplemental Figure 2). Furthermore, no correlation between the LMO2 and Ki67 expression was observed in 410 primary DLBCL cases analyzed by immunohistochemistry (not shown).
LMO2 effects on cell proliferation, and centrosome number. OCILy1 cells transduced with doxycycline inducible LMO2 lentivirus were left un-induced or were exposed to the indicated concentrations of doxycycline for specified time periods. (A) Proliferation analyzed by an MTS assay. Results are shown as the means ± SD and are representative of 3 independent experiments. Western blot confirms dose-dependent induction of LMO2 protein at 24 hours after addition of doxycycline. (B) The cells were spun down on slides, fixed and permeabilized as described in “Centrosome quantification,” and stained with γ-tubulin antibody and DAPI. Centrosome number was counted in at least 100 cells per experiment and percentage of cells with 2 or less, and 3 or more centrosomes per cell is shown. The presented results represent the average from 3 independent experiments (*P < .001 compared with control). (C) Examples of immunofluorescence centrosome staining visualized with a Zeiss Apotome microscope with 63×/1.4 NA plan apochromat objective lens, using software AxioVision LE. For details see “Centrosome quantification.”
LMO2 effects on cell proliferation, and centrosome number. OCILy1 cells transduced with doxycycline inducible LMO2 lentivirus were left un-induced or were exposed to the indicated concentrations of doxycycline for specified time periods. (A) Proliferation analyzed by an MTS assay. Results are shown as the means ± SD and are representative of 3 independent experiments. Western blot confirms dose-dependent induction of LMO2 protein at 24 hours after addition of doxycycline. (B) The cells were spun down on slides, fixed and permeabilized as described in “Centrosome quantification,” and stained with γ-tubulin antibody and DAPI. Centrosome number was counted in at least 100 cells per experiment and percentage of cells with 2 or less, and 3 or more centrosomes per cell is shown. The presented results represent the average from 3 independent experiments (*P < .001 compared with control). (C) Examples of immunofluorescence centrosome staining visualized with a Zeiss Apotome microscope with 63×/1.4 NA plan apochromat objective lens, using software AxioVision LE. For details see “Centrosome quantification.”
Notably, OCILy1 cells induced to express LMO2 for 20 days exhibited a statistically significant (P < .001) and dose-dependent increase in centrosome number associated with increases in centrosome size and shape. A similar effect was observed in the LMO2-expressing Rck8 cells (not shown). Overall, these observations suggest that LMO2 may be involved in regulation of centrosome number in DLBCL (Figure 2B-C).
LMO2 transcriptional complex in DLBCL cells
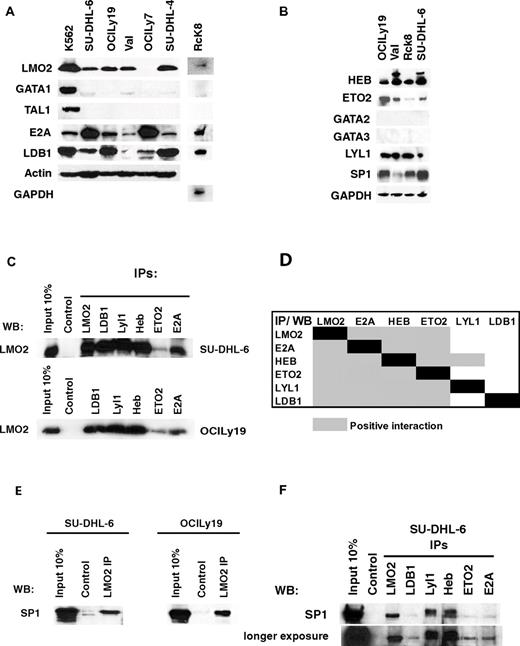
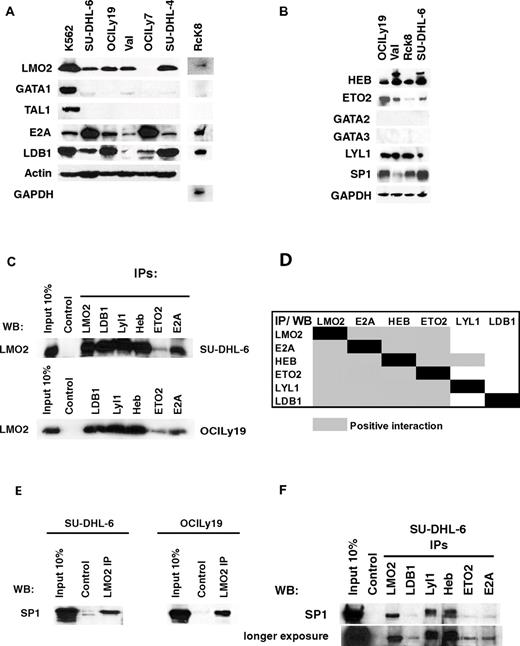
In non-B cells, LMO2 binds to TAL1 and GATA1 proteins and affects gene expression.9 However, TAL1, GATA1, GATA2, and GATA3 proteins were not expressed in any of the analyzed DLBCL cell lines (Figure 3A-B). LDB1 protein, a core component of all the reported LMO2 complexes,9 was expressed in all the analyzed DLBCL cell lines irrespective of LMO2 expression. We next examined expression of previously reported LMO2 interacting partners11,30 in DLBCL cell lines. E2A, Lyl1, HEB, and ETO2 proteins were expressed in all the analyzed DLBCL cell lines (Figure 3A-B). Furthermore, LMO2 coimmunoprecipitated with the LDB1, Lyl1, HEB, E2A, and ETO2 proteins in the SU-DHL-6 and OCILy19 DLBCL cell lines (Figure 3C). Because these proteins are part of a large complex, we next examined specific interactions between individual LMO2 complex partners in DLBCL cell lines by bidirectional coimmunoprecipitation experiments (Figure 3D, supplemental Figure 3). Most of these proteins showed interaction with all the other LMO2 binding partners in one way or bidirectional coimmunoprecipitation in at least one cell line. Although HEB and E2A abundantly heterodimerized with each other in the SU-DHL-6 cell line, interaction with other complex partners (LMO2, LDB1, Lyl1, and HEB) was less copious, as demonstrated by the requirement for longer blot exposure to detect specific coimmunoprecipitation (supplemental Figure 2). In the OCILy19 cell line, E2A and HEB proteins also heterodimerized and interacted with all the other partners. ETO2 coimmunoprecipitated with all the postulated LMO2 complex partners (LMO2, LDB1, Lyl1, HEB, E2A) in at least one of the analyzed DLBCL cell line. In contrast, Lyl1 immunoprecipitated only HEB protein, but was detected in the immunoprecipitates of all the other complex components with the exception of Ldb1 protein. Overall, these studies demonstrated that every protein in the complex interacted with all the other complex components in at least 1 analyzed cell line with the exception of interaction between Lyl1 and Ldb1 that was not detected in bidirectional coimmunoprecipitations.
LMO2 interacting proteins in DLBCL. (A-B) Protein expression in nuclear extracts of DLBCL cell lines was assayed by immunoblotting with specific antibodies. Immunoblotting for actin and GAPDH served as a loading control. (C) Nuclear extracts were prepared from the SU-DHL-6 and OCILy19 cell lines and used for immunoprecipitation of indicated proteins and immunoblotting with LMO2 antibody. (D) A table summarizing the immunoprecipitation results shown in supplemental Figure 2. Coimmunoprecipitation in at least one cell line was defined as positive interaction. (E) Nuclear extracts were prepared from the SU-DHL-6 and OCILy19 cell lines and used for immunoprecipitation with LMO2 antibody and immunoblotting with SP1 antibody. (F) Nuclear extracts were prepared from the SU-DHL-6 cell line and used for immunoprecipitation of indicated proteins and immunoblotting with SP1 antibody. Results in panels A through F are representative of 3 independent experiments.
LMO2 interacting proteins in DLBCL. (A-B) Protein expression in nuclear extracts of DLBCL cell lines was assayed by immunoblotting with specific antibodies. Immunoblotting for actin and GAPDH served as a loading control. (C) Nuclear extracts were prepared from the SU-DHL-6 and OCILy19 cell lines and used for immunoprecipitation of indicated proteins and immunoblotting with LMO2 antibody. (D) A table summarizing the immunoprecipitation results shown in supplemental Figure 2. Coimmunoprecipitation in at least one cell line was defined as positive interaction. (E) Nuclear extracts were prepared from the SU-DHL-6 and OCILy19 cell lines and used for immunoprecipitation with LMO2 antibody and immunoblotting with SP1 antibody. (F) Nuclear extracts were prepared from the SU-DHL-6 cell line and used for immunoprecipitation of indicated proteins and immunoblotting with SP1 antibody. Results in panels A through F are representative of 3 independent experiments.
SP1, a ubiquitous transcriptional factor, was also previously reported to bind to the LMO2 complex in hematopoietic precursors and to interact directly with LMO2 in vitro.10,31 SP1 was expressed in all the analyzed DLBCL cell lines (Figure 3B) and coimmunoprecipitated with LMO2 in the analyzed SU-DHL-6 and OCILy19 cell lines (Figure 3E). Furthermore, SP1 also coimmunoprecipitated with other known LMO2 interacting partners LDB1, Lyl1, HEB, ETO2, and E2A (Figure 3F), corroborating previous reports implicating SP1 as an integral part of the LMO2 complex.31 Overall, these studies suggest that LMO2 forms a multiprotein complex with LDB1, Lyl1, HEB, ETO2, E2A, and SP1 in DLBCL cells. A similar multiprotein complex may also be formed in normal GC B cells expressing LMO2. Indeed, LDB1, E2A, Lyl1, HEB, SP1, and ETO2 RNAs and proteins were also expressed in the GC B cells (supplemental Figure 4A-B).
LMO2 directly binds to DLEU2 promoter and regulates DLEU2 gene expression
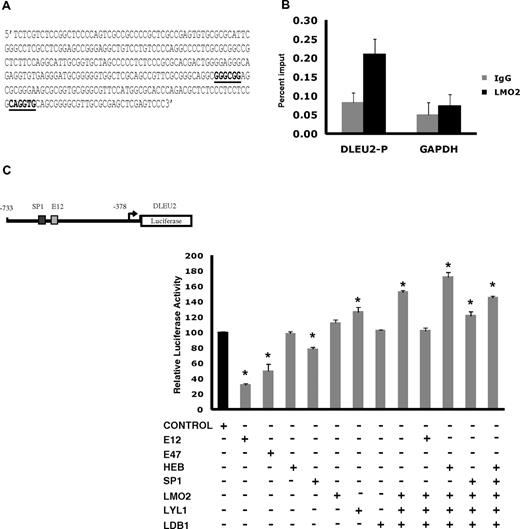
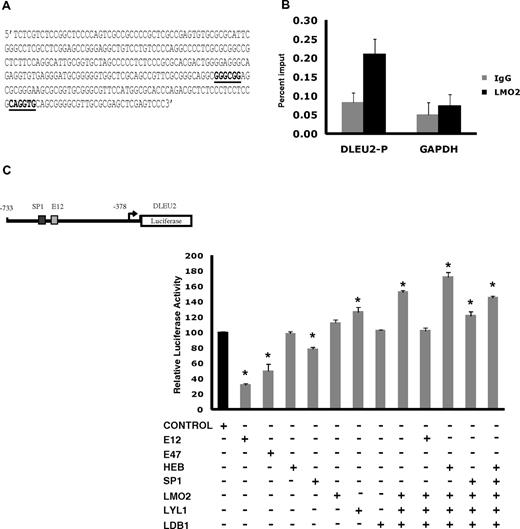
To corroborate the transcriptome and interactome findings in the DLBCL, we next examined in vivo LMO2 binding to the promoter of the DLEU2 gene, which was induced 1.94-fold on LMO2 overexpression. Interrogation of the DLEU2 promoter for putative LMO2 complex binding sites identified a region with at least 1 GC box and an E box binding motifs for SP1 and E2A proteins, respectively (Figure 4A). ChIP experiments confirmed in vivo binding of endogenous LMO2 to this DLEU2 promoter region (Figure 4B). To further evaluate the interaction between LMO2 and its partners in regulation of DLEU2 expression, we cloned a 355bp region of DLEU2 promoter containing these binding sites into a pGL3 basic plasmid and performed luciferase reporter assays in Raji cells (Figure 4C). Individual overexpression of E12, E47, and SP1 significantly repressed reporter activity. HEB and LDB1 alone did not affect luciferase activity, whereas expression of LMO2 and Lyl1 alone increased reporter activity. Concomitant overexpression of LMO2, LDB1 and Lyl1 significantly increased DLEU2 reporter activity and reverted the inhibitory effects of E12 and SP1, corroborating the observed DLEU2 increased expression after LMO2 overexpression in the Raji cells. Similar increase in the DLEU2 reporter activity was observed in DOHH2 cells concomitantly overexpressing LMO2, LDB1, and Lyl1 proteins in comparison with individual LMO2 overexpression (supplemental Figure 5A). Cotransfection of LMO2, Lyl1, LDB1, with HEB further increased luciferase reporter activity. Overall, these findings confirmed formation of functional multiprotein LMO2 transcriptional complex in the context of B-cell lymphoma cells.
A complex containing LMO2, Lyl1, LDB1, SP1, and HEB or E2A activates the DLEU2 promoter. (A) DLEU2 promoter sequence from position −733 to −378 that contains an SP1 and an E2A binding sites. The E-box and GC-box are indicated in bold and underlined. (B) LMO2 protein indirectly associates with the DLEU2 promoter in vivo. Chromatin extracts from the OCILy1 cell line induced to express LMO2 protein were subjected to immunoprecipitation with FLAG (LMO2) and GAPDH antibodies and control IgG. Enriched DNA fragments were quantified by real-time PCR using. The GAPDH gene was used as a negative control. The fold enrichment was expressed as the percentage of input. Results are representative of 3 independent experiments. (C) DLEU2 promoter region (A) was cloned in a pGL3 plasmid. Raji cells were transfected with the DLEU2 Luciferase promoter (10 μg), internal control plasmid pRLTK (10 ng), indicated expression vectors (E12, E47, HEB, SP1, LMO2, Lyl1, LDB1, 2 μg each) and control pCDNA3.1 plasmid that was used to keep constant the total amount of transfected DNA. Luciferase activity was determined 48 hours after transfection. The values are relative luciferase activities, with the average value obtained for the same amount of reporter and pcDNA3.1 control plasmid set at 1. Values are means + SE of 3 independent experiments, each performed in 3 replicate samples.
A complex containing LMO2, Lyl1, LDB1, SP1, and HEB or E2A activates the DLEU2 promoter. (A) DLEU2 promoter sequence from position −733 to −378 that contains an SP1 and an E2A binding sites. The E-box and GC-box are indicated in bold and underlined. (B) LMO2 protein indirectly associates with the DLEU2 promoter in vivo. Chromatin extracts from the OCILy1 cell line induced to express LMO2 protein were subjected to immunoprecipitation with FLAG (LMO2) and GAPDH antibodies and control IgG. Enriched DNA fragments were quantified by real-time PCR using. The GAPDH gene was used as a negative control. The fold enrichment was expressed as the percentage of input. Results are representative of 3 independent experiments. (C) DLEU2 promoter region (A) was cloned in a pGL3 plasmid. Raji cells were transfected with the DLEU2 Luciferase promoter (10 μg), internal control plasmid pRLTK (10 ng), indicated expression vectors (E12, E47, HEB, SP1, LMO2, Lyl1, LDB1, 2 μg each) and control pCDNA3.1 plasmid that was used to keep constant the total amount of transfected DNA. Luciferase activity was determined 48 hours after transfection. The values are relative luciferase activities, with the average value obtained for the same amount of reporter and pcDNA3.1 control plasmid set at 1. Values are means + SE of 3 independent experiments, each performed in 3 replicate samples.
Interrogation of potential novel LMO2 binding partners mediating LMO2-induced transcriptional changes
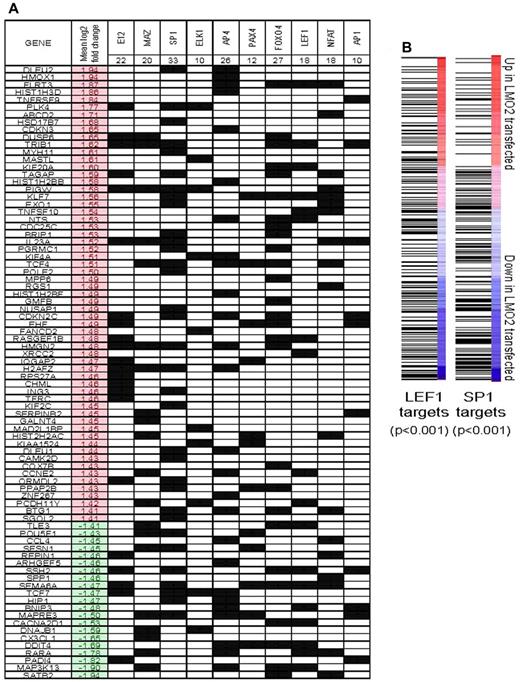
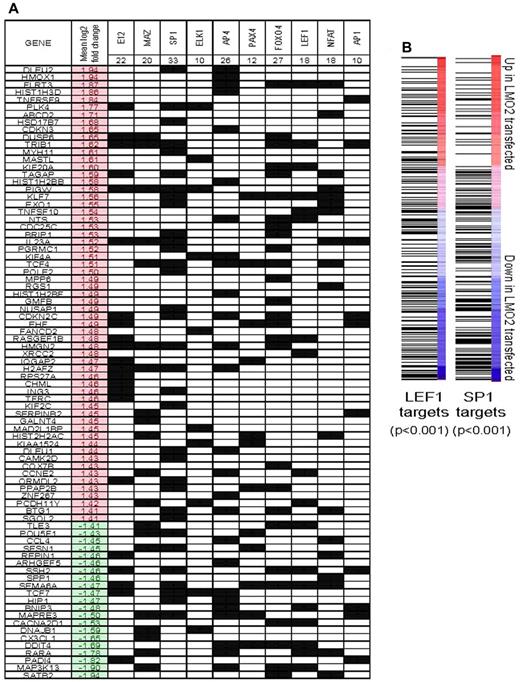
Because LMO2 does not directly bind to DNA but facilitates formation of multiprotein regulatory complexes with other transcriptional factors (TFs), we reasoned that the DEGs previously identified would be enriched for targets of specific TFs that represent potential binding partners of LMO2. Accordingly, we tested whether sets of genes that are related by the presence of specific trancription factor binding motifs in their promoter sequences were concordantly regulated by LMO2 expression using Nextbio, as described in supplemental Methods. We then evaluated whether these transcription factor targets shared known LMO2 binding motifs (Figure 5A). This analysis revealed that genes with motifs for several known DNA-binding proteins were over-represented among the genes regulated by LMO2 over-expression (P < .001 by hypergeometric test). The most significant overlap was with targets of E12, an E2A protein demonstrated to be part of the LMO2 complex,12 thus corroborating our coimmunoprecipitation and reporter results in B lymphoma cells. Enrichment for targets of SP1 and MAZ proteins that have similar binding sites and overlapping targets was also observed,32 further corroborating our studies examining LMO2 transcriptional complex in DLBCL. There was also a significant enrichment for DEGs harboring binding motifs for ELK1, AP4, PAXA4, NFAT, LEF1, and AP1 (uncorrected P < .001 in all cases).
Interrogation of binding site motifs associated with genes regulated by stable LMO2 transfection. (A) Differentially expressed genes were evaluated for enrichment of specific binding site motifs in their promoter region by comparison to the C3 motif gene sets that are part of mSigDB, using Nextbio, as described in supplemental Methods. The occurrence of a motif for specific binding factor in the promoter of a particular gene is indicated by a black square. The mean log2 expression change of each gene between control and LMO2-transfected experiments is shown (red indicates up-regulation by LMO2 transfection). Genes that did not have a binding motif for the factors found to be significant are not shown. (B) All genes were ranked by their mean change in expression between control and LMO2-transfected lines. Gene set enrichment analysis was used to evaluate the distribution of genes with predicted SP1 and LEF1 binding sites in their promoter regions. In both cases, SP1 and LEF1 target genes were generally down-regulated by LMO2 transfection (both P < .001 by Kolmogorov-Smirnov test).
Interrogation of binding site motifs associated with genes regulated by stable LMO2 transfection. (A) Differentially expressed genes were evaluated for enrichment of specific binding site motifs in their promoter region by comparison to the C3 motif gene sets that are part of mSigDB, using Nextbio, as described in supplemental Methods. The occurrence of a motif for specific binding factor in the promoter of a particular gene is indicated by a black square. The mean log2 expression change of each gene between control and LMO2-transfected experiments is shown (red indicates up-regulation by LMO2 transfection). Genes that did not have a binding motif for the factors found to be significant are not shown. (B) All genes were ranked by their mean change in expression between control and LMO2-transfected lines. Gene set enrichment analysis was used to evaluate the distribution of genes with predicted SP1 and LEF1 binding sites in their promoter regions. In both cases, SP1 and LEF1 target genes were generally down-regulated by LMO2 transfection (both P < .001 by Kolmogorov-Smirnov test).
Gene set enrichment analysis (GSEA) of the entire expression dataset of all the genes ranked by their mean fold change between control and transfected conditions revealed that SP1 and LEF1 targets were skewed toward down-regulated genes (P < .001 in both cases, Kolmogorov-Smirnov test; Figure 5B). This supplements the observed overrepresentation of genes with SP1 and LEF1 motifs in the list of DEGs (Figure 4C).
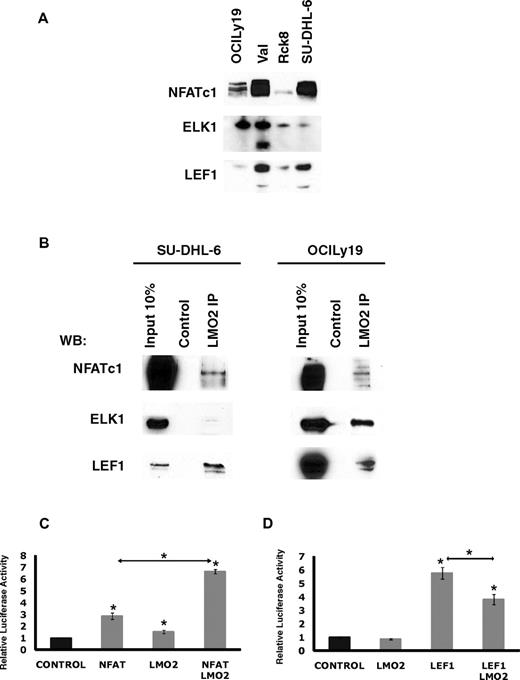
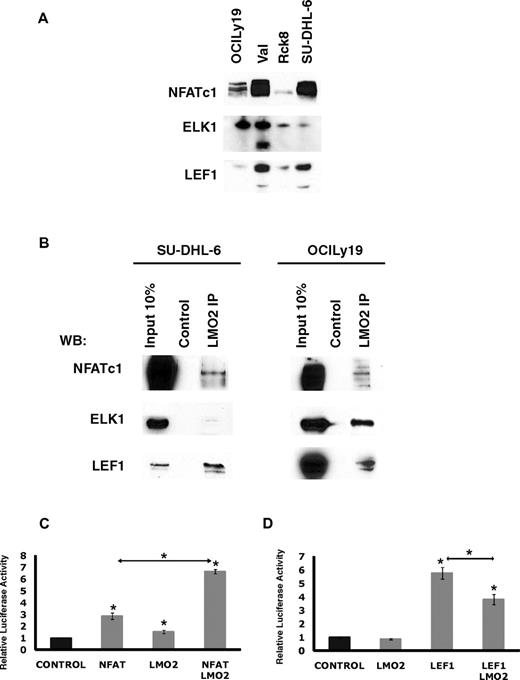
Overall, this analysis suggested that LMO2 may potentially interact with novel TFs in the context of DLBCL cells. To address this possibility, we focused on NFATc1, ELK1, and LEF1, because they were expressed in all the analyzed DLBCL cell lines (Figure 6A). Of note, the VAL cell line also expressed a short isoform of ELK1. LMO2 immunoprecipitation experiments in the OCILy19 and SU-DHL-6 cell lines confirmed interaction between LMO2 and NFATc1, ELK1, and LEF1 proteins, respectively (Figure 6B), confirming the validity of our microarray data and computational approach.
LMO2 interacts with LEF1, ELK1, and NFATc1 proteins. (A) Protein expression in nuclear extracts of DLBCL cell lines was assayed by immunoblotting with specific antibodies. Immunoblotting for Histone 3 served as a loading control. (B) Nuclear extracts were prepared from SU-DHL-6 and OCILy19 cell lines and used for immunoprecipitation with LMO2 antibody and immunoblotting with antibodies to indicated proteins. Results in panels A and B are representative of 2 independent experiments. (C) NFAT-driven luciferase reporter construct (10 μg), and either the NFATc1 (5 μg), LMO2 (5 μg), or both plasmids were coexpressed in the DOHH2 cells. Luciferase activity was determined 48 hours after transfection. The values are relative luciferase activities, with the average value obtained for the same amount of reporter and pcDNA3.1 control plasmid set at 1. Values are means + SE of 3 independent experiments, each performed in 3 replicate samples. (D) LEF1-driven luciferase reporter construct (3 μg), and either the LEF1 (10 μg), LMO2 (5 μg), or both plasmids were coexpressed in the Raji cells. Luciferase activity was determined 48 hours after transfection. The values are relative luciferase activities, with the average value obtained for the reporter using same amount of pcDNA3.1 control plasmid set at 1. Values are means plus standard error (error bars) of 3 independent experiments, each performed in 3 replicate samples.
LMO2 interacts with LEF1, ELK1, and NFATc1 proteins. (A) Protein expression in nuclear extracts of DLBCL cell lines was assayed by immunoblotting with specific antibodies. Immunoblotting for Histone 3 served as a loading control. (B) Nuclear extracts were prepared from SU-DHL-6 and OCILy19 cell lines and used for immunoprecipitation with LMO2 antibody and immunoblotting with antibodies to indicated proteins. Results in panels A and B are representative of 2 independent experiments. (C) NFAT-driven luciferase reporter construct (10 μg), and either the NFATc1 (5 μg), LMO2 (5 μg), or both plasmids were coexpressed in the DOHH2 cells. Luciferase activity was determined 48 hours after transfection. The values are relative luciferase activities, with the average value obtained for the same amount of reporter and pcDNA3.1 control plasmid set at 1. Values are means + SE of 3 independent experiments, each performed in 3 replicate samples. (D) LEF1-driven luciferase reporter construct (3 μg), and either the LEF1 (10 μg), LMO2 (5 μg), or both plasmids were coexpressed in the Raji cells. Luciferase activity was determined 48 hours after transfection. The values are relative luciferase activities, with the average value obtained for the reporter using same amount of pcDNA3.1 control plasmid set at 1. Values are means plus standard error (error bars) of 3 independent experiments, each performed in 3 replicate samples.
To further interrogate the consequences of LMO2 interaction with NFATc1 LEF1 and ELK1, the effects of LMO2 expression on NFATc1, LEF1, and ELK1 transcriptional activity were analyzed by reporter assays. LMO2 and NFATc1 coexpression led to a 6.6-fold increase in the NFAT reporter activity in the DOHH2 cell line not expressing endogenous LMO2 compared with 2.3-fold increase after expression of NFAT alone, and only minimal increase in the reporter activity after expression of LMO2 alone (Figure 6C). In contrast, in Raji cells not expressing endogenous LEF1, LMO2 significantly decreased transcriptional activity of the transfected LEF1, although having little effect on LEF1 reporter activity when transfected alone (Figure 6D). Coexpression of ELK1 encoding plasmid with the E743-tk80 luciferase plasmid in Raji cells led to an enormous decrease in luciferase activity, similar to a previous report.27 This decreased luciferase activity could not be affected by transfection of LMO2 (supplemental Figure 5B). These results support our finding that LMO2 can bind with LEF1 and NFATc1 and corroborate our gene expression data demonstrating predominant enrichment for down-regulated LEF1 target genes.
LEF1 expression in DLBCL
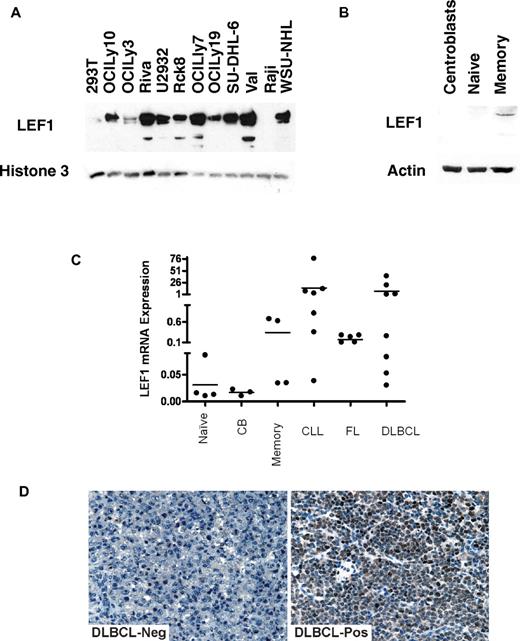
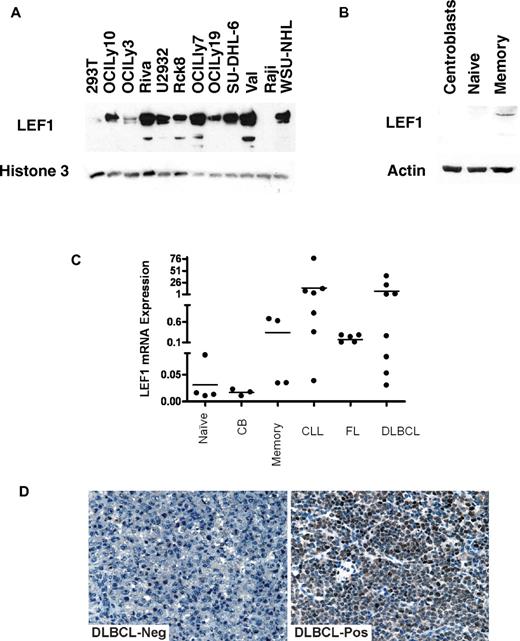
Although NFATc1 and ELK1 are known to be expressed in B cells and B-cell derived lymphomas, our finding of LEF1 expression and coimmunoprecipitation with LMO2 were unexpected, because LEF1 is a T-cell transcription factor mainly involved in T-cell differentiation. Although LEF1 is critical for B-cell development in bone marrow, it is not expressed in mature normal B cells.33 Consequently, to further validate our observations we evaluated LEF1 expression in nuclear extracts of 11 B-cell lymphoma cell lines (Figure 7A). Although LEF1 was not expressed in Raji Burkitt lymphoma cell line, it was expressed at various levels in all the analyzed DLBCL cell of both GC-like and ABC-like subtype. Analysis of LEF1 mRNA and protein expression in purified tonsil naive, GC, and memory cells revealed weak LEF1 protein expression only in normal memory cells but not in the other mature B-cell differentiation stages (Figure 7B-C). However, similar to chronic lymphocytic leukemia (CLL) cells in which LEF1 is known to be aberrantly expressed,34 we detected LEF1 mRNA expression also in most DLBCL, and follicular lymphoma (FL) cells (Figures 7C). To further validate our observations, LEF1 protein expression was analyzed by immunohistochemistry in a panel of 68 DLBCL samples (Figure 7D). A total of 71% of the evaluated DLBCL specimens expressed LEF1 at different levels, with 37 tumors (54%) exhibiting a strong expression. LEF1 expression was not associated with LMO2 expression or with GC-like and ABC-like subtypes (not shown).
LEF1 expression in B cells and B-cell derived lymphomas. (A) LEF1 protein expression in nuclear extracts of DLBCL cell lines was assayed by immunoblotting with LEF1 antibody using 293T and Raji cells as controls. Immunoblotting for Histone 3 served as a loading control. (B) LEF1 protein expression in cell extracts of normal B cells at different differentiation stages analyzed by immunoblotting with LEF1 antibody. (C) LEF1 mRNA expression in normal B cells at different differentiation stages and in primary CLL, FL, and DLBCL cell suspensions. (D) Immunohistochemistry of primary DLBCL specimens using LEF1 antibody. Representative examples of LEF1 negative (left) and positive (right) specimens are shown. Images were acquired using a Nikon Eclipse E1000 microscope (Nikon) equipped with 40× objective lenses. Images were captured with a SPOT flex mosaic 15.2 digital camera and software (Diagnostic Instruments). Digitized images were processed using Adobe Illustrator Version 10 software (Adobe Systems).
LEF1 expression in B cells and B-cell derived lymphomas. (A) LEF1 protein expression in nuclear extracts of DLBCL cell lines was assayed by immunoblotting with LEF1 antibody using 293T and Raji cells as controls. Immunoblotting for Histone 3 served as a loading control. (B) LEF1 protein expression in cell extracts of normal B cells at different differentiation stages analyzed by immunoblotting with LEF1 antibody. (C) LEF1 mRNA expression in normal B cells at different differentiation stages and in primary CLL, FL, and DLBCL cell suspensions. (D) Immunohistochemistry of primary DLBCL specimens using LEF1 antibody. Representative examples of LEF1 negative (left) and positive (right) specimens are shown. Images were acquired using a Nikon Eclipse E1000 microscope (Nikon) equipped with 40× objective lenses. Images were captured with a SPOT flex mosaic 15.2 digital camera and software (Diagnostic Instruments). Digitized images were processed using Adobe Illustrator Version 10 software (Adobe Systems).
Discussion
In this study we characterized the LMO2 interactome and transcriptome in DLBCL. Moreover, we defined components of the LMO2 multiprotein complex in DLBCL which includes LDB1, E2A, HEB, Lyl1, ETO2, and SP1 but not the classic interacting partners GATA1 and TAL1, which are not expressed in these cells. We demonstrated that this LMO2 complex regulates gene expression by binding to bipartite DNA motifs containing E2A and GC box binding sites. In addition, we identify novel LMO2 interaction partners, such as ELK1, NFATc1, and LEF1 that regulate expression of a distinct transcriptome in DLBCL cells. Finally, we demonstrate for the first time that LMO2 expression increases centrosome number, an effect that may lead to chromosome missegregation and contribute to oncogenesis.
Herein we demonstrate that overexpression of LMO2 affects expression of a transcriptome composed of 311 genes, both increasing and decreasing expression of specific genes. Interestingly, similar number of genes was shown to be down or up-regulated by LMO2 in embryonic stem cells (154 genes) and thymus T cells (113 genes),20,35 but the transcriptomes are quite unique in each of these cell types. This observation suggests existence of unique protein complexes in distinct cell types and/or cell-specific dynamic loss or addition of particular components that modulate activity and specificity of the LMO2-protein complex core. Differences in the composition of LMO2 complexes were indeed observed in the erythroid, vascular-endothelial, and T cells. We also demonstrated the formation of a specific LMO2 complex in DLBCL cells. Differences in up-regulation and down-regulation of specific genes by LMO2 complexes suggest that their effects probably depend on the particular context of gene promoters, resulting in sequestration of specific transcriptional factors or blocking their accessibility to the DNA, as was previously shown for LMO2-mediated regulation of E2A-HEB in T-ALL.21,36
Categorization of the LMO2-target genes by GO and IPA networks suggested that LMO2 protein may play a role in cell cycle, chromosome, nucleosome, and kinetochore structure. Although we did not observe LMO2 to have an effect on cell proliferation, LMO2 expression lead to numerical centrosome aberrations, accompanied by structural irregularities. Centrosomes are the microtubule-organizing centers controlling cell shape, polarity, and motility, and are crucial for chromosome segregation and cytokinesis. Centrosome abnormalities have long been related to aneuploidy and proposed to contribute to the development of cancer.37 Our findings that LMO2 leads to supernumerary centrosomes may at least partially explain its oncogenic potential in T-ALL and may suggest that LMO2 may also play an important role in DLBCL pathogenesis. Supernumerary centrosomes can arise as a result of centriole/centrosome duplication because of an aborted cell division and/or cell-cycle arrest or secondary to multiple rounds of centriole duplication or de novo centrosome generation within the same cell cycle. Absence of LMO2 effect on cell proliferation and cell cycle suggests that LMO2 most probably induces de novo centrosome generation. Although LMO2 increased the expression of Polo kinase 4, a positive regulator of centriole formation,38 the mechanism underlying LMO2 effect on centrosomes is currently unknown and is actively studied in our laboratory.
The LMO2 complex formed in the DLBCL cells was distinct from previously reported complexes in the erythroid, endothelial, and T cells. Several GATA family members were not expressed in all the analyzed DLBCL cell lines and normal GC B cells as also previously reported,39,40 and we did not find enrichment for GATA-binding motifs among LMO2-target genes in DLBCL. We also did not detect TAL1 expression in DLBCL, in concordance with previous reports on TAL1 down-regulation during terminal B-lymphoid lineage differentiation. However, Lyl1, a tissue specific member of the same basic helix-loop-helix (bHLH) family with highly homologous bHLH domain,41 was expressed in the DLBCL cells and coimmunoprecipitated with LMO2 and other components of the complex. Similar to TAL1, Lyl1 can heterodimerize with ubiquitously expressed class I bHLH family members HEB or E2A gene product E12 and interact with DNA.42 E2A is absolutely essential for B-lymphoid development. In cooperation with EBF1, E2A regulates the establishment of B-cell specific gene expression profile, is involved in IgH and IgL chain rearrangement and regulates AID expression in mature T cells.43,44 Herein we demonstrate that E2A is a component of the LMO2 complex in DLBCL. Whether LMO2 complex is involved in E2A-promoted immunoglobulin (Ig) mutations and class switch recombination is presently unknown and need to be addressed in future studies. However, it should be noted, that ABC-like DLBCL characterized by low or absent LMO2 expression frequently express nonswitched surface IgM, in contrast to GCB-like DLBCL that commonly express LMO2 and switched Ig receptors.45
Another member of the class I bHLH family, HEB protein, was also expressed in DLBCL cells, and coimmunoprecipitated with LMO2 and other classic components of the LMO2-protein complex. Previous studies suggested that HEB is necessary for early B-cell development but is dispensable in mature B cells in which only E2A homodimers exist.46 Moreover, previous studies suggested that HEB is not expressed in GC B-cells.44,47 However, we demonstrated HEB expression in GC B cells and in GCB-like DLBCL and showed its interaction with E2A and other components of the LMO2 complex.
ETO2, a non-DNA binding protein inhibiting differentiation of erythroid cells and megakaryocytes by interaction with LMO2 complex,11,13 was also expressed and coimmunoprecipitated with LMO2 in DLBCL cells. Its function in DLBCL is currently unknown. SP1 protein, previously reported to be part of the LMO2 protein complex in hematopoietic progenitor cells where it regulates c-kit,31 was also part of the LMO2 complex in the DLBCL cells.
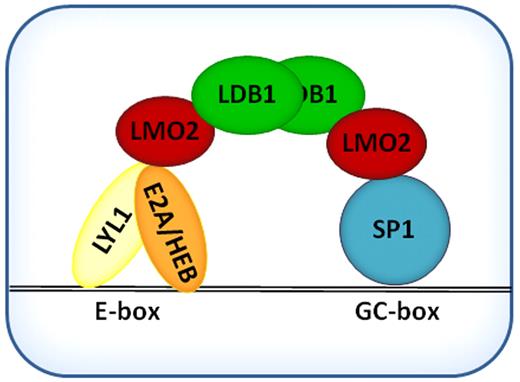
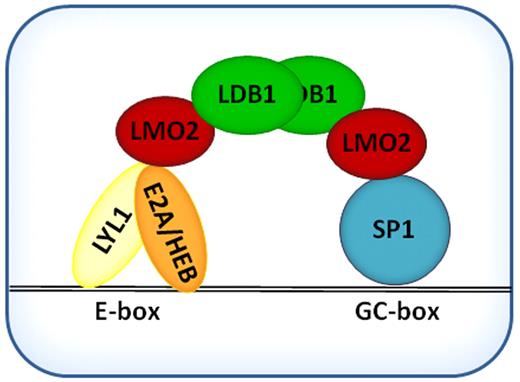
Taking into account previous data on function of LMO2 complexes in different cells and using the promoter of DLEU2 gene, whose expression was up-regulated after LMO2 expression, we tried to refine the function of the LMO2 complex in the context of B cell lymphoma. These studies suggest that in DLBCL the LMO2 complex is composed of LDB1, acting as a scaffold protein that recruits LMO2. LMO2 binds through its LIM domains with SP1 and with the bHLH domain of Lyl1,31 which heterodimerizes to class I bHLH proteins E2A and/or HEB and thereby bind DNA through the GC box (GGGCGGR) and Ebox (CAGGTG) motifs (Figure 8). Sequence divergence between the distinct gene promoters containing GC boxes and/or variation in the composition of the LMO2 complex on different promoters may contribute to the observed increased or decreased gene expression and require dedicated studies of regulations of specific genes. For example, the complex may bind ETO2, a reported repressor, and potentially additional proteins.
Schematic representation of basic LMO2 protein complex in DLBCL cells.
Indeed, based on TF motif-enrichment studies, we demonstrated that LMO2 interacts with ELK1, LEF1, and NFATc1 transcription factors, novel binding partners, contributing to regulation of LMO2 target gene expression. ELK1 is a member of the ETS family of transcription factors that can regulate LMO2 expression.48 Furthermore, recent study in T cells showed that LMO2 interacts with ETS family members (ERG, FLI1) regulating expression of HHEX/PRH, implicated in development of T-ALL.48 Although our results suggest that LMO2 is not affecting ELK1 transcriptional activity as assessed by the E743-tk80 reporter vector, they are not totally excluding the possibility that LMO2 may affect ELK1 transcriptional activity in vivo and in the setting of specific promoter sequences and/or in the presence of additional interacting proteins. It should be noted that in a previous study,27 only transfection of large quantities of constitutively active RAF-1 kinase could alleviate ELK1-mediated repression of the E743-tk80 reporter used herein. Therefore, further studies are needed to fully assess effects of LMO2 on ELK1 transcriptional activity.
NFATc1 is a member of the NFAT family of transcription factors that regulates expression of several cytokines in T cells. It is activated on B-cell receptor signaling and is implicated in regulating and maintaining survival of DLBCL cells49 Herein we demonstrate that LMO2 coimmunoprecipitates with NFATc1 and increased NFATc1 reporter activity. Further studies are ongoing to investigate the significance of LMO2 and NFATc1 interaction in DLBCL pathobiology.
LEF1 is involved in WNT–β-catenin signaling regulating cell differentiation. It was previously reported to be expressed in T cells and is implicated in the pathogenesis of acute leukemias.50,51 In mature B-cell lineage, LEF1 protein was previously reported to be expressed only in chronic lymphocytic leukemia.34 Herein we demonstrate that LEF1 is also expressed in normal memory B cells as well as in a fraction of primary DLBCL independent of their cell of origin. Furthermore, we show that LMO2 interacts with LEF1 and decreases its reporter activity. These observations suggest that although LEF1 can be expressed in both GCB-like and ABC-like DLBCL, its transcriptional activity may be reduced in the GCB-like tumors expressing high levels of the LMO2 protein. Further studies are needed to elucidate the role of LEF1 in DLBCL and other B-cell tumors.
Overall, our studies elucidated the LMO2 transcriptome and identified novel protein complex and interacting partners in DLBCL cells. These findings also provided important clues and raised multiple critical questions regarding potential functions of LMO2 in this disease that are currently actively investigated in our laboratory.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Cornelis Murre (University of California, San Diego), Dr Luc Sabourin (University of Ottawa, Ottawa, ON), Dr Jonathan Horowitz (North Carolina State University), Dr Serban San-Marina (Ontario Cancer Institute, ON), Dr Trang Hoang (IRCM, Montreal, QC), and Dr Ralf Janknecht (University of Oklahoma Health Sciences Center) for providing valuable reagents that were used in this paper. They thank Dr George McNamara for technical support with microscopy.
E.C. is partially supported by the Fundacion Alfonso Martin Escudero. I.S.L. is supported by National Institutes of Health (NIH) grants CA109335, CA122105, and U56 CA112973, and the Dwoskin Family and Recio Family foundations. I.S.G. is supported by MICINN (SAF2009-08 803), and by MEC OncoBIO Consolider-Ingenio 2010 (Ref. CSD2007-0017). All Spanish funding is cosponsored by the European Union FEDER program.
National Institutes of Health
Authorship
Contribution: E.C. designed the study, performed experiments, analyzed the data, and wrote the paper; A.J.G. designed the study, performed experiments, analyzed the data, and wrote the paper; C.H., S.B., X.L., X.J., A.F., S.Z., and C.E.B. performed experiments; Y.N. performed experiments, analyzed the data, and wrote the paper; I.R.-C., J.A.M.-C., and I.S.-G. analyzed the data; A.M. analyzed the data and wrote the paper; I.S.L. designed the study, performed experiments, analyzed the data, and wrote the paper; and all authors reviewed the paper and agree with its content.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Izidore S. Lossos, Professor of Medicine, Sylvester Comprehensive Cancer Center, University of Miami, 1475 NW 12th Ave (D8-4), Miami, FL 33136; e-mail: ilossos@med.miami.edu.