Abstract
Targeted T-cell therapy is a potentially less toxic strategy than allogeneic stem cell transplantation for providing a cytotoxic antileukemic response to eliminate leukemic stem cells (LSCs) in acute myeloid leukemia (AML). However, this strategy requires identification of leukemia-associated antigens that are immunogenic and exhibit selective high expression in AML LSCs. Using microarray expression analysis of LSCs, hematopoietic cell subpopulations, and peripheral tissues to screen for candidate antigens, cyclin-A1 was identified as a candidate gene. Cyclin-A1 promotes cell proliferation and survival, has been shown to be leukemogenic in mice, is detected in LSCs of more than 50% of AML patients, and is minimally expressed in normal tissues with exception of testis. Using dendritic cells pulsed with a cyclin-A1 peptide library, we generated T cells against several cyclin-A1 oligopeptides. Two HLA A*0201-restricted epitopes were further characterized, and specific CD8 T-cell clones recognized both peptide-pulsed target cells and the HLA A*0201-positive AML line THP-1, which expresses cyclin-A1. Furthermore, cyclin-A1–specific CD8 T cells lysed primary AML cells. Thus, cyclin-A1 is the first prototypic leukemia-testis-antigen to be expressed in AML LSCs. The pro-oncogenic activity, high expression levels, and multitude of immunogenic epitopes make it a viable target for pursuing T cell–based therapy approaches.
Introduction
It is well established that acute myeloid leukemia (AML) is organized hierarchically, initiated and maintained by a small population of cells referred to as leukemia stem cells (LSCs) that are characterized not only by unlimited reproductive capacity but also by enhanced resistance to chemotherapy and radiation. This primitive cell population, which is usually contained within a subpopulation of leukemic cells that are CD34+ but lack expression of CD38 and lineage markers, is essential and adequate for long-term engraftment of primary AML cells in NOD/SCID transplantation models.1-3 The LSC model suggests that, for a therapeutic anti-AML effect to be curative in patients, it will be necessary to identify strategies that efficiently eliminate the LSC compartment, which is often resistant to conventional chemotherapy.
In patients with intermediate-risk, high-risk, or relapsed AML, the allogeneic T cell–mediated graft-versus-leukemia effect after hematopoietic cell transplantation (HCT) or infusion of donor-derived lymphocytes in the post-HCT period has been shown to be essential for achievement of long-term remissions.4-7 However, allogeneic HCT and unselected donor lymphocyte infusions are associated with significant toxicity because of both the conditioning regimen and the graft-versus-host activity of donor lymphocytes. An alternative strategy to provide anti-LSCs cytotoxic T lymphocytes to treat AML patients would use more targeted T-cell therapy, consisting of either adoptive transfer of T cells specific for, or vaccination against, leukemia associated antigens (LAAs).8,9 The ability of such antigen-specific T cells to eliminate AML LSCs has been demonstrated in NOD/SCID transplantation models.10-12
For targeted T-cell therapy to achieve maximal efficacy against AML with minimal toxicity, identified LAAs need to have not only high expression in and presentation by leukemic cells but also lack significant expression in healthy tissues. Several AML LAAs have been described, but only Wilms tumor protein 1 (WT1), which is currently being targeted in clinical trials both with adoptive T-cell transfer and peptide vaccination, has been shown to be expressed in LSCs of the majority of AML patients at levels significantly higher than the physiologic levels in hematopoietic stem cells (HSCs).12-15 Although objective responses/remissions have been observed in some treated patients, in many no anti-WT1 T-cell response can be elicited, and in others WT1 is not detected at levels in leukemic cells sufficiently distinct from HSCs to be targeted. Thus, additional candidate LAAs expressed in AML LSCs are greatly needed.
In this study, analyses of differential gene expression identified cyclin-A1 as a candidate new T-cell target. Cyclin-A1 is selectively expressed normally in testis, regulating progression of male germ cells through meiosis I.16,17 CCNA1−/− mice are viable and phenotypically normal, with the exception of male infertility.18,19 Cyclin-A1 is aberrantly expressed in AML as well as other malignancies.16,20 In AML, it can sustain the malignant phenotype through pro-proliferative and antiapoptotic activities,21-23 and overexpression of cyclin-A1 in mice causes dysplastic myelopoiesis with 15% of mice developing transplantable myeloid leukemias.24 We now show that cyclin-A1 is a testis-leukemia-antigen that harbors a multitude of immunogenic MHC class I epitopes, which can be used to generate T cells from healthy donors that recognize and lyse leukemic cells.
Methods
Human samples
Mononuclear cells of patients with AML, chronic myeloid leukemia (CML), and myelodysplastic syndrome (MDS) from peripheral blood and bone marrow (BM) were isolated by leukopheresis or Ficoll-Hypaque (Biochrom). AML samples contained more than 60% malignant cells. Cells were collected at Fred Hutchinson Cancer Research Center and Charité Campus Benjamin Franklin. For generation of T-cell lines, leukapheresis products were obtained from 2 healthy donors at Fred Hutchinson Cancer Research Center. All samples were collected after written informed consent in accordance with the Declaration of Helsinki and with approval of the institutional review boards of both participating institutions.
Cell lines
Epstein-Barr virus–transformed lymphoblastoid cell lines (LCLs) were generated as described.25 The T-cell/B-cell hybrid cell line T2 used to present epitopes is TAP-deficient and expresses only HLA A*0201. LCL 721.221 expresses no endogenous HLA class I because of radiation-induced deletion of the respective alleles and was stably transfected with the retroviral vector pLBPC containing the indicated HLA alleles.26 Cell lines K562 (CML), THP-1, HL60, KG1 (AML), and U937 (monocytic cell line) were maintained in RPMI 1640 supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin (Invitrogen), and 10% FBS, with 50μM β-mercaptoethanol (Sigma-Aldrich) also added for THP-1. T cells and dendritic cells (DCs) were maintained as described.27
Microarray data analysis
Two panels of microarray datasets (Affymetrix) were used: (1) 9 AML LSC samples (lineage−, CD34+, CD38−, CD90−),14 7 corresponding leukemic blast samples (lineage−, CD34−),28 4 HSC samples (lineage−, CD34+, CD38−, CD90+),14 and datasets from PBMCs, CD34+ BM mononuclear cells, and tissues (National Center for Biotechnology Information Gene Expression Omnibus [GEO] server GSM279585-279588, 414970, 414972, 414975, 419165-419174, 457175-457177, 483480-483496, 80576, 80582, 80602, 80615, 80619, 80653, 80689, 80712, 80734, 80738, 80739, 80759, 80792, 80824, 80826, 80867, 80869, HG U133 plus 2.0 format); and (2) 30 AML samples (> 75% malignant cells, 8 CD34+ BM samples, 9 PBMCs samples and 2 testis samples (HG U133A format). This microarray data is available at the GEO database under the accession number GSE73707. Samples were normalized using the invariant set method (dChip 2.0 software29 ). Before analysis at the single probe set level, unsupervised hierarchical clustering was performed to rule out clustering because of the sample origin rather than the biologic background. Samples exceeding the mean expression plus 3 standard deviations (SDs) of the HSC samples were considered “positive.” Expression values in LSCs were compared with other cell types using a 2-tailed Mann-Whitney test. Expression values from 7 LSCs with their corresponding paired leukemic blasts were compared using a 2-tailed Wilcoxon signed rank test.
Quantitative real-time PCR
Total RNA was extracted using Trizol reagent (Invitrogen). Reverse transcription was performed using Superscript III (Invitrogen). A panel of cDNAs from pooled healthy tissues was purchased from Clontech, and 5 samples of healthy BM were purchased from Cambrex. Quantitative 2-step real-time PCR (RT-PCR) was performed on an ABI 7500 machine (Applied Biosystems) with TA = 60°C using the following primers/probes: glyceraldehyde-3-phosphate-dehydrogenase (GAPDH)_fwd: GAGTCAACGGATTTGGTCGT; GAPDH_probe: 6FAM-GATATTGTTGCCATCAATGACCCCT-TAMRA; GAPDH_rev: GACAAGCTTCCCGTTCTCAG; CyclinA1_fwd: CATGAAGAAGCAGCCAGACA; cyclin-A1_probe: 6FAM-TTCGAGCAGAGACCCTGTATCTGG-TAMRA; cyclin-A1_rev: TTCGAAGCCAAAAGCATAGC. Crossing points were plotted against standard curves of pCR4-TOPO plasmids (Invitrogen) containing the respective PCR product.30 Reactions were performed in duplicates, and expression was presented as copies per copies of GAPDH. Samples exceeding the mean expression plus 3 SDs of the BM samples were considered “positive.”
Cytokines and peptides
Recombinant human IL-1β, IL-4, IL-7, IL-15, and TNF-α were obtained from R&D Systems, IL-2 and GM-CSF from Chiron, prostaglandin E2 from MP Biomedicals, and IL-21 from PeproTech. A peptide library of a total of 103 15-mers with an overlap of 11 amino acids (AA) spanning cyclin-A1 (isoform c, NM_001111046) was purchased from Sigma-Aldrich.
Generation of cyclin-A1–specific T-cell clones
T-cell lines were generated as described with minor modifications.27 Briefly, DCs were derived from plastic adherent PBMCs after culture for 2 days (day −2 to day 0) in DC media (CellGenix) supplemented with GM-CSF (800 U/mL) and IL-4 (1000 U/mL). On day −1, maturation cytokines TNF-α (1100 U/mL), IL-1β (2000 U/mL), IL-6 (1000 U/mL), and prostaglandin E2 (1 μg/mL) were added. On day 0, DCs were harvested and pulsed with peptide (single peptides at 10 μg/mL, peptide pools at 2 μg/mL). T cells were isolated from PBMCs using anti-CD8 microbeads (Miltenyi Biotec) and stimulated with DCs at an E:T ratio of 1:5 to 1:10 in the presence of IL-21 (30 ng/mL). On day 3, IL-2 (12.5 U/mL), IL-7 (5 ng/mL), and IL-15 (5 ng/mL) were added.
Cells were restimulated between days 10 and 14 with the plastic adherent fraction of irradiated autologous PBMCs as antigen presenting cells after pulsing with peptide. IL-21 was added on day 0; cells were supplemented from day 1 on with IL-2, IL-7, and IL-15.
T-cell clones were generated in 2 ways: (1) unselected, by plating T cells from library-specific cell lines at limiting dilution and expanding with TM-LCLs coated with OKT3 (Ortho Biotech) and allogeneic PBMCs as feeders (REP protocol) as described27 ; and (2) selected for a single specificity, in which T cells were stimulated with autologous LCLs pulsed with the specific peptide for 4 hours at an E:T ratio of 1:2. IFN-γ secreted during the next 45 minutes was bound to the surface of the specific cells by the catch matrix reagent of the IFN-γ–secretion assay (Miltenyi Biotec). Cells were stained with IFN-γ–PE (Miltenyi) and CD8-FITC (BD Biosciences) and sorted on an Aria III instrument (BD Biosciences). T-cell clones were then generated/expanded as described.
Four T-cell clones were analyzed in this study: 2264.E30 is specific for epitope 341-351, clones 2196.D9, 2196.D11a and 2196.D11b are specific for epitope 227-235, and D11a and D11b are sister clones sharing the same TCR.
ICS and IF
For IFN-γ staining, antigen-presenting cells were pulsed with 10 μg/mL peptide overnight and washed once. Effector cells were coincubated with these cells for 6 hours in the presence of monensin. Cells were stained with anti-CD8–FITC, permeabilized using the BD Cytofix/Cytoperm kit, and stained with anti–IFN-γ–APC (BD Bioscience).
Immunofluorescence (IF) staining of AML cells and normal BM was performed after fixation with 4% formaldehyde followed by permeabilization/erythrocyte lysis with 0.1% Triton X-100. Air-dried cytospin preparations were permeabilized with 95% and 70% ethanol and immunostained with anti-cyclin-A1 (Novus Biologicals). Slides were then incubated with secondary goat anti–rabbit IgG–AlexaFluor-488 (Invitrogen), washed, dried, and mounted with 4,6-diamidino-2-phenylindole Vectashield (Vector Laboratories).
For FACS intracellular staining (ICS) staining of cyclin-A1, BM cells from AML patients were stained with anti-CD45–V450 and either anti-CD34–APC or anti-CD33–PE-Cy5 (BD Biosciences). Cells were fixed and permeabilized in lysis/permeabilization buffer (0.233% Triton X-100 in PBS, calcium, and magnesium free). Cyclin-A1 was detected using human-specific anti-cyclin-A1 antibody (Novus Biologicals) followed by indirect detection with AlexaFluor-480–conjugated goat anti–rabbit F(ab′)2 fragment (Invitrogen).
HLA stabilization assay
T2 cells were pulsed with 100 μg/mL peptide in serum-free RPMI containing 1 μg/mL β2-microglobulin (Sigma-Aldrich) for 16 hours. Cells were then incubated for 4 hours in the presence of brefeldin A (Sigma-Aldrich), and stained with anti-HLA A/B/C-FITC (W6/32; BD Biosciences).
Caspase-3 assay
Target cells were membrane-labeled with PKH26 (Sigma-Aldrich). T-cell clones were used at the end of the REP cycle (day 12 or later). Targets and T cells were incubated at an E:T ratio of 3 to 5:1 for 4 hours. As a negative control, targets were incubated without effectors; as a positive control, targets were incubated in the presence of 4μM camptothecin or 1μM staurosporine (Sigma-Aldrich). Cells were fixed and permeabilized using the BD Cytofix/Cytoperm kit and stained with anti-active caspase-3–antibody conjugated to FITC or AlexaFluor-647 (C92-625; BD Biosciences).
51Chromium release assay
Standard 51Cr release assays were performed as described27 using 5000 targets cells at E:T ratios of 10 to 1.25:1 in triplicates. Spontaneous release was assessed by incubating targets in the absence effectors. Percentage specific lysis was calculated using the formula: 100 × (experimental release − spontaneous release)/(maximum release − spontaneous release).
Results
Cyclin-A1 is selectively expressed in AML LSCs, leukemic blasts, and testis
To systematically screen for candidate genes selectively expressed in the AML LSC compartment for targeting with T cells, we analyzed 9 LSC microarray datasets with samples of different hematopoietic cell subsets and nonhematopoietic tissues. Suitable candidate genes were identified by mathematical filtering and manual vetting of the model-based expression values (Figure 1). Based on the microarray expression data, as well as published data on oncogenicity and cellular location, and after validating that the probe sets were homologous to coding mRNA, 7 candidate probe sets were identified (Figure 2A-B). For all of the probe sets, the levels detected in LSCs were not statistically different from the levels detected in the corresponding more differentiated leukemic blast samples (Figure 2C; data not shown). The overexpression of the probe sets in AML samples was confirmed in a second independent panel of microarrays for all probe sets, with the exception of 233734_s_at and 1554298_a_at, which were not represented on the HG U133A format (data not shown).
Systematic approach to identify potential target genes in AML LSCs based on 3′IVT expression microarray data. The microarray panel was created combining data files from 5 independent studies. Datasets were filtered for present calls in more than 10% of the samples and overall variability (reject probe sets with an SD/mean < 2). Hierarchical cluster analysis was performed to confirm clustering in accordance to sample biology. Target candidates were identified by mathematical filtering followed by visual inspection of the respective expression patterns. Targeted sequences of the probe sets were then analyzed to make sure that they actually represented a coding mRNA sequence. In case of several probe sets representing the same gene, expression patterns of all probe sets were inspected for consistency. Finally, target candidates, which were known to be membrane bound or secreted, were rejected.
Systematic approach to identify potential target genes in AML LSCs based on 3′IVT expression microarray data. The microarray panel was created combining data files from 5 independent studies. Datasets were filtered for present calls in more than 10% of the samples and overall variability (reject probe sets with an SD/mean < 2). Hierarchical cluster analysis was performed to confirm clustering in accordance to sample biology. Target candidates were identified by mathematical filtering followed by visual inspection of the respective expression patterns. Targeted sequences of the probe sets were then analyzed to make sure that they actually represented a coding mRNA sequence. In case of several probe sets representing the same gene, expression patterns of all probe sets were inspected for consistency. Finally, target candidates, which were known to be membrane bound or secreted, were rejected.
Identification of cyclin-A1 as target candidate byin silicoexpression analysis and RT-PCR quantification. (A) Seven candidate target genes predicted in silico: Heatmap of normalized probe sets. Each row represents one probe set, and each column represents one sample dataset. Red represents increased levels of expression; and blue, decreased levels of expression. The darkest shade of red/blue represents average expression ± 3 SDs. (B-C) Model-based expression of probe set 205899_at representing cyclin-A1. (B) Expression in AML LSCs compared with HSCs/CD34+ BM mononuclear cells, PBMCs, and nonhematopoietic somatic tissues. The dashed line represents the cut-off value of 69.2 (mean plus 3 SDs of the HSC samples). *P < .001 (explorative). (C) Expression in AML LSCs and corresponding blasts (P = .297). (D) Cyclin-A1 expression quantified by quantitative RT-PCR. The dashed line represents the cut-off value of 0.048 (mean plus 3 SDs of the BM samples). (E) Immunofluorescence staining of cyclin-A1 in primary AML samples (representative example). Shown is a healthy BM (top row) and primary leukemic blasts (bottom row). Cells were stained with 4,6-diamidino-2-phenylindole (DAPI; left) and indirectly for cyclin-A1 (AlexaFluor-488, right). Negative controls without primary antibody and nonspecific isotype showed no fluorescence in both samples (not shown). Images were captured using a Nikon Eclipse E800 microscope. (F) Intracellular FACS staining of cyclin-A1 in primary AML blasts (representative example). BM mononuclear cells of a patient with cyclin-A1-expressing AML after gating out doublets in forward scatter (FSC)-area/height (A/H) and SSC-A/H plots. Top row: gating on blasts based on SSC and CD45 expression. Bottom row: gating on CD33-positive cells. Histograms show cyclin-A1 (black line) and isotype control (shaded) in the blast population.
Identification of cyclin-A1 as target candidate byin silicoexpression analysis and RT-PCR quantification. (A) Seven candidate target genes predicted in silico: Heatmap of normalized probe sets. Each row represents one probe set, and each column represents one sample dataset. Red represents increased levels of expression; and blue, decreased levels of expression. The darkest shade of red/blue represents average expression ± 3 SDs. (B-C) Model-based expression of probe set 205899_at representing cyclin-A1. (B) Expression in AML LSCs compared with HSCs/CD34+ BM mononuclear cells, PBMCs, and nonhematopoietic somatic tissues. The dashed line represents the cut-off value of 69.2 (mean plus 3 SDs of the HSC samples). *P < .001 (explorative). (C) Expression in AML LSCs and corresponding blasts (P = .297). (D) Cyclin-A1 expression quantified by quantitative RT-PCR. The dashed line represents the cut-off value of 0.048 (mean plus 3 SDs of the BM samples). (E) Immunofluorescence staining of cyclin-A1 in primary AML samples (representative example). Shown is a healthy BM (top row) and primary leukemic blasts (bottom row). Cells were stained with 4,6-diamidino-2-phenylindole (DAPI; left) and indirectly for cyclin-A1 (AlexaFluor-488, right). Negative controls without primary antibody and nonspecific isotype showed no fluorescence in both samples (not shown). Images were captured using a Nikon Eclipse E800 microscope. (F) Intracellular FACS staining of cyclin-A1 in primary AML blasts (representative example). BM mononuclear cells of a patient with cyclin-A1-expressing AML after gating out doublets in forward scatter (FSC)-area/height (A/H) and SSC-A/H plots. Top row: gating on blasts based on SSC and CD45 expression. Bottom row: gating on CD33-positive cells. Histograms show cyclin-A1 (black line) and isotype control (shaded) in the blast population.
To confirm the microarray data, all 7 candidates were quantified by quantitative RT-PCR in a third independent sample set (Figure 2D; supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). All but 1 of the candidates were eliminated because of either lack of correlation between microarray data and quantitative RT-PCR results (1 gene), significant expression in healthy tissues and/or HSCs (5 genes), or low total copy number less than 10−2 copies/copies GAPDH (1 gene).
Only cyclin-A1 displayed selective expression in LSCs and/or AML blasts in all 3 datasets. The respective probe set had been validated by quantitative RT-PCR in an earlier study.20 In the first microarray panel, cyclin-A1 was overexpressed in 6 of 9 analyzed LSC samples (Figure 2B). No overexpression was found in any non-AML sample except testis. Expression values of cyclin-A1 were significantly higher in LSCs than in all other samples (P < .001, Figure 2B), although this P value has to be considered exploratory because the statistical testing was performed on the array set used for target selection. As no significant difference in cyclin-A1 expression was observed between the LSCs and leukemic blasts derived from the same patients (P = .297, Figure 2C), the expression pattern of cyclin-A1 was then confirmed in an additional panel that included AML cells not selected for LSCs, BM normal CD34+, and PBMCs, parts of which have already been published.20 We now analyzed these AML datasets with 2 datasets of testis tissue. Using a cut-off value of mean plus 3 SDs of the BM samples, no BM CD34+ or PBMC sample achieved positivity, whereas 21 of 30 AML samples displayed cyclin-A1 expression. In both testis samples, the cyclin-A1 expression was higher than the average expression in the AML samples (data not shown).
To confirm the expression detected in arrays with probe set 205899_at, cyclin-A1 was quantified in AML samples, other hematopoietic cell subsets, and nonhematopoietic tissues using quantitative RT-PCR. Cyclin-A1 over-expression was detected in 24 of 44 analyzed AML samples not selected for LSCs (55%). No over-expression of cyclin-A1 was detected in BM, G-CSF mobilized CD34+ cells or proliferating T cells (Figure 2D).
Next, we analyzed the frequencies and expression levels of cyclin-A1 in different French-American-British (FAB) AML subtypes and BM samples from patients with CML and MDS. Highest expression levels were observed in acute promyelocytic leukemia (APL), as previously described.31 Moreover, the frequencies of cyclin-A1 positivity did vary with FAB subtype, with 100% positivity in APL (3/3) and M4 (5/5), > 50% positivity in M0 (3/5) and M5 (7/13), and < 50% in M1 (3/8), M2 (1/3) and unspecified and/or secondary AML (2/7, Figure 2D). No over-expression was found in patients with MDS or CML. The median copy number of cyclin-A1 per GAPDH in the AML samples was approximately 10-fold higher than the copy number of WT1 in the same sample set (supplemental Figure 2).
To confirm a uniform cyclin-A1 expression within the malignant blast population, IF was performed in 3 AML samples, which expressed cyclin-A1 in quantitative RT-PCR and in healthy BM. Whereas the BM showed no fluorescence, we observed a strong homogenous fluorescence in all 3 AML samples (Figure 2E; data not shown). In addition, cyclin-A1 ICS was performed in 4 primary AML samples. Again, we observed a uniform bright staining in the blasts in all samples tested, both gated on blasts in the CD45/side scatter (SSC) plot and after staining CD33 or CD34 (Figure 2F; data not shown).
Mapping of multiple immunogenic oligopeptides on cyclin-A1
For identification of MHC class I-restricted T-cell epitopes, a reverse immunology approach was used. Three different isoforms for cyclin-A1 have been described with isoform C distinguishable by having a shorter N-terminus. As no functional domains have been identified on the longer N-termini of isoforms A and B, and the respective transcripts for these isoforms could not be amplified by nested PCR either from testis or AML samples (data and primer sequences not shown), we constructed a peptide library representing the shorter isoform C so that immune escape from targeting epitopes in the N-termini could not occur if cells express only this shorter isoform.
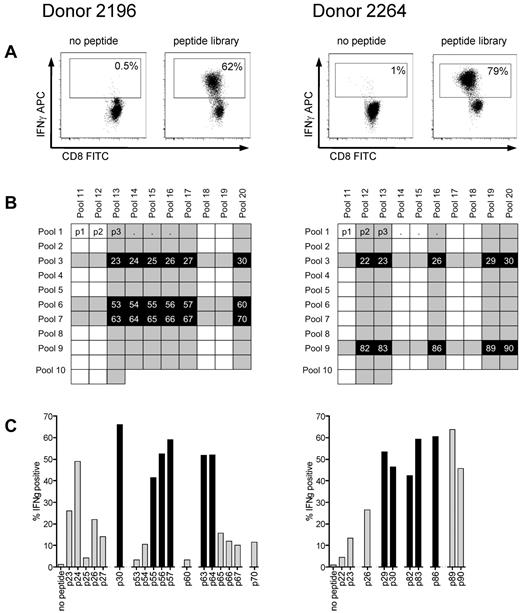
T-cell lines were generated from donors 2196 and 2264, who both express HLA A*0201, the most common class I allele found in whites, by 4 stimulations with autologous antigen presenting cells pulsed with the peptide library. Using autologous LCLs pulsed with the library as stimulators for an ICS assay, more than 60% of cells in both T-cell lines appeared specific for cyclin-A1 based on IFN-γ production (Figure 3A). To identify the immunogenic 15-mers within the library, the 2 T-cell lines were stimulated with 20 peptide pools containing 10 to 13 15-mers, with every 15-mer being contained in 2 peptide pools (Figure 3B) and tested for IFN-γ production. We identified 18 peptides for donor 2196 and 10 peptides for donor 2264 that were present in corresponding pools and recognized by the respective T-cell lines, and these 15-mers were tested individually (Figure 3C). After identifying the immunogenic 15-mers, the minimal immunogenic AA sequence of each epitope was determined by stimulating with shorter peptides. With this approach, 8 immunogenic oligopeptides were mapped (Table 1).
Reverse immunology strategy for epitope mapping in cyclin-A1 using ICS for IFN-γ. (A) After 4 stimulations with the peptide library, T-cell lines from both donors consisted of more than 60% specific cells (gated on CD8+ cells). (B) The cell lines were subsequently tested against 20 peptide pools with each 15-mer being represented in 2 different pools in the peptide matrix. Shaded rows/columns indicate pools with more than 10% IFN-γ-positive cells from each donor. (C) Peptides, which tested positive in both of its pools (> 10% specificity, marked black), were further analyzed as individual peptides. Black columns represent the peptides for which the minimal immunogenic AA sequence from the initial 15-mer was determined.
Reverse immunology strategy for epitope mapping in cyclin-A1 using ICS for IFN-γ. (A) After 4 stimulations with the peptide library, T-cell lines from both donors consisted of more than 60% specific cells (gated on CD8+ cells). (B) The cell lines were subsequently tested against 20 peptide pools with each 15-mer being represented in 2 different pools in the peptide matrix. Shaded rows/columns indicate pools with more than 10% IFN-γ-positive cells from each donor. (C) Peptides, which tested positive in both of its pools (> 10% specificity, marked black), were further analyzed as individual peptides. Black columns represent the peptides for which the minimal immunogenic AA sequence from the initial 15-mer was determined.
Immunogenic peptides on cyclin-A1 isoform c
| AA position . | AA sequence . | Donor . | HLA restriction . | Clone available . |
|---|---|---|---|---|
| 118-127 | YEVDTGTLKS | 2264 | B*4001 | Yes |
| 120-131 | VDTGTLKSDLHF | 2196 | B*5701 | Yes |
| 167-175 | YAEEIYQYL | 2264 | B*4001 | Yes |
| 218-226 | AETLYLAVN | 2196 | A*0201 | No |
| 227-235 | FLDRFLSCM | 2196 | A*0201 | Yes |
| 253-261 | ASKYEEIYP | 2196 | ND | No |
| 330-339 | LEADPFLKYL | 2264 | B*4001 | No |
| 341-351 | SLIAAAAFCLA | 2264 | A*0201 | Yes |
| AA position . | AA sequence . | Donor . | HLA restriction . | Clone available . |
|---|---|---|---|---|
| 118-127 | YEVDTGTLKS | 2264 | B*4001 | Yes |
| 120-131 | VDTGTLKSDLHF | 2196 | B*5701 | Yes |
| 167-175 | YAEEIYQYL | 2264 | B*4001 | Yes |
| 218-226 | AETLYLAVN | 2196 | A*0201 | No |
| 227-235 | FLDRFLSCM | 2196 | A*0201 | Yes |
| 253-261 | ASKYEEIYP | 2196 | ND | No |
| 330-339 | LEADPFLKYL | 2264 | B*4001 | No |
| 341-351 | SLIAAAAFCLA | 2264 | A*0201 | Yes |
The HLA restriction of the HLA B epitopes was determined using ICS for IFN-γ production with 721.221 stimulator cells transfected with a single HLA allele or allogeneic LCLs sharing one or 2 HLA class I alleles with the respective donor.
ND indicates not determined.
To identify epitopes that are HLA A*0201-restricted, the T-cell lines were tested for recognition of K562 directly, which does not express any endogenous HLA class I molecules on the surface, or K562 stably transfected with HLA A*0201 as APCs pulsed with the respective peptide (supplemental Figure 3A; data not shown). Epitopes 218-226, 227-235, and 341-351 stimulated responses in the context of HLA A*0201. Both donors expressed HLA A*0201, which was the only shared MHC class I allele; by regenerating T-cell lines to each epitope from fresh cells obtained from both donors, we subsequently confirmed both the immunogenicity and the HLA A*0201-restriction of these epitopes (data not shown). The HLA restriction of other epitopes was determined using 721.221 cells transfected with a single HLA allele or allogeneic LCLs sharing defined HLA class I alleles with the respective donor (supplemental Figure 3B; data not shown).
Characterization of 2 HLA A*0201 restricted epitopes and generation/analysis of T-cell clones
The minimal immunogenic sequence of the 15-mer 341-355 was determined in the T-cell line to be the 11-mer SLIAAAAFCLA (Figure 4A), which was surprising because of both its length and the absence of a characteristic AA residue in the carboxy-terminal HLA A*0201 anchor position, particularly because one of the 10-mers (SLIAAAAFCL, 10-mer 1) had the appropriate AA residues in both anchor positions. We therefore tested whether the 11-mer could form stable peptide-MHC complexes by performing an HLA stabilization assay. For this, T2 cells were pulsed with 100 μg/mL of an irrelevant peptide or the 11-mer, 10-mer 1, or 10-mer 2 (LIAAAAFCLA) overnight. After washing and incubation with brefeldin for 4 hours, HLA surface expression was assessed by FACS. An increase in MFI was observed for both the 11-mer and 10-mer 1, but the 11-mer/MHC complex did not appear as stable as the 10-mer 1/MHC complex (Figure 4B).
HLA A*0201-restricted epitope 341-351. Mapping the minimal immunogenic AA sequence and HLA stabilization. (A) The position of the different peptides in cyclin-A1. IFN-γ ICS of T-cell line (donor 2264) stimulated by autologous LCLs pulsed with the 10-mer 1, 10-mer 2, and 11-mer. Only pulsing with the 11-mer results in IFN-γ production in the T-cell line. (B) HLA stabilization assay: both 10-mer 1 and the 11-mer peptide stabilize HLA A*0201 on T2 cells. Negative controls are T2 cells pulsed with an irrelevant 15-mer (shaded).
HLA A*0201-restricted epitope 341-351. Mapping the minimal immunogenic AA sequence and HLA stabilization. (A) The position of the different peptides in cyclin-A1. IFN-γ ICS of T-cell line (donor 2264) stimulated by autologous LCLs pulsed with the 10-mer 1, 10-mer 2, and 11-mer. Only pulsing with the 11-mer results in IFN-γ production in the T-cell line. (B) HLA stabilization assay: both 10-mer 1 and the 11-mer peptide stabilize HLA A*0201 on T2 cells. Negative controls are T2 cells pulsed with an irrelevant 15-mer (shaded).
To analyze lysis of AML samples, we initially generated T-cell clones by limiting dilution from both of the poly-specific T-cell lines and screened the derived clones for specificity. This approach only yielded clones specific for epitopes not restricted to A*0201. Therefore, to facilitate isolation of clones specific for the A*0201 epitopes, we had identified from analysis of the poly-specific CD8 T-cell lines the lines were first enriched for reactive T cells by stimulation with the A*0201-binding peptides and sorting of the cells secreting IFN-γ with an IFN-γ–capture reagent before cloning. This approach produced several clones specific for epitope 227-235 from donor 2196 and 341-351 from donor 2264.
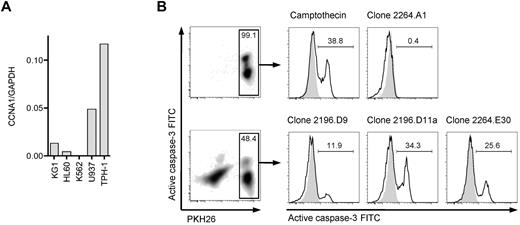
To identify a suitable cyclin-A1–expressing leukemic cell line that could be used as a target cell to assess processing and presentation of cyclin-A1 epitopes, we quantified cyclin-A1 in 5 myeloid leukemia cell lines (Figure 5A). THP-1, a HLA A*0201-positive M5b AML line, expressed the highest levels of cyclin-A1. Therefore, THP-1 cells were coincubated with HLA-A*0201-restricted T-cell clones, and lysis from recognition of presented cyclin-A1 epitopes assessed by measuring caspase-3 activation/cleavage. Using an E:T ratio of 3:1, no significant caspase-3 cleavage was detected in THP-1 cells cultured without effectors or after 4-hour coincubation with clone 2264.A1, which is specific for the B*4001-restricted epitope 118-127. However, caspase-3 cleavage was observed in THP-1 cells incubated with clone 2196.D9 or 2196.D11a (both specific for epitope 227-235) and 2264.E30 (specific for epitope 341-351; Figure 5B). Thus, both epitopes are processed from endogenously expressed protein and presented in the context of HLA A*0201.
T-cell clones recognize endogenous processed epitopes 227-235 and 341-351. (A) Expression of cyclin-A1 in several myeloid cell lines quantified by quantitative RT-PCR. (B) Clones 2196.D9 and D11a (both specific for epitope 227-235) and clone 2264.E30 (specific for epitope 341-351) were tested for apoptosis induction in the THP-1 cell line using a caspase-3 assay. As negative controls, targets alone (shaded) and clone 2264.A1 specific for epitope 118-127 (HLA B*4001 restricted, THP-1 is B*4001-negative) were used. As a positive control, targets were incubated with 4μM camptothecin. The histograms correspond to the gates on the PKH26-prelabeled target cells.
T-cell clones recognize endogenous processed epitopes 227-235 and 341-351. (A) Expression of cyclin-A1 in several myeloid cell lines quantified by quantitative RT-PCR. (B) Clones 2196.D9 and D11a (both specific for epitope 227-235) and clone 2264.E30 (specific for epitope 341-351) were tested for apoptosis induction in the THP-1 cell line using a caspase-3 assay. As negative controls, targets alone (shaded) and clone 2264.A1 specific for epitope 118-127 (HLA B*4001 restricted, THP-1 is B*4001-negative) were used. As a positive control, targets were incubated with 4μM camptothecin. The histograms correspond to the gates on the PKH26-prelabeled target cells.
CD8 T cells specific for epitope 227-235 recognize and lyse primary AML cells
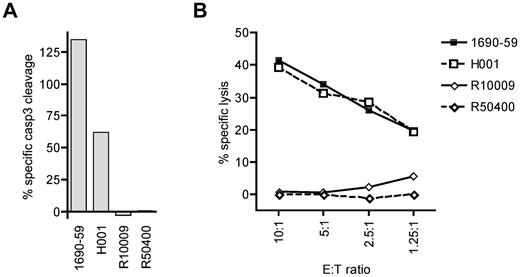
To determine whether cytotoxic T lymphocytes specific for a cyclin-A1 epitope recognize primary AML cells, we tested cyclin-A1–expressing blasts from 2 A*0201-positive and 2 A*0201-negative patients. Because of very limited amounts of cells available, the analysis was performed only with clone 2196.D11b, which recognized epitope 227-235. The clone 2196.D11b was first tested for induction of apoptosis in a 4-hour caspase-3 assay. For maximal apoptosis of these targets, staurosporine was used rather than camptothecin because it induced apoptosis in a higher percentage of cells. As the different AML samples showed different rates of spontaneous apoptosis, the data were normalized by calculating specific caspase-3 cleavage as: 100 × (experimental − spontaneous)/(staurosporine − spontaneous). Using an E:T ratio of 5:1, 2196.D11b induced significant apoptosis of the A*0201-positive AML specimens, but not A*0201-negative ones (Figure 6A). To determine whether the observed caspase-3 cleavage reflected classic lytic activity, we performed a standard 4-hour 51Cr release assay over a range of E:T ratios. Significant lysis of the A*0201-positive specimens was observed at an E:T as low as 1.25:1, whereas no specific lysis was detectable in the A*0201-negative targets. Thus, primary AML cells were killed in an HLA-restricted fashion.
CD8 T-cell clone 2196.D11b exhibits specific cytotoxic activity for cyclin-A1–expressing primary AML samples. (A) Four-hour caspase-3 assay. Specific caspase-3 cleavage is shown after subtraction of spontaneous caspase-3 activation, and determined relative to staurosporine-induced cleavage as 100%. (B) Four-hour 51Cr release assay at a range of E:T ratio with the same effector and target cells. H001 and 1690-59 are HLA A*0201 positive, and R10009 and R50400 are A*0201-negative.
CD8 T-cell clone 2196.D11b exhibits specific cytotoxic activity for cyclin-A1–expressing primary AML samples. (A) Four-hour caspase-3 assay. Specific caspase-3 cleavage is shown after subtraction of spontaneous caspase-3 activation, and determined relative to staurosporine-induced cleavage as 100%. (B) Four-hour 51Cr release assay at a range of E:T ratio with the same effector and target cells. H001 and 1690-59 are HLA A*0201 positive, and R10009 and R50400 are A*0201-negative.
Discussion
In the present study, we found that cyclin-A1 is an LAA expressed in AML stem cells that appears to be an attractive target for T cell-based therapy. To identify target candidates selectively expressed in LSCs, a systematic analysis of microarray data was performed, including from sorted LSC specimens. Using a reverse immunology approach, several class I epitopes were characterized, including 3 HLA-A*0201–restricted epitopes, and T-cell responses generated to 2 of these epitopes were capable of killing an AML cell line in vitro. Moreover, a CD8 clone specific for at least one of these epitopes effectively killed primary AML cells.
The identification of an appropriate target antigen is a critical and commonly limiting step for development of T cell–based immunotherapy. AML stem cell microarray data were used to identify probe sets with a suitable expression pattern. Although more than 100 probe sets passed the mathematical selection step, only one gene was found as a suitable target candidate after quantitative RT-PCR. The selection process displayed some of the drawbacks of a pure in silico approach. First, not all probe sets identified by the mathematical selection steps displayed the desired expression pattern. Second, after revising the target sequences of the probe sets, we found a high percentage of sequences to not represent a coding transcript. Finally, we found large differences between in silico data predictions and quantitative RT-PCR results. This might be because, as opposed to quantitative RT-PCR, microarrays do not provide linear data. Furthermore, the absence of a standardized protocol for sample processing and hybridization impacts the comparability of the different datasets.
Cyclin-A1 was highly expressed in AML cells, including the stem cell compartment of more than 50% of patients, and cyclin-A1 expression was highly restricted to malignant cells, a fact that was predescribed only based on semiquantitative data.31 We previously noted that both WT1 and cyclin-A1 are expressed at significantly higher levels in LSCs than in HSCs,14 but WT1 can also be detected in several nonhematopoietic organs, such as kidney, spleen, and ovary at levels potentially higher than in leukemic blasts (supplemental Figure 1C). In the present study, which is the first providing linear expression data of cyclin-A1 in AML, healthy hematopoiesis, and tissues in direct comparison, we now found that, except for AML, cyclin-A1 is expressed only in germ cells from testis, which is considered to be an immune-privileged site.32 Minimal cyclin-A1 staining in healthy BM as described in immunohistochemistry in an earlier study31 was not observed in our IF staining and might be the result of unspecific binding of the polyclonal antibodies used. Consequently, toxicities from recognition of normal cells while targeting cyclin-A1 appear unlikely.
Based on the expression pattern, cyclin-A1 falls into a group of antigens called cancer-testis-antigens (CTAs), which are characterized by expression in normal tissues restricted to immature germ cells in testis and by aberrant expression in cancers. There have been at least 2 systematic genome-wide studies to detect new cancer-testis genes (CTGs), but neither identified CCNA1 as such a gene.33,34 This is probably because hematologic malignancies, compared with solid tumors, have been found to infrequently express any of the known CTGs35 and were therefore either not included or underrepresented in the cancer panels used in the 2 studies. Recently, a classification of CTGs based on a systematic analysis of in silico expression data differentiating CTGs as testis-restricted and testis-selective CTGs was proposed, with the latter displaying minimal expression in non-immune privileged tissues. Interestingly, testis-selective CTGs had a high probability of being autosomal, not belonging to a multigene family, and being associated with a known function in gametogenesis and/or meiosis.33 Most of these characteristics of a testis-selective CTA apply to CCNA1, as it is located on chromosome 13q, is a single-copy gene, and has a known function in meiosis. The only predescribed leukemia-associated CTA PRAME was neither expressed in any LSC sample of our microarray datasets nor found overexpressed by quantitative RT-PCR in any of the AML samples (not shown). Thus, we think that cyclin-A1 can now be classified as the first described CTA associated with AML LSCs.
Cyclin-A1 is an alternative CDK2-associated type-A cyclin with an essential role in male gametogenesis, positively regulating the G2/M phase transition in meiosis I.17,36-38 The expression of meiotic genes in nonmeiotic non–germ cell malignancies has been observed with other CTGs, such as SYCP-1 and SPO11, and expression of meiosis-associated genes has been hypothesized to contribute to chromosomal aberrations and therefore support the initiation and further cytogenetic clonal evolution of a malignancy.39,40 In the case of cyclin-A1 in AML, this mechanism may not apply because, even though expression of cyclin-A1 has been shown to sustain the malignant phenotype,21-23 our recently published data indicated higher cyclin-A1 expression in AML specimens with normal rather than aberrant karyotypes.20 Furthermore, in case of APL, cyclin-A1 appears to be a target of the RAR-α fusion proteins rather than causal for the translocation.41 Several lines of evidence suggest that cyclin-A1 can play a role in the cell cycle regulation of mitotic and especially malignant cells, with cyclin-A1 positively regulating the G1/S transition independent of the particular cell type.37,42 Therefore, the major leukemogenic mechanism resulting from overexpression of cyclin-A1 in myeloid cells might be accelerated S-phase entry rather than induction of chromosomal instability.37
Besides the expression pattern and physiologic function of a gene, another important characteristic of a broadly useful T-cell target is having a large number of available epitopes, and overexpressed LAAs would be predicted to have a much larger number of targetable AA sequences than the very limited distinct sequences present in unique leukemia antigens generated by translocations or mutations. Using 2 normal donors, we identified 8 immunogenic peptides restricted to at least 3 different HLA class I molecules. Such broad immunogenicity is probably facilitated by limited tolerance induction to CTAs.
The characterization of epitope 341-351 demonstrates one limitation of silico epitope prediction approaches for identifying immunogenic peptides. Whereas the observed binding to A*0201 of the 10-mer(341-350) from the immunogenic 15-mer could be predicted retrospectively by 3 of 3 tested prediction algorithms (SYFPEITHI,43 BIMAS,44 IEDB analysis resource45 ), no binding was predicted for the 11-mer; therefore, this epitope would have never been identified without the use of a peptide library. A possible explanation for the fact that all clones generated to the larger 15-mer peptide were reactive exclusively against the 11-mer might be that T cells reactive with the 10-mer had been eliminated by negative selection because of cross-reactivity to a similar epitope on a high-abundance self-protein. Indeed, for the shorter epitope 341-350, which did bind to A*0201 more stably, similar though not identical epitopes were identified in cyclin-A2 and cyclin-B1, which are broadly expressed self-antigens.
The efficient cytotoxic activity of cyclin-A1-specific T-cell clones demonstrated in this study may reflect several factors: Cyclin-A1 expression appears significantly higher than other AML associated antigens such as WT1, PRAME, HMMR, and PRTN3 (Li et al46 ; S.O., unpublished data, May 2011). Moreover, the dramatic periodicity of cyclin-A1 expression is accompanied by high degradation rates regulated by the ubiquitin-proteasome-mediated pathway.47 This suggests that epitopes are likely being made available for presentation. Finally, although the testis appears to provide peripheral immune privilege to germline cells,32 little is known about negative selection during development of T cells reactive with gametogenesis-associated antigens, especially CTAs. However, reduced deletion and/or central tolerance for testicular antigens would explain both the high incidence of autoimmune mediated infertility observed in patients after traumatic disruption of the blood-testis barrier32 and the high cytotoxic activity of the T-cell clones generated in this study. Further studies analyzing both the functionality and the TCR affinity of spontaneous T-cell responses in patients with cyclin-A1 expressing diseases are initiated to further investigate this assumption.
Recently, a National Cancer Institute pilot project to prioritize cancer antigens made an effort to not only develop a priority-ranked list of the known cancer antigens but to also provide a list of weighted “ideal” antigen criteria/characteristics for evaluating other antigens.13 According to these criteria, cyclin-A1 would appear to be a highly suitable antigen for targeting AML based on: a very selective oncofetal expression pattern with uniformly high expression in leukemia in a large fraction of patients, being leukemogenic when overexpressed in mice, expression in the LSC compartment, having many epitopes available, and having an intracellular location. Thus, applying the weighted criteria of the National Cancer Institute pilot project to cyclin-A1 resulted in a ranking for cyclin-A1 between 3 to 5 among the 75 antigens that were ranked.13 Therefore, cyclin-A1 appears to now be the first described non–X-linked leukemia-testis-antigen with optimal features for targeting of AML LSCs by T cell–based therapy approaches, such as vaccination and/or adoptive T-cell transfer.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Hieu Nguyen, Louise Badenhorst, and Rebecca Yoda for technical assistance.
This work was supported by the National Institutes of Health/National Cancer Institute (grants PO1 CA018029 and RO1 CA 33084) and Leukemia & Lymphoma Society (grant FND7008-00). K.R.L. is supported by the National Institutes of Health/National Institute of Diabetes, Digestive and Kidney (grant P30 DK056465-13). S.O. is a scholar of Deutsche Krebshilfe, Germany (Mildred-Scheel-Stipendium 108995).
National Institutes of Health
Authorship
Contribution: S.O. designed and performed the experiments and prepared the final manuscript; R.M. and I.L.W. provided and analyzed microarray data; T.S. performed FACS assays; D.S. provided and analyzed microarray data and provided patient samples; U.K., M.B., E.H.W., and B.W. provided patient samples; K.R.L. and Y.E.C. performed IF and FACS assays; M.H. performed FACS sorting; Y.A. performed molecular cloning and retroviral transfection; P.D.G. designed the concept, reviewed the data, and prepared the final manuscript; and all authors reviewed and edited the manuscript.
Conflict-of-interest disclosure: A patent application for the use of cyclin-A1 as a T-cell target antigen has been submitted by S.O., P.D.G., R.M., and I.L.W. The remaining authors declare no competing financial interests.
Correspondence: Philip D. Greenberg, Clinical Research Division, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, D3-100, Seattle, WA 98109; e-mail: pgreen@u.washington.edu.