Abstract
Gene regulatory networks that govern hematopoietic stem cells (HSCs) and leukemia-initiating cells (L-ICs) are deeply entangled. Thus, the discovery of compounds that target L-ICs while sparing HSC is an attractive but difficult endeavor. Presently, most screening approaches fail to counter-screen compounds against normal hematopoietic stem/progenitor cells (HSPCs). Here, we present a multistep in vitro and in vivo approach to identify compounds that can target L-ICs in acute myeloid leukemia (AML). A high-throughput screen of 4000 compounds on novel leukemia cell lines derived from human experimental leukemogenesis models yielded 80 hits, of which 10 were less toxic to HSPC. We characterized a single compound, kinetin riboside (KR), on AML L-ICs and HSPCs. KR demonstrated comparable efficacy to standard therapies against blast cells in 63 primary leukemias. In vitro, KR targeted the L-IC–enriched CD34+CD38− AML fraction, while sparing HSPC-enriched fractions, although these effects were mitigated on HSC assayed in vivo. KR eliminated L-ICs in 2 of 4 primary AML samples when assayed in vivo and highlights the importance of in vivo L-IC and HSC assays to measure function. Overall, we provide a novel approach to screen large drug libraries for the discovery of anti–L-IC compounds for human leukemias.
Introduction
Acute myeloid leukemia (AML) is organized as a hierarchy where most of the cells are postmitotic leukemia blasts that are replenished from a pool of progenitor cells ultimately derived from the leukemia-initiating cell (L-IC).1-3 L-ICs are defined by 3 criteria: prospective purification, leukemia-initiating function, and capacity for self-renewal. Several correlative studies have linked cancer stem cell (CSC) properties with patient outcomes, especially for AML L-IC.4-7 Our recent data establishing that L-IC–specific gene expression signatures are highly significant predictors of patient survival provides strong evidence for the hierarchical organization of AML according to the CSC model and firmly links the properties that underlie stemness as influencing clinical outcome.8 The molecular basis that underlies the relative resistance of L-ICs to chemotherapy and radiation compared with non–L-IC progenitors or blasts is not well defined but might be explained by biological properties of L-ICs including quiescence, altered DNA damage response, and expression of drug transporters.9-13 However, our studies also show that L-IC and normal hematopoietic stem cells (HSCs) strongly share gene expression signatures, and the common signature is highly enriched for known stem cell regulators pointing to the importance that the determinants of stemness play in governing patient survival.8 Nevertheless, L-ICs also display distinct differences in gene expression leading to altered cell-surface markers, transcription factors, and enzymes that could be targeted.8,14 Indeed, we have found that differential expression of the cell-surface marker CD123 could be exploited as a novel AML–L-IC therapeutic.15 Guzman et al identified parthenolide (PTL) as an agent able to selectively target L-ICs over HSCs because of an over-dependence on NF-κB signaling in L-IC versus HSC cells.10,16,17 Thus, identifying agents that target L-ICs but spare HSCs remains a difficult but an attractive target; however, no rational, high throughput methods have been developed to screen for such agents.
Traditional high-throughput screens have measured overall cell growth in common leukemia cell lines, such as HL-60, which has identified many agents (ie, cytarabine, mitoxantrone, etc) that mainly target AML blast cells, but not the more quiescent L-IC. Common cell lines also often exhibit gene alterations that arise from long-term cell culture making them less reflective of primary leukemia samples. Whereas an ideal high-throughput assay would examine pure L-IC and HSC populations, the low L-IC frequency within primary samples,1-3,8 an inability to purify L-ICs to homogeneity, cytogenetic variability between patients, and difficulty maintaining L-ICs in vitro precludes the use of L-ICs in primary screens. Until recently, it was also not possible to directly isolate and screen against normal HSCs because of their rarity and the absence of definitive purification markers.18 To bypass the limitations of using common leukemia cells lines or primary L-ICs, we used 2 novel leukemia cell lines, TEX and M9-ENL1,19,20 as surrogate screening populations because of their unique biological properties. Moreover, for counter-screening against normal hematopoietic cells, we used cell fractions that are highly enriched for both HSC and primitive progenitor cells, because the vastly more numerous progenitor cells are still highly reflective of hematopoietic toxicity. Both leukemia lines were derived from an experimental human leukemogenesis approach we developed, in which normal HSPC cells are transduced with single oncogenes and the transformed cells rapidly appear. In vitro and in vivo limiting dilution experiments suggest that TEX cells, derived from TLS-ERG transduced Lin− CB cells, are functionally heterogeneous with a minority of cells (1 in 120 cells, in vitro; 1 in 3.8 × 105 in vivo) possessing L-IC function. Erg expression regulates mouse HSC self-renewal and function, and forced Erg expression promotes T-cell leukemia.21,22 Genome-wide mapping of transcription factor binding in hematopoietic progenitor cells identified a core network, including Erg, Tall, Lyl1, Gata2, Runx1, Lmo2, and Fli-1 that regulate ∼ 930 genes highly enriched in HSC fractions.23 Furthermore, TEX cells require exogenous SCF and IL3 in the presence of serum to maintain a relatively undifferentiated state and terminally differentiate on removal of either cytokine,19 suggesting that TEX cells have functioning and responsive regulation of differentiation, an attribute of stem cells. The M9-ENL1 line has extremely frequent L-ICs, although still being able to adopt altered differentiation fates on changes in cytokine stimulation.20,24 Thus, most cells in this line maintain all the functions necessary to act as L-ICs in vivo. Together, these lines provide an opportunity to screen for agents that may target L-ICs because they proliferate in culture conditions, yet they have not been cultured for decades and they maintain some aspects of the machinery of hierarchically-organized tissue and stem cells, unlike most common leukemia cell lines.
Here, we present a new multistep approach for the identification of anti–L-IC compounds by screening these primitive hematopoietic cell lines, counter screening primary hits on normal HSPCs to remove cytotoxic compounds, and then in vitro and in vivo verification with primary AML cells. From a relatively small collection of 4000 compounds, we identified kinetin riboside, an adenosine derivative, which was active on L-ICs as assayed in NOD/SCID mice.
Methods
Primary and secondary screens
Primary human AML and chronic myelogenous leukemia (CML) cells were isolated from peripheral blood according to procedures approved by the University Health Network Research Ethics Board (Toronto, ON). Cord blood samples were obtained according to procedures approved by the University Health Network and Trillium Hospital (Mississauga, ON) Research Ethics Boards. TEX (3000/well) and M9-ENL1 (10 000/well) cells were plated in 100 μL in 96-well clear nontreated plates using a Biomek FX liquid handler. Within 1 hour after cell seeding, compounds (5 μL/well) were added for 1μM (Prestwick) or 5μM (LOPAC, Spectrum) final concentration (0.05% DMSO vehicle). Cells were cultured for 72 hours and then 10 μL/well of Alamar Blue was added. Alamar Blue fluorescence was measured the following day by exciting at 535 nm and reading emission at 595 nm using a PHERAstar reader (BMG Labtech). Normalized data were used to calculate the Z-score and B-score for each compound.25 Two-hundred unique compounds were identified by either Z- or B-score. Eighty compounds reduced the Alamar Blue signal 75% of control cells and were cherry picked for dose-response secondary screens on TEX and M9-ENL1 cells (2-fold dilution from 10μM to ∼ 20nM). Twenty-five structurally redundant or known cytotoxic compounds were removed and the remaining 55 were screened on 3 separate pools of Lin− CB cells at 10 000 cells/well in 100 μL in 96-well plates at 5, 1.67, and 0.556μM.
Flow cytometry
Apoptosis was measured using FITC annexin-V staining. Treated cells were resuspended in 100 μL PBS/5% FBS and stained with 1 μL FITC–annexin-V, 5 μL 100 μg/mL propidium iodide, CD34-APC, and CD38-PE for 30-60 minutes at room temperature (RT). Proliferation was measured by BrdU incorporation. Cells were plated in 24-well plates in 1 mL media and 10 μL of 1mM BrdU was added for the last 2 hours of culture. Cells were harvested, fixed in 1.6% paraformaldehyde, permeabilized with 70% methanol, treated with 0.3 mg/mL DNase, and stained in 100 μL PBS/5% FBS with 100 ng FITC-mouse anti-BrdU and 7AAD. Cells were analyzed on BD LSR2 and at least 10 000 events were acquired. In some experiments, leukemia cells were treated for 16 hours at various doses, harvested and stained with CM-H2DCFDA (10μM final), mBBR (40μM final), TMRM (250nM final), and APC–annexin-V (3 μL per 100 μL cells) for 30 minutes at RT protected from light.26,27 Cells for intracellular FACS were harvested, immediately fixed in ice-cold 1.6% PFA for 30 minutes, permeabilized with ice-cold 90% methanol/9.5% PBS/0.5% FBS for 30 minutes, washed with PBS/5% FBS, and then stained in 100 μL of volume with 10-25 ng phospho-H2A.X antibody. FACS analysis was performed on a BD LSR2 equipped with 488 nm, 633 nm, 405 nm, and 355 nm lasers and appropriate filters. Typically, 30 000-50 000 events were acquired.
Immunocompromised mouse model
Immunocompromised mice were derived from the crossing of NOD/ShiLtSz-Prkdcscid (NOD.CB17-Prkdcscid) to Beige/Nude/X-linked SCID, termed nude/NOD/SCID and previously described.20 All mice were bred at the Ontario Cancer Institute (Toronto, ON) and housed in ventilated micro-isolator cages with autoclaved water and irradiated food ad libitum in a high-barrier facility. For each AML sample or Lin− CB pool, a frozen aliquot was thawed and cultured in 24-well plates with 2 × 106 AML cells in 1 mL per well (12 wells total) or 0.25 × 106 Lin− CB cells in 1 mL per well in the presence of 10μM KR or 5μM PTL. After 16 hours, cells were harvested from plates, counted, and a small aliquot processed for CD34, CD38, and annexin-V staining. Cells were normalized to viable cell counts after drug treatment and injected into the right femur of 5 to 8 mice, 20-24 weeks old, irradiated ∼ 24 hours previously with 208 cGy.28,29 Eight weeks after injection, the mice were killed and the right femur removed and flushed. Cells were stained in 100 μL PBS/5% FBS for 30-60 minutes with human-specific CD45 antibody. When possible, 1000 human CD45+ cells were recorded, otherwise between 30 000-100 000 live cells were acquired. Data analysis was performed using FlowJo Version 8.8.4 (TreeStar).
Supplemental materials and methods
Reagents, cell culture, primary cells, methylcellulose colony formation, Alamar Blue, microarray (accession GSE33892), bioinformatic analysis, chemical synthesis of KR monophosphate, and immunoblotting are described in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Results
High-throughput identification of kinetin riboside
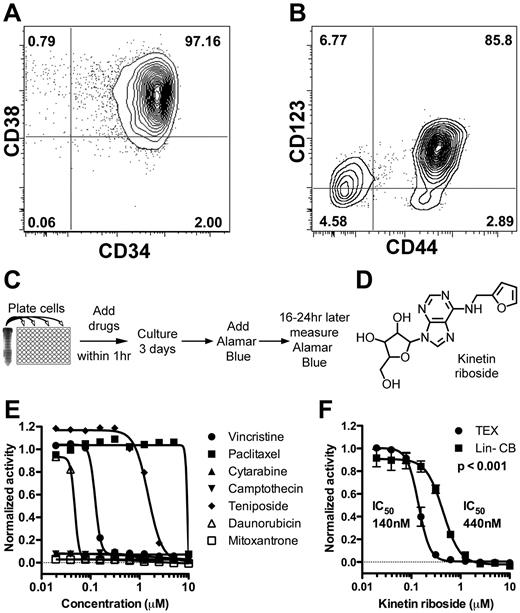
We previously developed 2 novel leukemia cell lines, TEX and M9-ENL1, by transduction of leukemogenic fusion oncogenes into normal HSPC from lineage-depleted CB (Lin− CB).19,20 TEX cells are derived from TLS-ERG transduced Lin− CB cells that underwent step-wise transformation and immortalization through acquisition of additional genetic changes, such as expression of hTERT, expression of L-IC markers CD34, CD4430 and CD12315 (Figure 1A-B), and the stem cell gene BMI1.31 Gene set enrichment analysis showed enrichment of known AML L-IC gene sets8,9 (supplemental Figure 1) in TEX compared with HL60 and THP1 cell lines suggesting that TEX cells could function as a surrogate cell population for L-ICs in high-throughput screens. M9-ENL1 cells are derived from MLL-ENL transduced Lin− CB cells injected into NOD/SCID mice that subsequently developed a reproducible, rapid, and fatal pro-B cell acute lymphoblastic leukemia (ALL) disease.20 Multiple cell lines, grown in the presence of SCF, IL3, IL7, and FLT3 were generated, which maintained the ability to initiate disease in NOD/SCID mice. The cells maintained a significant ability to shift fate between myeloid and lymphoid lineages after changes in cytokine combinations, which indicates that the cells reflect a primitive stem-like state.24,32 Accordingly, M9-ENL1 cells showed an enrichment of embryonic and B-cell progenitor gene sets (supplemental Figure 1). Thus, by well-established criteria of both in vitro cell line initiation and in vivo leukemia initiation, these 2 lines possess hallmarks and gene expression patterns of stem cell properties.
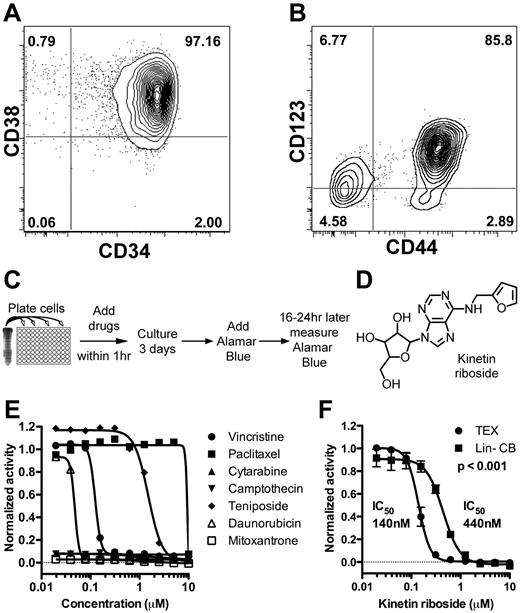
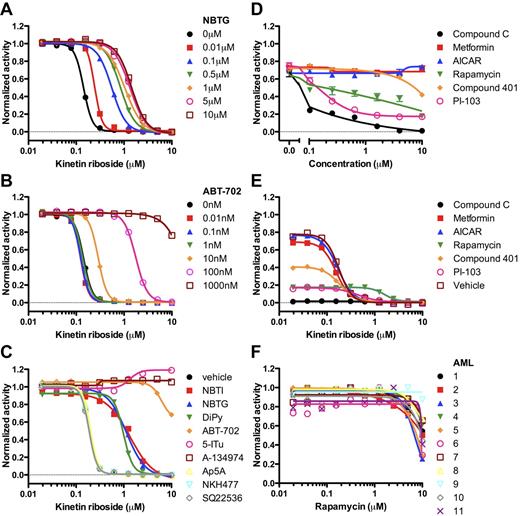
Identification of kinetin riboside from screen of novel leukemia cell lines. (A-B) FACS staining of TEX cells for CD34, CD38, CD44, and CD123. (C) Schematic of primary growth screen of TEX and M9-ENL1 cells using Alamar Blue. (D) Structure of kinetin riboside verified by NMR. (E) Seven chemotherapeutics were evaluated on TEX cells. (F) Kinetin riboside inhibited cell growth in TEX cells at submicromolar concentrations, but was not as effective on Lin− CB cells (P < .001; F-distribution).
Identification of kinetin riboside from screen of novel leukemia cell lines. (A-B) FACS staining of TEX cells for CD34, CD38, CD44, and CD123. (C) Schematic of primary growth screen of TEX and M9-ENL1 cells using Alamar Blue. (D) Structure of kinetin riboside verified by NMR. (E) Seven chemotherapeutics were evaluated on TEX cells. (F) Kinetin riboside inhibited cell growth in TEX cells at submicromolar concentrations, but was not as effective on Lin− CB cells (P < .001; F-distribution).
To begin the process to identify compounds that potentially target L-ICs, we conducted growth inhibition screens (Figure 1C) of ∼ 4000 small molecules from 3 libraries (supplemental Figure 2, supplemental Table 1) using TEX19 and M9-ENL120,24 cells and identified 200 compounds as initial “hits” using a Z-score or B-score more than 3 standard deviations from the mean.25 To filter the primary 200 hits, we selected the 80 compounds with a 75% reduction in growth, as measured by Alamar Blue, compared with DMSO treated cells (supplemental Figure 2B). Nineteen of the top 80 hits included known antileukemia drugs, such as cytarabine and mitoxantrone (Figure 1E; supplemental Table 2). One-half of the 80 compounds passed the Lipinski Rule of 533 indicating potential oral bioavailability (supplemental Table 2).
In an effort to remove compounds that are potentially cytotoxic to normal hematopoietic cells in general, 55 were counter-screened on Lin− CB cells as a source of normal HSPCs. Ten compounds were toxic to TEX and M9-ENL1 cells, but not Lin− CB cells, and further tested (supplemental Figure 2, supplemental Table 3). Kinetin riboside, N6-furfuryladenosine (Figure 1D), killed TEX cells, but toxicity was 3-fold less on Lin− CB cells (Figure 1F). Thus, our experimental design was successful in identifying small molecules that are selectively effective in L-IC–driven cell line models.
KR kills primary leukemia cells
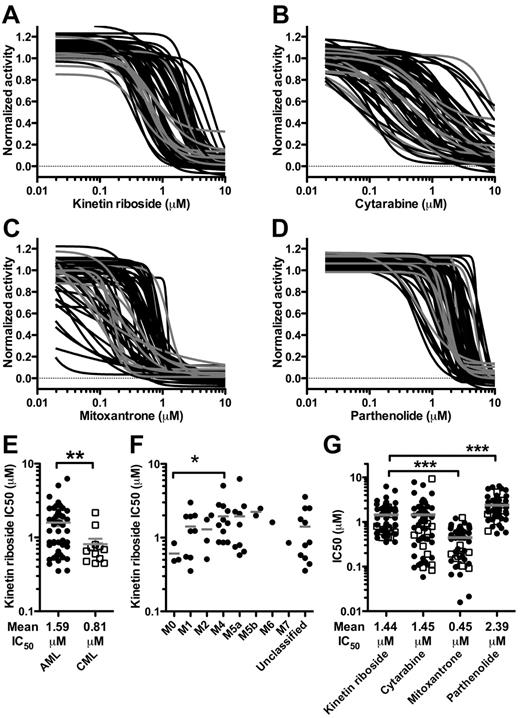
AML patients exhibit significant heterogeneity in drug response and survival outcomes, and as with all preclinical models, our cell lines are unlikely to reflect such wide functional heterogeneity. Thus, we evaluated the efficacy of KR on a wide spectrum of 63 primary AML and CML samples in short-term (4 day) culture assays (supplemental Table 4). KR was compared with standard drugs used for induction AML therapy (cytarabine, mitoxantrone) as well as PTL (Figure 2A-D). KR killed bulk AML and CML cells at low micromolar concentrations (Figure 2A-E). In addition, KR was equally effective against all AML FAB subtypes except for M3, which did not grow in these conditions (Figure 2F). KR was similar to cytarabine, less effective than mitoxantrone, but more effective than PTL at killing primary AML and CML cells (Figure 2G). We can conclude that primary samples showed a heterogeneous response to KR induced killing with KR showing significant efficacy against the majority of samples in this in vitro assay.
Kinetin riboside kills primary leukemia cells. The effect of treatment with (A) KR, (B) cytarabine, (C) mitoxantrone, or (D) PTL on bulk leukemia cells from 51 primary AML (black) and 12 CML (gray) samples. (E) KR was more effective against CML than AML. (F) KR was effective against all AML FAB subtypes tested; some samples were unclassified. (G) The IC50 (μM) for each sample for KR, cytarabine, mitoxantrone, and PTL. Gray bar: mean IC50. Open squares indicate CML; closed circles indicate AML (*P < .05, **P < .01, and ***P < .001).
Kinetin riboside kills primary leukemia cells. The effect of treatment with (A) KR, (B) cytarabine, (C) mitoxantrone, or (D) PTL on bulk leukemia cells from 51 primary AML (black) and 12 CML (gray) samples. (E) KR was more effective against CML than AML. (F) KR was effective against all AML FAB subtypes tested; some samples were unclassified. (G) The IC50 (μM) for each sample for KR, cytarabine, mitoxantrone, and PTL. Gray bar: mean IC50. Open squares indicate CML; closed circles indicate AML (*P < .05, **P < .01, and ***P < .001).
KR induces apoptosis in L-ICs and not HSCs
To determine whether KR targeted the L-ICs present in primary AML samples, we used an assay based on the induction of apoptosis in the CD34+CD38− cell fraction, which is highly enriched for L-ICs.1-3,8 Both KR and PTL induced apoptosis in primitive CD34+ (Figure 3A) and phenotypic L-IC–enriched CD34+CD38− fractions (Figure 3B) in 11 primary AML samples (supplemental Table 5), in agreement with earlier reports for PTL.17 KR and PTL induced apoptosis in additional CD34/CD38 fractions (supplemental Figure 3), including CD34+CD38+, which also contain L-ICs.8,34 We can conclude that KR kills all phenotypic subpopulations of AML including those with L-IC activity. Brief treatment with PTL also decreased the fraction of CD14+ cells in these AML samples (Figure 3C). To directly assess the potential toxicity of KR against HSPC, similar drug concentrations were tested. KR did not induce apoptosis in fractions containing normal progenitor (CD34+) or HSPC (CD34+CD38−) cell populations (Figure 3D-E) or alter CD34+CD38− levels (Figure 3F). Myeloid and erythroid progenitor cell activity in methylcellulose was comparable with PTL (Figure 3G-H). Thus, KR preferentially targets phenotypic L-ICs, but not HSPCs in this in vitro assay.
Kinetin riboside targets phenotypic CD34+CD38− L-ICs. KR induced apoptosis (annexin-V+) in phenotypic CD34+CD38− L-ICs, but not CD34+CD38− HSCs after overnight treatment. Eleven primary AML samples (A-C) or 9-12 separate pools of Lin− CB cells (D-F) were cultured for 16 hours with DMSO vehicle (black), 10μM KR (white), or 5μM PTL (gray). The frequency of apoptotic cells in CD34+ (A,D) and CD34+CD38− (B,E) fractions. (C) PTL reduced the frequency of CD14+ AML cells. (F) KR or PTL did not reduce the frequency of phenotypic Lin− CB HSC (CD34+CD38−). (G-H) Progenitor cell activity was measured by plating 3 separate pools of Lin− CB cells in methylcellulose in the presence of KR or PTL and counting myeloid (G) or erythroid (H) colonies (*P < .05, **P < .01, and ***P < .001).
Kinetin riboside targets phenotypic CD34+CD38− L-ICs. KR induced apoptosis (annexin-V+) in phenotypic CD34+CD38− L-ICs, but not CD34+CD38− HSCs after overnight treatment. Eleven primary AML samples (A-C) or 9-12 separate pools of Lin− CB cells (D-F) were cultured for 16 hours with DMSO vehicle (black), 10μM KR (white), or 5μM PTL (gray). The frequency of apoptotic cells in CD34+ (A,D) and CD34+CD38− (B,E) fractions. (C) PTL reduced the frequency of CD14+ AML cells. (F) KR or PTL did not reduce the frequency of phenotypic Lin− CB HSC (CD34+CD38−). (G-H) Progenitor cell activity was measured by plating 3 separate pools of Lin− CB cells in methylcellulose in the presence of KR or PTL and counting myeloid (G) or erythroid (H) colonies (*P < .05, **P < .01, and ***P < .001).
KR prevents L-IC engraftment in NOD/SCID mouse model
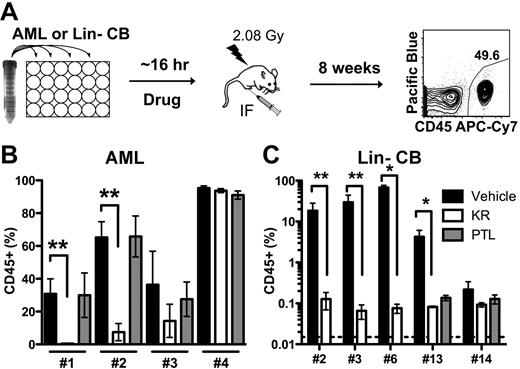
Because KR induced apoptosis in cell populations enriched for primary AML L-ICs, we directly determined drug efficacy on L-IC functionally assayed using NOD/SCID xenotransplantation. We have previously shown that only L-ICs are able to engraft NOD/SCID mice,1-3,20,30 and that antibodies (CD44 and CD123)15,30 that target L-ICs can be identified on the basis of an assay, in which AML cells are exposed ex vivo to the agent and their effect on L-ICs is measured on the basis of in vivo leukemia initiation. Cells from 4 primary AML samples that showed efficacy in short-term in vitro assays (supplemental Figure 4, supplemental Table 6) were treated in vitro with KR or PTL and then injected intra-femorally into irradiated NOD/SCID mice (Figure 4A). After 8 weeks, leukemia engraftment in the injected right femur was quantified using a human-specific CD45 antibody and with lineage-specific antibodies (Figure 4B, supplemental Figure 5). L-ICs in 2 of the 4 samples were markedly inhibited by brief KR treatment (AML 1, 2) indicating that there is inter-sample variation in response among individual primary samples. In contrast to the equivalent efficacy found in in vitro assays (Figure 3B), PTL did not inhibit engraftment of any sample at the dose tested, which is slightly less (5μM vs 7.5μM) than previously published doses.17 This is not likely because of slight differences in the number of cells injected per mouse between KR and PTL (supplemental Table 6). Following drug treatment, we normalized cell numbers prior to transplantation and did not carry out limiting dilution analysis making the engraftment assay a relative, and not absolute, measure of drug effects on L-IC number. This combined with the enhanced detection of L-IC that our optimized transplant method affords could explain differences with prior work.17,29 Collectively, this data demonstrates the importance of incorporating direct L-IC analysis when assessing drug efficacy and not relying solely on phenotypically defined cell populations and short-term in vitro assays.
KR inhibits AML engraftment in NOD/SCID model. (A) Experimental design of ex vivo treatment. AML or Lin− CB cells were treated overnight with DMSO vehicle (black), 10μM KR (white), or 5μM PTL (gray), and injected into irradiated NOD/SCID mice. Eight weeks later, the surviving mice were killed and human leukemia or normal hematopoietic cell engraftment (CD45+) was measured in the injected right femur. (B) Leukemic engraftment by 2 of 4 AML samples was inhibited by KR treatment. (C) Normal hematopoietic engraftment from 5 Lin− CB pools was inhibited by KR and PTL treatment (*P < .05 and **P < .01).
KR inhibits AML engraftment in NOD/SCID model. (A) Experimental design of ex vivo treatment. AML or Lin− CB cells were treated overnight with DMSO vehicle (black), 10μM KR (white), or 5μM PTL (gray), and injected into irradiated NOD/SCID mice. Eight weeks later, the surviving mice were killed and human leukemia or normal hematopoietic cell engraftment (CD45+) was measured in the injected right femur. (B) Leukemic engraftment by 2 of 4 AML samples was inhibited by KR treatment. (C) Normal hematopoietic engraftment from 5 Lin− CB pools was inhibited by KR and PTL treatment (*P < .05 and **P < .01).
To test the effect of KR on functionally-defined HSCs, we treated 5 separate pools of Lin− CB cells in vitro with KR and/or PTL, and then injected cells intra-femorally into NOD/SCID mice at near limiting cell doses (supplemental Table 6).28 In contrast to our in vitro results showing a lack of effect on apoptosis induction in the HSPC populations (Figure 3E), both KR and PTL markedly inhibited human engraftment at the doses tested (Figure 4C). Loss of human engraftment correlated with the loss of myeloid and progenitor cell activity at similar doses (Figure 3G-H). This suggests that the vastly more numerous progenitor cells within the HSPC fraction are less sensitive to KR than the rare HSCs present in the CD34+CD38− fractions. Our data establish that KR can target functional L-ICs in a subset of primary AML patients, but that under the ex vivo treatment conditions normal HSCs are also targeted even though HSPCs were not affected in vitro. This indicates that L-ICs and normal progenitors might respond differently to KR.
KR induces apoptosis in the absence of DNA damage, ROS, or p53
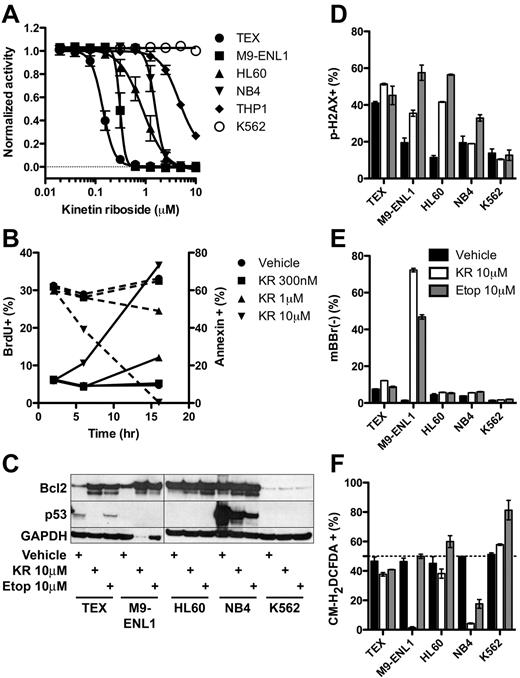
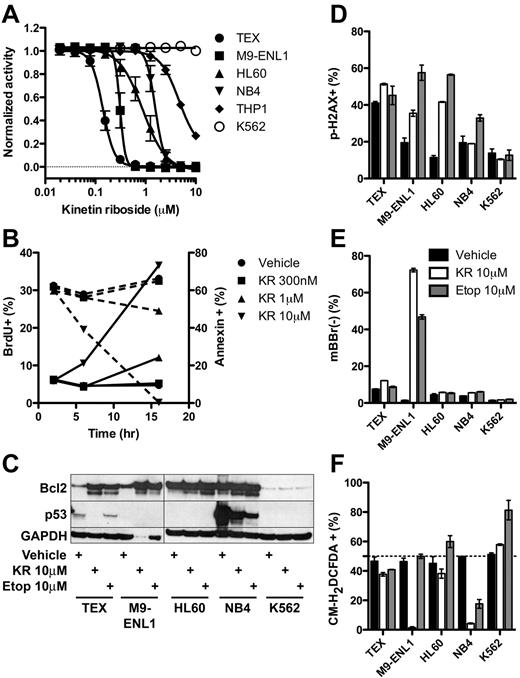
To gain more insight into the mechanism of KR killing, we carried out a series of studies on known pathways and biological processes. KR was effective at low micromolar levels against additional leukemia cell lines tested except K562 (Figure 5A). Within 16 hours, KR reduced proliferation and induced apoptosis via caspase-3 cleavage and loss of mitochondrial membrane potential, assayed by loss of TMRM staining (Figure 5B, supplemental Figure 6). Accordingly, KR treatment induced cleavage of Bcl2 (Figure 5C), which may switch it from antiapoptotic to proapoptotic.35 Given the structural similarity to adenosine, we reasoned that KR incorporation into DNA may induce DNA damage and activate p53; however, KR did not consistently induce DNA damage (phospho-H2A.X staining) and it reduced p53 levels in TEX and NB4 cells instead of raising them as many chemotherapeutic agents do (Figure 5C-D). PTL and TDZD-8 have been shown to kill L-ICs in part by reactive oxygen species (ROS) formation17 or depletion of free thiol groups (loss of monobromobimane staining),16 respectively. Except for M9-ENL1 cells, KR did not deplete free thiol groups (Figure 5E), and actually reduced ROS formation in multiple cell lines (Figure 5F). Accordingly, addition of the antioxidant N-acetyl-cysteine did not inhibit KR-induced cell death (supplemental Figure 7A). Thus, KR induces apoptosis through mechanisms distinct from other L-IC specific compounds PTL and TDZD-8, namely via cleavage of caspase-3 in the absence of DNA damage, ROS formation, or modulation of free thiol groups.
Molecular effects of KR treatment of leukemia cells. (A) The growth of KR treated leukemia cell lines was measured using Alamar Blue from 4-9 separate experiments. (B) TEX cells were treated with KR for indicated times and doses. BrdU was added for the final 2 hours of culture. Proliferation (BrdU+; dashed lines) and apoptosis (annexin-V+; solid lines) was measured by FACS in triplicate. (C) Bcl2 and p53 Western blot of 10μM KR or 10μM etoposide treated leukemia cells. (D-F) Leukemia cells were treated overnight in triplicate with DMSO vehicle (black), 10μM KR (white), or 10μM etoposide (gray). Harvested cells were stained for (D) DNA damage (phoshpo-H2A.X), (E) loss of intracellular free thiol groups (mBBr−); or (F) reactive oxygen species (CM-H2DCFDA).
Molecular effects of KR treatment of leukemia cells. (A) The growth of KR treated leukemia cell lines was measured using Alamar Blue from 4-9 separate experiments. (B) TEX cells were treated with KR for indicated times and doses. BrdU was added for the final 2 hours of culture. Proliferation (BrdU+; dashed lines) and apoptosis (annexin-V+; solid lines) was measured by FACS in triplicate. (C) Bcl2 and p53 Western blot of 10μM KR or 10μM etoposide treated leukemia cells. (D-F) Leukemia cells were treated overnight in triplicate with DMSO vehicle (black), 10μM KR (white), or 10μM etoposide (gray). Harvested cells were stained for (D) DNA damage (phoshpo-H2A.X), (E) loss of intracellular free thiol groups (mBBr−); or (F) reactive oxygen species (CM-H2DCFDA).
KR requires adenosine kinase activity
Structurally, KR is generated by the addition of a furan moiety to adenosine. Adenosine can signal through 4 cell-surface G-protein–coupled receptors or be rapidly metabolized by adenosine kinase (ADK).36 Various derivatives of adenosine (eg, fludarabine) are clinically used in treating hematologic malignancies and require intracellular conversion to the triphosphate metabolite for efficacy.37 We reasoned that KR also required intracellular activation and used chemical probes that modulate adenosine metabolism and signaling to dissect the molecular pathways modulated by KR. A competitive antagonist for P2×7 (A-438079), a cell-surface purinoreceptor for ATP, did not prevent KR-induced killing of TEX cells (supplemental Figure 7B), indicating that KR is transported into cells without phosphate modification. In addition, primary AML and cell lines expressed low levels of CD39 and CD73 (supplemental Figure 8), which are cell-surface ectonucleotidases that modulate extracellular signaling by adenine nucleotides.36 Inhibition of cell-surface nucleobase transporters (NBTG, NBTI, and DiPy) prevented KR-induced killing, which suggests that KR acts intracellularly (Figure 6A,C). As a first step in adenosine metabolism, ADK transfers a phosphate to adenosine to generate adenosine monophosphate. Highly specific ADK inhibitors (ABT-702,38 5-Itu, and A-13497439 ) were very potent at inhibiting KR-induced killing (Figure 6B-C) showing KR requires ADK activity. Higher intracellular ADK levels were correlated with higher KR IC50, but not etoposide or PTL IC50 (supplemental Figure 9). Modulators of adenylate cyclase (NKH477, SQ22536) and adenylate kinase (Ap5A)40 did not inhibit KR-induced killing (Figure 6C). Because ADK activity was required for KR-induced cell death, we synthesized kinetin riboside monophosphate (KR-P; supplemental Figure 10A)41 and tested whether KR-P would be resistant to ADK inhibitors. The IC50 for KR-P was approximately 2-fold higher than KR (supplemental Figure 10B). Unexpectedly, KR-P induced killing was reduced by ADK inhibitors (supplemental Figure 10C). Although ADK activity is required for KR and KR-P–induced cell killing, it remains possible that other pathways are important in modulating KR-induced cell killing. Thus, our study has uncovered a novel mechanism of KR cell killing that involves ADK, opening a new pathway of drug discovery to identify additional L-IC specific drugs.
KR acts intracellularly and requires adenosine kinase activity to inhibit leukemia cell growth. (A) NBTG and (B) ABT-702 prevented the ability of KR to inhibit TEX cell growth. (C) Inhibitors of nucleobase uptake (NBTI-red squares; NBTG-blue triangles; DiPy-green inverted triangles), adenosine kinase (ABT-702-orange diamonds; 5-ITu-pink open circles; A-134974-brown open squares), adenylate kinase (Ap5A-open yellow triangles), adenylate cyclase (SQ22536-open gray diamonds), and an adenylate cyclase activator (NKH477-open blue inverted triangles) were tested at 500nM in the presence of KR. (D) TEX cells were cultured in the presence of Compound C (black circles), metformin (red squares), AICAR (blue triangles), rapamycin (green inverted triangles), compound 401 (orange diamonds), or PI-103 (pink open squares). Growth inhibition was measured using Alamar Blue. (E) Synergism between kinetin riboside and 10μM of Compound C (black circles), metformin (red squares), AICAR (blue triangles), rapamycin (green inverted triangles), compound 401 (orange diamonds), PI-103 (pink open squares), or DMSO vehicle (brown open squares) was tested using Alamar Blue. (F) Rapamycin was tested on 11 primary AML samples using Alamar Blue.
KR acts intracellularly and requires adenosine kinase activity to inhibit leukemia cell growth. (A) NBTG and (B) ABT-702 prevented the ability of KR to inhibit TEX cell growth. (C) Inhibitors of nucleobase uptake (NBTI-red squares; NBTG-blue triangles; DiPy-green inverted triangles), adenosine kinase (ABT-702-orange diamonds; 5-ITu-pink open circles; A-134974-brown open squares), adenylate kinase (Ap5A-open yellow triangles), adenylate cyclase (SQ22536-open gray diamonds), and an adenylate cyclase activator (NKH477-open blue inverted triangles) were tested at 500nM in the presence of KR. (D) TEX cells were cultured in the presence of Compound C (black circles), metformin (red squares), AICAR (blue triangles), rapamycin (green inverted triangles), compound 401 (orange diamonds), or PI-103 (pink open squares). Growth inhibition was measured using Alamar Blue. (E) Synergism between kinetin riboside and 10μM of Compound C (black circles), metformin (red squares), AICAR (blue triangles), rapamycin (green inverted triangles), compound 401 (orange diamonds), PI-103 (pink open squares), or DMSO vehicle (brown open squares) was tested using Alamar Blue. (F) Rapamycin was tested on 11 primary AML samples using Alamar Blue.
AMPK pathway and KR
Various enzymes, including ADK, tightly regulate levels of adenosine, AMP, ADP, and ATP. Cellular stress, including chemotherapy, is known to increase the ratio of AMP:ATP via inhibition of ATP production and activation of AMPK via phosphorylation by the tumor suppressor LKB1. Activated AMPK phosphorylates TSC2 leading to inhibition of Rheb and mTORC1 activity, which regulates protein translation and ribosome biogenesis.42 We hypothesized that KR treatment would increase the AMP:ATP ratio, induce AMPK and TSC2 phosphorylation, and ultimately inhibit cell proliferation. However, overnight treatment of TEX cells with KR led to decreased phosphorylated AMPK (supplemental Figure 11B). We used the AMPK activators metformin and AICAR,42 the AMPK inhibitor Compound C,43 the mTOR inhibitor rapamycin,42 the DNA-PK/PI3K/mTOR inhibitor PI 103,44 and the DNA-PK/mTOR inhibitor compound 40144 to further test if AMPK/mTOR/PI3K signaling was required for KR-induced cell killing. Compound 401 was mostly ineffective whereas PI 103 killed cells at low micromolar levels (Figure 6D). Metformin, AICAR, and Compound C did not alter KR-induced cell killing compared with single-agent treatment (Figure 6E). Higher concentrations of metformin and AICAR did not consistently affect apoptosis induced by KR (supplemental Figure 11C). In addition, rapamycin was generally ineffective on primary AML cells (Figure 6F). Thus, KR does not appear to require AMPK signaling to mediate the cell killing effects.
Discussion
Here, we developed a new approach combining high-throughput screening of leukemia cell lines driven by L-IC–like cells, counter-screening hits on normal HSPCs, and then in vitro and in vivo validation of hits on primary AML to identify compounds that target L-ICs. We propose this approach to be useful for discovering CSC-specific compounds in other tissues. In lieu of directly using L-ICs from primary AML samples, which are exceedingly rare, difficult to purify to homogeneity, and hard to maintain in vitro for extended periods of time, we used 2 novel leukemia cell lines driven by L-IC–like cells, able to differentiate in vitro on removal of cytokines, and importantly, retain a hierarchal organization unlike traditional leukemia cell lines (ie, HL-60, THP1). In addition, these lines, unlike HL-60 and THP1, express many genes identified in L-ICs as well as within the shared L-IC/normal HSC signature, indicating these novel experimentally generated cell lines use some of the same stem cell regulatory networks and may reflect a more representative population for L-ICs in high-throughput screens. Because most high-throughput screens have used other leukemia cell lines, these screens probably missed agents with anti–L-IC activity. By using cells transformed with 2 distinctly different oncogenes, TLS-ERG and MLL-ENL, our screen was designed to identify agents with broad activity. As such, 76 of the most potent 80 compounds targeted both TEX and M9-ENL1 cells showing the screen did identify broadly active agents.
BM toxicity and suppression is a common side effect of current chemotherapeutics because they often target both normal stem and progenitor cells. In an attempt to identify agents with reduced side effects, we tested the majority of our top hits on Lin− CB cells as a source of HSPCs. Indeed, some compounds (33%) were more toxic to Lin− CB cells or showed no specificity (49%), indicating that a counter-screen on normal HSPCs is a useful step in a preclinical setting. From the remaining 10 compounds, some are unlikely to move forward because of structural issues (thiram, erysolin, and ammonium pyrrolidinedithiocarbamate), some have been or are currently used in the clinic (etoposide and mechloroethamine), and some have research or industrial uses (puromycin, 8-hydroxyquinoline, and pararosaniline pamoate). Ciclopirox olamine, which was identified in our screen, is able to target AML L-ICs in a preclinical setting.45 Ciclopirox olamine acts as an intracellular iron chelator that inhibits iron-dependent enzymes including ribonucleotide reductase, and is a topical antifungal approved for dermatogical uses. Because of a favorable prior safety record and pharmacokinetics, ciclopirox olamine is currently under investigation for the treatment of AML in a phase 1 clinical trial.46 Overall, these results suggest that our approach is valid and can identify viable candidates for phase 1 clinical trials.
As we report here, the remaining compound, kinetin riboside, showed the greatest selectivity between Lin− CB and TEX. Because parthenolide was previously shown to target L-ICs and not HSCs,17 we also included a comparison of KR and PTL. Based on the apoptosis induction in CD34+CD38− AML cells, we were surprised that PTL appeared to be ineffective at preventing engraftment of primary AML samples. It is important to consider that PTL kills L-ICs and blasts equally as shown in Figure 3 and Guzman et al.17 However, because our study was not designed to quantify L-IC numbers after treatment, we could not determine whether PTL had no effect on L-IC numbers even though large numbers of bulk blasts were killed in 16 hours of ex vivo treatment, or that many L-ICs were killed but enough remained to still contribute to repopulation. Given that we used the sensitive IF xenotransplantation assay and that Guzman et al have shown that L-ICs are indeed killed, we favor the latter suggestion.17 Therefore, a positive response in an in vitro apoptosis assay does not necessarily predict a positive response in an in vivo functional assay indicating that quantitative L-IC measurements form an essential component of our approach.
The counter-screen on HSPCs from Lin− CB cells represents an important component of the screen because it effectively removed a significant number of the original 55 candidate compounds that would have high likelihood of hematopoietic toxicity. As our subsequent functional data showed, the in vitro counter-screen was less effective in ensuring that the compounds did not target HSCs. This is probably because of the fact that functional HSCs represent a minor fraction within the HSPC population, and are vastly outnumbered by more committed progenitors that would be expected to contribute to the major read-out in the Alamar Blue and apoptosis screens.18 Indeed progenitors, as assayed by clonogenic assay, are several fold more resistant to KR killing than HSCs, an effect potentially because of the greater sensitivity of HSCs than progenitor cells to genotoxic stress.47 Many effective cancer drugs induce some degree of myelosuppression by acting on HSCs and progenitors, but their toxicity is managed clinically so this result alone should not preclude further evaluation of KR or its analogues. Future studies at lower KR doses are warranted to identify concentrations that inhibit AML engraftment and maintain HSC engraftment. However, we would propose a refinement of our original screening strategy to also include an additional counter-screen against CD34+CD38−CD45RA−CD90+CD49f+ cells because we recently showed that human HSCs are highly enriched with this set of markers.18 Such a step might be valuable in eliminating compounds without any HSC toxicity.
Although KR is effective on numerous primary AML patient samples in in vitro screening, L-IC activity as measured by in vivo transplantation was not inhibited by pretreatment with KR for all patients, indicating that there is heterogeneity in response. These results point to the need to evaluate a broad spectrum of patient samples with both in vitro and in vivo assays to identify patients whose L-ICs are sensitive. We expect the extent of correlation between in vitro and in vivo assays to be drug specific. For example, ∼ 30% of AML patients responded to etoposide in vitro and this correlated well with anti–L-IC activity, indicating some anti–L-IC compounds may be effective in a limited patient cohort, and that our approach could also be used to rapidly identify which patient would benefit the most from treatment (S.P.M., K.E., F.N., M.D.M, and J.E.D., Etoposide targets leukemia stem cells in a subset of acute myeloid leukemia patients, in preparation).
Recently, it was shown that engineered breast cancer cells with an EMT (“stem cell”) phenotype were sensitive to salinomycin, abamectin, nigericin, and etoposide.48 These compounds were more selective on tumor cells than parental breast cells. 3 of these compounds, etoposide, nigericin, and salinomycin, were identified in our screens in addition to narasin, which is structurally similar to salinomycin. Salinomycin was effective on TEX, but not M9-ENL1 cells and therefore not tested on normal HSCs. Narasin and nigericin were included among the 80 most potent hits in our screen and tested on Lin− CB cells. At doses studied by Gupta et al,48 narasin and nigericin were more toxic to normal hematopoietic cells and therefore not further tested on primary AML cells (supplemental Figure 12). Although in vitro hematopoietic toxicity does not always correlate with toxicity in humans, our findings clearly indicate that additional toxicity experiments on normal HSCs are warranted for those compounds. Indeed, our in vitro HSPC apoptosis (Figure 3E) and in vivo transplantation (Figure 4C) results with KR and PTL point to a dissociation between results using CD34+CD38− as a phenotypic HSC marker and functional assays, such as colony formation (progenitor cell activity) and NOD/SCID engraftment (stem cell activity). Together, these results highlight the need for all CSC screens to incorporate a central feature of our screening strategy, that of counter-screening using functional assays on normal HSPCs.
Here, we propose a new approach for identification of anti-CSCs compounds: (1) screening of surrogate cancer cell populations that reflect CSC biology; (2) incorporation of counter-screens on normal hematopoietic cells and potentially intestinal, skin, and neural cells to remove potentially toxic compounds; (3) validation on primary tumor tissue in vitro and in NOD/SCID xenograft assays designed to test CSC function; (4) comparison to standard treatment; and (5) dissection of the molecular pathways targeted by the compound(s). Such a strategy would raise the probability that compounds passing this screen would not only be effective on CSCs, but also have reduced side effects in a clinical setting.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Dick laboratory and Craig Jordan for critical input regarding this work and paper.
This work was supported by a fellowship from the Leukemia & Lymphoma Society (K.E.) and grants from the Canadian Institutes for Health Research; the Stem Cell Network of Canadian National Centres of Excellence; the Canadian Cancer Society and the Terry Fox Foundation; Genome Canada through the Ontario Genomics Institute; Ontario Institute for Cancer Research with funds from the province of Ontario; the Leukemia & Lymphoma Society; the Canadian Institutes for Health Research; and a Canada Research Chair. This research was funded in part by the Ontario Ministry of Health and Long Term Care (OMOHLTC). The views expressed do not necessarily reflect those of the OMOHLTC.
Authorship
Contribution: S.P.M. designed experiments, performed research, analyzed data and microarray data, and wrote the paper; K.E. performed research (injected cells into mice), analyzed microarray data, and wrote the paper; F.N., performed research (injected cells into mice) and wrote the paper; M.I. provided new reagents (synthesized KR-monophosphate); A.D. performed research (primary and secondary screens); M.D.M. provided primary leukemia samples; R.A. and J.W. provided research support; and J.E.D. analyzed data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John E. Dick, Toronto Medical Discovery Tower, Rm 8-301, 101 College St, Toronto, ON Canada M5G 1L7; e-mail: jdick@uhnres.utoronto.ca.