Abstract
The positively charged chemokine platelet factor 4 (PF4) forms immunogenic complexes with heparin and other polyanions. Resulting antibodies can induce the adverse drug effect heparin-induced thrombocytopenia. PF4 also binds to bacteria, thereby exposing the same neoantigen(s) as with heparin. In this study, we identified the negatively charged lipopolysaccharide (LPS) as the PF4 binding structure on Gram-negative bacteria. We demonstrate by flow cytometry that mutant bacteria with progressively truncated LPS structures show increasingly enhanced PF4 binding activity. PF4 bound strongest to mutants lacking the O-antigen and core structure of LPS, but still exposing lipid A on their surfaces. Strikingly, PF4 bound more efficiently to bisphosphorylated lipid A than to monophosphorylated lipid A, suggesting that phosphate residues of lipid A mediate PF4 binding. Interactions of PF4 with Gram-negative bacteria, where only the lipid A part of LPS is exposed, induce epitopes on PF4 resembling those on PF4/heparin complexes as shown by binding of human anti-PF4/heparin antibodies. As both the lipid A on the surface of Gram-negative bacteria and the amino acids of PF4 contributing to polyanion binding are highly conserved, our results further support the hypothesis that neoepitope formation on PF4 after binding to bacteria is an ancient host defense mechanism.
Introduction
Besides their pivotal role in hemostasis, platelets are involved in host defense against pathogens and in modulation of immune reactions. This function of platelets occurs either indirectly through their interaction with endothelial cells and leukocytes1,2 or directly by secretion of antimicrobial substances from platelet storage granules and lysosomes.3,4
Recently, we have shown that the chemokine platelet factor 4 (PF4), which is stored within platelet α-granules, plays a role in bacterial host defense by inducing a humoral immune response to PF4-coated bacteria.5 During bacterial infections, platelets are activated6,7 and release positively charged PF4, which can bind in a charge-dependent manner to the bacterial surface, thereby inducing neoepitopes. The formation of antigenic PF4 clusters is probably the result of neutralization of the positive charge of PF4 by polyanions,8 which allows narrowing of the distance between single PF4 tetramers down to 3 to 5 nm. This creates linear, ridge-like complexes and exposes new antigenic epitopes on PF4.9 Antibodies to PF4/polyanion complexes bind to PF4 on the bacterial surface, leading to opsonization and increased phagocytosis of PF4-coated bacteria.5 As PF4 is capable of binding to a large variety of bacteria, the antibody response to PF4/polyanion complexes constitutes a very broad reactive defense mechanism and could represent an evolutionary interface between innate and specific immunity. Antibodies induced by PF4 clusters would be an example of antibodies with a limited target antigen repertoire that nevertheless could result in binding to a large variety of bacteria when these bacteria are coated with PF4.5
In medicine, research on the immune reaction to PF4/polyanion complexes to date has primarily focused on its role in causing an adverse reaction to the anticoagulant heparin as PF4 forms immunogenic complexes with heparin on platelet surfaces. Anti-PF4/polyanion antibodies bind to these PF4/heparin complex-coated platelets and induce Fc-receptor–dependent platelet activation,10,11 leading to intravascular consumption of platelets, associated potentiation of in vivo thrombin generation, and the prothrombotic syndrome, heparin-induced thrombocytopenia (HIT).
We and others have recently demonstrated the prevalence of anti-PF4/heparin antibodies of the IgM class in up to 20% and of the IgG class in up to 6% of the general population and in a slightly lower number of normal blood donors.5,12 These antibodies are highly significantly associated with periodontitis, one of the most prevalent human infections, often associated with transient bacteremia.13 The major bacterial species in periodontitis are the Gram-negative bacteria Aggregatibacter actinomycetemcomitans and Porphyromonas gingivalis.14 Both bacteria bind PF4 and consequently expose epitopes recognized by anti-PF4/heparin antibodies.13
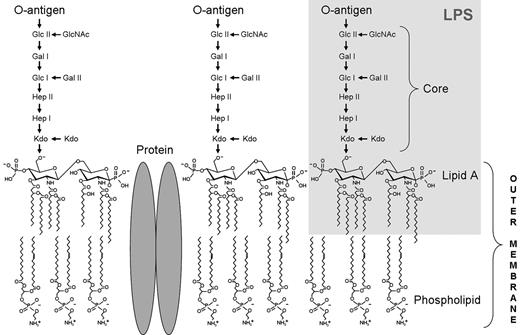
The outer leaflet of the outer membrane of Gram-negative bacteria is mainly composed of negatively charged lipopolysaccharide (LPS), interspersed with proteins (Figure 1).15 LPS, a complex glycolipid, is anchored in the outer membrane by the highly conserved lipid A. The central core oligosaccharide is linked to lipid A and can be divided into an inner core composed of heptoses and 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo), and an outer core consisting of hexoses, followed by the most variable part of the LPS molecule, the O-specific polysaccharide chain.16
In this study, we have identified the phosphate groups of the highly conserved lipid A as the binding site for PF4 on the surface of Gram-negative bacteria. In accordance with our hypothesis that opsonization of bacteria by PF4 might represent an ancient host defense mechanism, we found the anion binding site of PF4 and PF4-like proteins conserved among different vertebrate species.
Methods
Bacterial strains
Bacterial strains used in this study are listed in Table 1. Escherichia coli K-12 wild-type strain BW30270 and E coli K-12 mutant strains17,18 were kindly provided by R. W. Woodard (University of Michigan, Ann Arbor, MI). Salmonella enterica sv Typhimurium SL3770 waa+ and the isogenic Salmonella LPS mutants were obtained from K. E. Sanderson (Salmonella Genetic Stock Center, Calgary, AB). Bacteria were cultured at 37°C to the exponential growth phase (A600 of 0.7-1.2) in Todd-Hewitt broth supplemented with 0.5% yeast extract (Roth).19
LPS and derivatives
LPS-biotin was purchased from InvivoGen. Lipid A substituted with 2 Kdo residues (Kdo2-lipid A) and lipid A were isolated from E coli F515 (Re chemotype). Separation and purification of lipid A from Kdo was performed as described.20 LPS isolated from E coli O55:B5, mono-(4′-P)-phosphoryl lipid A (MPLA) isolated from E coli F583 (Rd mutant) and Kdo were purchased from Sigma-Aldrich.
Purification of PF4 and PF4 biotinylation
Human PF4 was isolated from platelets (Chromatec) and biotinylated as described.5 The concentration of biotinylated PF4 was determined by a bicinchoninic acid protein assay kit using BSA as standard (Sigma-Aldrich).
Binding of LPS to immobilized PF4 and binding of PF4 to immobilized LPS derivatives
To investigate binding of LPS to immobilized PF4, wells of a microtiter plate (MaxiSorp, Nunc) were coated with PF4 (10 μg/mL), BSA, or buffer (carbonate-bicarbonate buffer, pH 9.6; Medicago) at 4°C overnight. In the reverse approach, wells (PolySorp, Nunc) were coated with LPS, Kdo2-lipid A, lipid A (bisphosphorylated), MPLA (monophosphorylated), or Kdo in carbonate-bicarbonate buffer pH 9.6 (each with 50 μg/mL, 4°C, overnight). Before the incubation with increasing concentrations of LPS-biotin (0.6, 1.3, 2.5, 5.0, 10, 20, and 40 μg/mL; 60 minutes, room temperature), PF4-biotin (0.06, 0.13, 0.25, 0.5, 1.0, 2.0, and 4.0 μg/mL; 60 minutes), or buffer as a control, microtiter plates were washed 5 times with PBS + 0.1% Tween and blocked with PBS + 0.1% Tween + 2% BSA (60 minutes, room temperature). Plates were washed and incubated with peroxidase-conjugated streptavidin (1:4000, 60 minutes, room temperature; Jackson ImmunoResearch Laboratories). After final washing steps, tetramethylbenzidine was added to the wells, the reaction was stopped with 1M H2SO4, and absorbance was measured at 450 nm. In addition, binding of PF4-biotin (2 μg/mL) to immobilized lipid A was measured in the presence of increasing concentrations (0.5, 1, 2, 4, and 8 μg/mL) of bacterial permeability increasing protein (BPI; Alpha Diagnostic International), polymyxin B sulfate (Sigma-Aldrich), or buffer.
PF4 binding to bacterial LPS mutants
E coli BW30270, KPM53, and KPM121, and S enterica sv Typhimurium strains SL3770, SL3749, SL3750, SL3748, SL3769, SL3789, and SL1102 (Table 1) were incubated (30 minutes, 4°C) with PF4-biotin, 20 μg/mL; or buffer, washed with PBS/0.05% BSA (3000g, 5 minutes, 4°C) and incubated (30 minutes, 4°C) with peridinin chlorophyll protein-Cy5.5 conjugated streptavidin (BD Biosciences). Bacteria were washed and fixed with 1% paraformaldehyde (20 minutes, 4°C). PF4 binding was analyzed by flow cytometry (Cytomics FC 500, Beckman Coulter). The geometric mean fluorescence intensity multiplied by the percentage of labeled bacteria constituted binding activity.
In inhibition assays, PF4-biotin (40 μg/mL) was preincubated (30 minutes, 4°C) with 200 μg/mL LPS, Kdo, Kdo2-lipid A, lipid A, MPLA, or buffer before assessing PF4 binding to the E coli ΔwaaA mutant KPM121 by flow cytometric analysis.
Furthermore, E coli strains BW30270, KPM53 and KPM121 (Table 1) were pretreated (15 minutes, 37°C) with pronase E (100 μg/mL; Merck), trypsin (100 μg/mL; Sigma-Aldrich), or buffer, washed 3 times with PBS, pH 7.4 (3700g, 6 minutes, room temperature), incubated (30 minutes, 4°C) with PF4-biotin (20 μg/mL) or buffer and analyzed for PF4 binding. Activity of pronase E and trypsin was controlled by reduction of E coli Antigen 43.
Anti-PF4/heparin antibody binding to PF4-coated bacterial LPS mutants
Binding of human anti-PF4/heparin antibodies to bacteria was assessed by adsorption and elution experiments as described.5 In brief, LPS mutant strains E coli KPM53 and KPM121 were incubated with PF4 (20 μg/5 × 107 bacteria, 30 minutes, 4°C) or buffer (PBS, pH 7.4), washed (3000g, 5 minutes, 4°C) and incubated (30 minutes, 4°C) with diluted human serum of patients known to contain anti-PF4/heparin IgG antibodies (n = 3 per strain), and washed again to remove unbound antibodies. Bound antibodies were eluted with glycine buffer (0.1M, pH 2.7; 5 minutes, room temperature), bacteria removed by centrifugation, and supernatants (eluates: containing the antibodies) neutralized with Tris buffer (1M, pH 9). Untreated sera and eluates were tested by PF4/heparin IgG ELISA,21 including inhibition by unfractionated heparin (100 IU/mL; Braun) and binding to PF4 alone. In addition, heparin-induced platelet activation test was performed as described22 to test the ability of eluates to activate platelets.
Phagocytosis assay
Phagocytosis assays were performed as described5 with the following modifications. The E coli K12 wild-type strain BW30270 and its isogenic ΔwaaA mutant KPM121 were self-labeled with FITC by adding 100mM NaHCO3, pH 9, and incubating with 370 μg/mL FITC dissolved in DMSO (60 minutes, 37°C; AppliChem). Bacteria were washed 3 times with PBS/0.05% BSA (3000g, 5 minutes, 4°C) and incubated with PF4 (20 μg/5 × 107 bacteria, 30 minutes, 4°C). The washed samples (3000g, 5 minutes, 4°C) were incubated (30 minutes, 4°C) with human serum (1:50) of patients known to contain anti-PF4/heparin IgG antibodies (n = 4 per strain). The sera had been preadsorbed with each of both strains (non–PF4-coated; 15 minutes, 4°C; 4 times). Bacteria were washed again to remove unbound antibodies. Finally, the whole blood phagocytosis assay was performed as described5 using pretreated bacteria.
PF4 amino acid sequence survey
For the bioinformatics analysis, different resources, including the Universal Protein Resource (UniProt, http://www.uniprot.org), the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov), and the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING, http://string-db.org/), were used. The survey was started with BLAST23 searches, and the complete human PF4 amino acid sequence was used for protein sequence comparisons against the genome sequence databases available for other species. Although the databases generally mirror each other, additional sequences were provided by the databases of NCBI and STRING. The multiple sequence alignment was performed using ClustalW2 (www.ebi.ac.uk).
Statistical analysis
Data are plotted as mean ± SD. We compared samples of bacterial binding studies, affinity purification experiments, and phagocytosis assay by paired Student t test. Differences between samples of ELISA data were calculated by paired t test, except for PF4 binding to bisphosphorylated lipid A compared with MPLA, which was analyzed by ANOVA test. P < .05 was considered statistically significant.
Results
Identification of the PF4 binding site on LPS
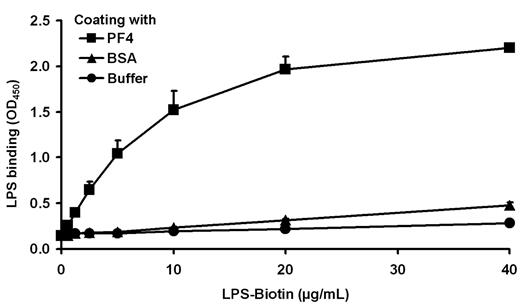
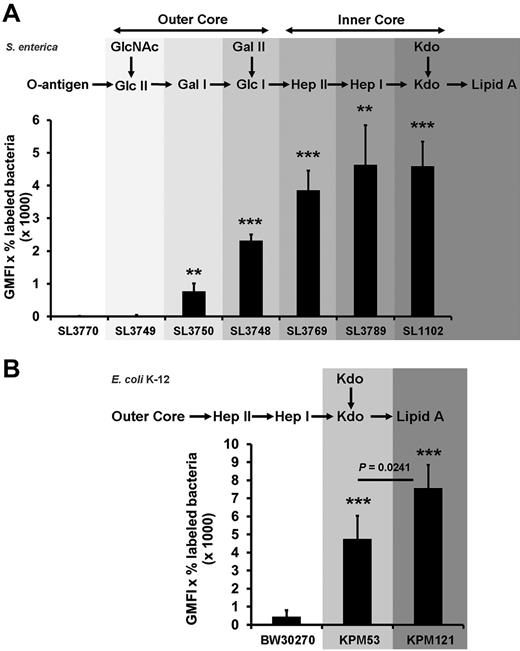
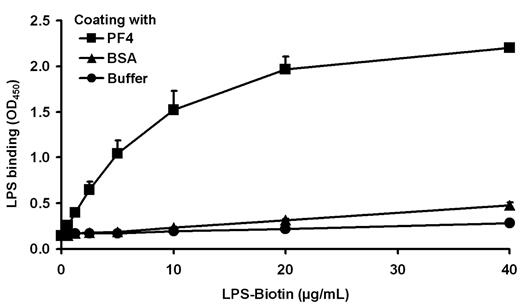
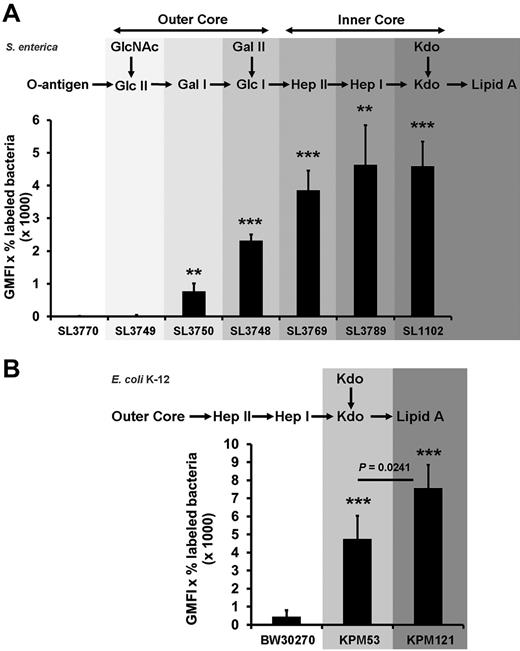
A negatively charged microbial component has been suggested to recruit PF4 to the Gram-negative bacterial cell surface. As PF4 binding is a general characteristic for Gram-negative bacteria, a conserved surface structure, such as LPS (Figure 1), may be involved in binding of PF4. To test this hypothesis, binding of LPS to immobilized PF4 was investigated. The results show that LPS binds dose-dependently to PF4 (Figure 2). To decipher in more detail the PF4 binding part in LPS, we performed PF4 binding assays using different mutants of S enterica sv Typhimurium with gradually truncated LPS structures. Surprisingly, the wild-type strain SL3770 with smooth (S)-form LPS (waa+) showed a low degree of PF4 binding (geometric mean fluorescence intensity [GMFI], 8.5 ± 2.9; Figure 3A). However, PF4 binding capacity increased step-wise in mutants with increasing truncations of the LPS outer and inner core oligosaccharides. The first mutant which was able to recruit PF4 in significant amounts was S enterica sv Typhimurium SL3750 (waaJ417), lacking the O-antigen and GlcII of the LPS outer core (GMFI, 768.2 ± 243.1 vs rfa+ GMFI, 8.5 ± 2.9; P = .0056). Further deletions of consecutive sugars of the outer and inner core to the level of Kdo2-lipid A, as represented by S enterica sv Typhimurium SL1102 (hldE543; GMFI, 4587.3 ± 754.3; P = .0005), gradually increased PF4 binding (Figure 3A).
Schematic architecture of the LPS embedded in the outer membrane of Gram-negative bacteria. LPS is a complex glycolipid composed of a highly conserved lipid A anchor, the more variable core oligosaccharide, and the hypervariable antigenic O-polysaccharide (O-antigen) consisting of several copies of oligosaccharide repeating units. Glc indicates glucose; Gal, galactose, GlcNAc, N-acetyl-d-glucosamine; and Hep, heptose.
Schematic architecture of the LPS embedded in the outer membrane of Gram-negative bacteria. LPS is a complex glycolipid composed of a highly conserved lipid A anchor, the more variable core oligosaccharide, and the hypervariable antigenic O-polysaccharide (O-antigen) consisting of several copies of oligosaccharide repeating units. Glc indicates glucose; Gal, galactose, GlcNAc, N-acetyl-d-glucosamine; and Hep, heptose.
LPS binds dose-dependently to PF4. Binding of biotinylated LPS (0.6, 1.3, 2.5, 5.0, 10, 20, and 40 μg/mL) to immobilized PF4 (10 μg/mL) was detected with peroxidase-conjugated streptavidin, followed by the addition of tetramethylbenzidine. Binding of PF4 to BSA or buffer (each with 10 μg/mL) served as controls to exclude nonspecific binding of LPS-biotin to proteins or to the plastic surface. Data are mean OD450 ± SD of 3 independent experiments.
LPS binds dose-dependently to PF4. Binding of biotinylated LPS (0.6, 1.3, 2.5, 5.0, 10, 20, and 40 μg/mL) to immobilized PF4 (10 μg/mL) was detected with peroxidase-conjugated streptavidin, followed by the addition of tetramethylbenzidine. Binding of PF4 to BSA or buffer (each with 10 μg/mL) served as controls to exclude nonspecific binding of LPS-biotin to proteins or to the plastic surface. Data are mean OD450 ± SD of 3 independent experiments.
Bacterial mutants with progressively truncated LPS backbones show increasing PF4 binding activity.S enterica sv Typhimurium wild-type strain SL3770 waa+, the isogenic LPS mutants SL3749 waaL446, SL3750 waaJ417, SL3748 waaI432, SL3769 waaG471, SL3789 waaF511, and SL1102 hldE543 (A), the E coli K-12 wild-type strain BW30270, and the LPS mutants KPM53 (ΔwaaC) and KPM121 (ΔwaaA; B) were incubated with biotinylated PF4 (20 μg/mL). PF4 binding was detected with peridinin chlorophyll protein-Cy5.5 conjugated streptavidin using flow cytometry and expressed as geometric mean fluorescence intensity (GMFI) multiplied by the percentage of labeled bacteria. Data represent mean ± SD of at least 3 independent experiments (S enterica sv Typhimurium wild-type and mutants, n = 3; E coli BW30270, n = 5; E coli KPM53, n = 5; and E coli KPM121, n = 3). **P < .01 versus wild-type. ***P < .001 versus wild-type.
Bacterial mutants with progressively truncated LPS backbones show increasing PF4 binding activity.S enterica sv Typhimurium wild-type strain SL3770 waa+, the isogenic LPS mutants SL3749 waaL446, SL3750 waaJ417, SL3748 waaI432, SL3769 waaG471, SL3789 waaF511, and SL1102 hldE543 (A), the E coli K-12 wild-type strain BW30270, and the LPS mutants KPM53 (ΔwaaC) and KPM121 (ΔwaaA; B) were incubated with biotinylated PF4 (20 μg/mL). PF4 binding was detected with peridinin chlorophyll protein-Cy5.5 conjugated streptavidin using flow cytometry and expressed as geometric mean fluorescence intensity (GMFI) multiplied by the percentage of labeled bacteria. Data represent mean ± SD of at least 3 independent experiments (S enterica sv Typhimurium wild-type and mutants, n = 3; E coli BW30270, n = 5; E coli KPM53, n = 5; and E coli KPM121, n = 3). **P < .01 versus wild-type. ***P < .001 versus wild-type.
To test whether lipid A alone is sufficient for PF4 binding, we switched to E coli KPM121 (ΔwaaA), which predominantly expresses the nonglycosylated, tetra-acylated lipid A precursor lipid IVA.17 Compared with S enterica sv Typhimurium SL3770, E coli K-12 wild-type strain BW30270 showed at least some PF4 binding (Figure 3B; GMFI, 445.0 ± 359.0). (It is noteworthy that the E coli K-12 is a strain with rough-type LPS, which lacks the O16-antigen because of an IS5 insertion mutation in the wbbL gene for a rhamnosyltransferase involved in O-antigen synthesis,24 whereas S enterica sv Typhimurium SL3770 expresses the S-form of LPS.) In accordance with the results obtained for the S enterica sv Typhimurium mutants, the E coli K-12 ΔwaaC mutant KPM53 with 2 Kdo residues attached to lipid A (corresponding to the hldE543 mutant of S enterica sv Typhimurium SL1102) showed increased PF4 binding compared with the wild-type (GMFI, 4738.5 ± 1304.6, P = .0001). When the 2 Kdo residues were also deleted in the E coli ΔwaaC mutant KPM121 (now exposing predominantly the tetraacylated lipid A precursor lipid IVA), PF4 binding further increased (GMFI, 7581.3 ± 1285.5; P = .0241; Figure 3B). This suggests that lipid A is a major PF4 binding partner on the surface of Gram-negative bacteria.
Phosphate groups of lipid A mediate PF4 binding
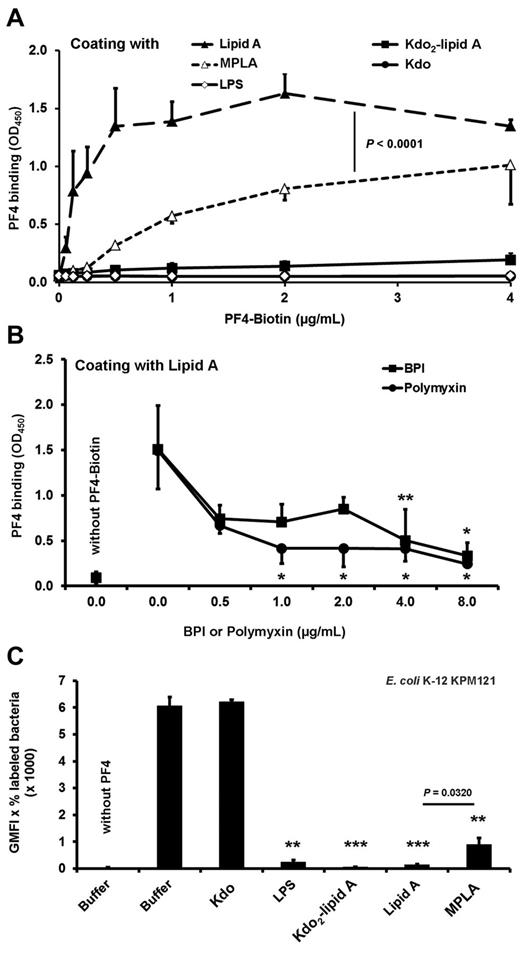
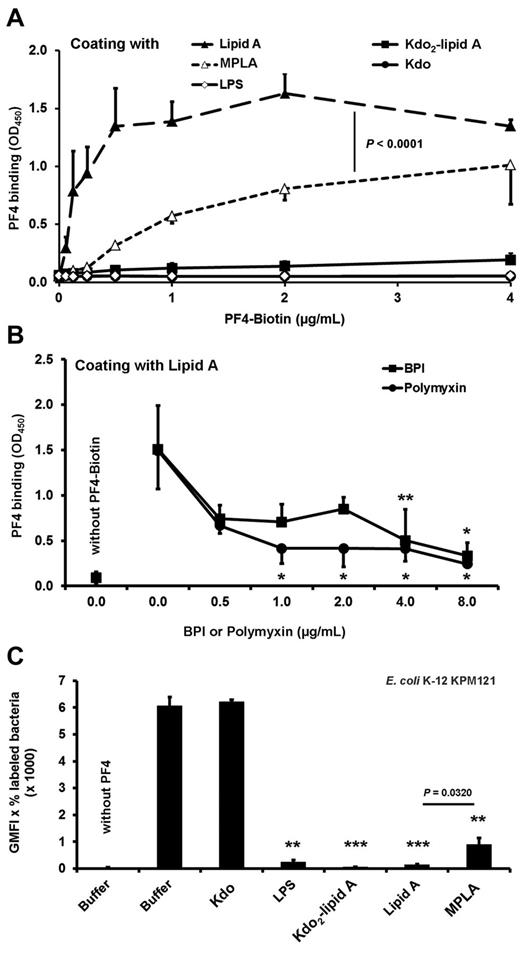
To assess whether PF4 directly interacts with lipid A, we analyzed binding of PF4 to isolated lipid A and Kdo2-lipid A. The binding experiments demonstrated that PF4 binds directly and in a dose-dependent manner to lipid A (Figure 4A). To show specificity, we coincubated lipid A with either BPI or polymyxin. These lipid A binding compounds inhibit binding of PF4 to lipid A in a dose-dependent manner, starting at 0.5 μg/mL and reaching significance at a concentration of 4 μg/mL (P = .0096) and 1 μg/mL (P = .0192), respectively (Figure 4B). PF4 bound to immobilized lipid A, but not to immobilized LPS or Kdo2-lipid A, probably because of steric hindrance (Figure 4A). This is consistent with the higher PF4 binding activity to nonglycosylated lipid IVA of E coli KPM121 compared with E coli KPM53 with 2 Kdo residues attached to lipid A (Kdo2-lipid A; P = .0241; Figure 3B). PF4 did not bind to immobilized Kdo (Figure 4A).
PF4 interaction with isolated lipid A relies on the phosphate groups. (A) PF4 binds dose-dependently to lipid A but not to LPS, Kdo2-lipid A, or to Kdo alone. Binding of PF4 to MPLA (mono-(4′-P)-phosphoryl lipid) is reduced compared with binding to bisphosphorylated lipid A (Lipid A). Binding of biotinylated PF4 (0.06, 0.13, 0.25, 0.5, 1, 2, and 4 μg/mL) to immobilized LPS, Lipid A, Kdo2-lipid A, MLPA, or Kdo (50 μg/mL) was detected with peroxidase-conjugated streptavidin followed by addition of tetramethylbenzidine. Data are mean OD ± SD of 3 independent experiments. (B) Binding of PF4 to Lipid A is inhibited by BPI or polymyxin B sulfate. Binding of biotinylated PF4 (2 μg/mL) in the presence of BPI or polymyxin (0.5, 1, 2, 4, and 8 μg/mL each) or buffer to immobilized Lipid A (50 μg/mL) was detected with peroxidase-conjugated streptavidin followed by addition of tetramethylbenzidine. Data are mean OD ± SD of 3 independent experiments. *P < .05 versus PF4-biotin + buffer. **P < .01 versus PF4-biotin + buffer. (C) Preincubation of PF4 with LPS, lipid A, Kdo2-lipid A, but not Kdo alone, inhibits PF4 binding to the E coli KPM121 displaying only lipid IVA on its surface, whereas the inhibitory effect of MPLA is reduced. PF4-biotin (40 μg/mL) was preincubated with Kdo, LPS, Kdo2-lipid A, Lipid A, MPLA (each with 200 μg/mL), or buffer before assessing PF4 binding to E coli KPM121 by flow cytometry. The results are expressed as geometric mean fluorescence intensity (GMFI) multiplied by the percentage of labeled bacteria. Data represent mean ± SD of 3 independent experiments. **P < .01 versus PF4-biotin preincubated with buffer. ***P < .001 versus PF4-biotin preincubated with buffer.
PF4 interaction with isolated lipid A relies on the phosphate groups. (A) PF4 binds dose-dependently to lipid A but not to LPS, Kdo2-lipid A, or to Kdo alone. Binding of PF4 to MPLA (mono-(4′-P)-phosphoryl lipid) is reduced compared with binding to bisphosphorylated lipid A (Lipid A). Binding of biotinylated PF4 (0.06, 0.13, 0.25, 0.5, 1, 2, and 4 μg/mL) to immobilized LPS, Lipid A, Kdo2-lipid A, MLPA, or Kdo (50 μg/mL) was detected with peroxidase-conjugated streptavidin followed by addition of tetramethylbenzidine. Data are mean OD ± SD of 3 independent experiments. (B) Binding of PF4 to Lipid A is inhibited by BPI or polymyxin B sulfate. Binding of biotinylated PF4 (2 μg/mL) in the presence of BPI or polymyxin (0.5, 1, 2, 4, and 8 μg/mL each) or buffer to immobilized Lipid A (50 μg/mL) was detected with peroxidase-conjugated streptavidin followed by addition of tetramethylbenzidine. Data are mean OD ± SD of 3 independent experiments. *P < .05 versus PF4-biotin + buffer. **P < .01 versus PF4-biotin + buffer. (C) Preincubation of PF4 with LPS, lipid A, Kdo2-lipid A, but not Kdo alone, inhibits PF4 binding to the E coli KPM121 displaying only lipid IVA on its surface, whereas the inhibitory effect of MPLA is reduced. PF4-biotin (40 μg/mL) was preincubated with Kdo, LPS, Kdo2-lipid A, Lipid A, MPLA (each with 200 μg/mL), or buffer before assessing PF4 binding to E coli KPM121 by flow cytometry. The results are expressed as geometric mean fluorescence intensity (GMFI) multiplied by the percentage of labeled bacteria. Data represent mean ± SD of 3 independent experiments. **P < .01 versus PF4-biotin preincubated with buffer. ***P < .001 versus PF4-biotin preincubated with buffer.
However, when the experiments were performed with soluble lipid A, soluble Kdo2-lipid A, and soluble LPS, all inhibited PF4 binding to E coli KPM121 (GMFI, 143.2 ± 17.5; P = .0002, 56.4 ± 8.2; P = .0002 and GMFI, 242.6 ± 74.8; P = .0022, respectively, vs buffer GMFI, 6071.8 ± 139.0; Figure 4C). As expected, soluble Kdo did not inhibit PF4 binding to E coli KPM121 (GMFI, 6213.5 ± 82.9 vs buffer GMFI, 6095.0 ± 435.9; P = .6763; Figure 4C).
The lipid A backbone [4′-P-β-D-GlcpN-(1→6)-α-D-GlcpN-1′-P] carries 2 phosphate groups in positions 4′ and 1′. These negatively charged phosphate groups are important for PF4 binding, as binding of PF4 to immobilized MPLA (monophosphorylated) was significantly lower than to its native (bisphosphorylated) lipid A counterpart (Figure 4A; P < .0001). Concordantly, soluble MPLA had reduced inhibitory effects on PF4 binding to E coli KPM121 compared with lipid A (GMFI, 893.9 ± 240.2 vs GMFI, 143.2 ± 17.5; P = .0320; Figure 4C).
Surface exposed proteins play no major role in PF4 binding
Besides LPS, negatively charged proteins decorate the bacterial surface. Possible binding of PF4 to proteins unmasked by the lack of LPS was not changed by treatment of E coli BW30270, KPM53, and KPM121 with proteolytic enzymes. Pretreatment of the wild-type strain (BW30270), the waaC mutant (KPM53), and the waaA mutant (KPM121) with pronase E or trypsin did not alter PF4 binding (P > .2 for all comparisons; data not shown).
PF4 bound to lipid A of Gram-negative bacteria exposes PF4/heparin-like epitopes
To test whether anti-PF4/heparin antibodies recognize PF4 bound to lipid A of Gram-negative bacteria, we used LPS mutant strains only displaying lipid A and Kdo (E coli KPM53), or lipid IVA without Kdo (E coli KPM121) in adsorption and elution experiments. Affinity-purified IgG antibodies eluted from PF4-coated E coli strains KPM53 and KPM121 reacted with PF4/heparin complexes in the PF4/heparin-ELISA (Figure 5A), suggesting that PF4 bound to lipid A on bacterial surfaces exposes epitopes, which are also present in PF4/heparin complexes. In contrast, no antibodies with PF4/heparin specificity were eluted from native bacteria (not preincubated with PF4; Figure 5A). As further controls, the affinity-purified antibodies did not react with PF4 alone, and the addition of excess of heparin (disrupting PF4/heparin complexes) abrogated binding of eluted anti-PF4/heparin antibodies in the PF4/heparin ELISA, indicating the specificity of antibodies eluted from PF4-coated bacteria for PF4/heparin complexes.
PF4-coated E coli LPS mutants are recognized by anti-PF4/heparin IgG from sera of patients with HIT. (A) PF4-coated E coli LPS mutants ΔwaaC (KPM53) or ΔwaaA (KPM121) were incubated with sera of 3 patients with HIT known to contain anti-PF4/heparin antibodies, and bound IgG was affinity purified. Symbols represent reactivities of IgG antibodies eluted from PF4-coated E coli KPM53 (squares) or KPM121 (triangles); means are presented as horizontal lines. Binding of the purified antibodies to PF4/heparin complexes (first column) was inhibited by excess heparin (100 IU/mL unfractionated heparin [UFH], column 2), which disrupts PF4/heparin complexes. Antibodies did not react with PF4 alone (column 3). PF4-untreated bacteria served as control for unspecific binding of the antibodies to bacteria alone (column 4). (B) E coli K12 wild-type strain BW30270 and its isogenic ΔwaaA mutant KPM121 were labeled with FITC and preincubated with PF4 and additionally with heat-inactivated human serum containing anti-PF4/heparin IgG (preadsorbed with bacteria alone). After preincubation, the bacteria were subjected to phagocytosis. The figure shows the MFI multiplied by the percentage of FITC-positive polymorphonuclear leukocytes (PMNs) as a measure for bacterial phagocytosis. Data are mean ± SD of 4 different sera.
PF4-coated E coli LPS mutants are recognized by anti-PF4/heparin IgG from sera of patients with HIT. (A) PF4-coated E coli LPS mutants ΔwaaC (KPM53) or ΔwaaA (KPM121) were incubated with sera of 3 patients with HIT known to contain anti-PF4/heparin antibodies, and bound IgG was affinity purified. Symbols represent reactivities of IgG antibodies eluted from PF4-coated E coli KPM53 (squares) or KPM121 (triangles); means are presented as horizontal lines. Binding of the purified antibodies to PF4/heparin complexes (first column) was inhibited by excess heparin (100 IU/mL unfractionated heparin [UFH], column 2), which disrupts PF4/heparin complexes. Antibodies did not react with PF4 alone (column 3). PF4-untreated bacteria served as control for unspecific binding of the antibodies to bacteria alone (column 4). (B) E coli K12 wild-type strain BW30270 and its isogenic ΔwaaA mutant KPM121 were labeled with FITC and preincubated with PF4 and additionally with heat-inactivated human serum containing anti-PF4/heparin IgG (preadsorbed with bacteria alone). After preincubation, the bacteria were subjected to phagocytosis. The figure shows the MFI multiplied by the percentage of FITC-positive polymorphonuclear leukocytes (PMNs) as a measure for bacterial phagocytosis. Data are mean ± SD of 4 different sera.
The affinity purified (using PF4-coated E coli K-12 mutants KPM53 and KPM121) anti-PF4/heparin antibodies also activated platelets at low (0.2 IU/mL) but not at high heparin concentrations (100 IU/mL) in a functional assay for platelet activating anti-PF4/heparin antibodies (heparin-induced platelet activation; 3 of 3 sera, each).
To assess the impact of exposed lipid A on opsonization, bacterial mutants lacking LPS and wild-type bacteria were pretreated with PF4 and anti-PF4/heparin antibodies, and phagocytosis of these bacteria by polymorphonuclear leukocytes was measured by flow cytometry. E coli K-12 mutant KPM121 was more efficiently phagocytosed than its isogenic wild-type strain BW30270 (MFI, 18 267.4 ± 1449.3 vs wild-type MFI, 6118.5 ± 1449.3; P = .0054), demonstrating that the enhanced opsonization resulted in increased phagocytosis (Figure 5B).
Amino acids of PF4 important for polyanion binding are highly conserved among species
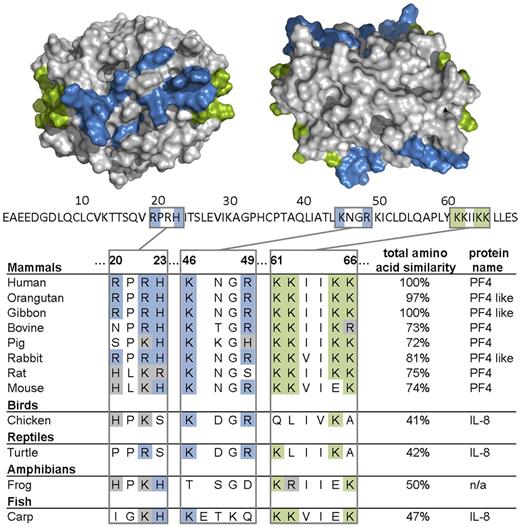
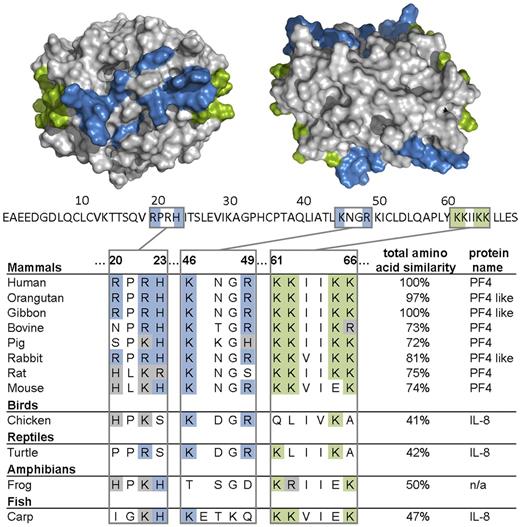
Finally, we analyzed the presence of PF4 and PF4-related proteins among vertebrate species in a PF4 amino acid sequence survey. The genomes encoding PF4, PF4-like protein, IL-8, or an uncharacterized protein with high similarities to human PF4 showed highly conserved amino acid residues among species, which represent essential motifs of the heparin binding site in human PF4 (Figure 6). These are lysine residues of the C-terminus (K61, K62, K65, and K66), arginine residues (R20, R22, and R49), histidine residues (H23), and lysine residue (K46).25–29 Even in the case of amino acid substitutions, one cationic amino acid is often replaced by another positively charged amino acid (Figure 6).
Amino acids of PF4 important for heparin binding are largely conserved among species. Alignment of the heparin binding region (R20, R22, H23, K46, R49, K61, K62, K65, and K66) of PF4, IL-8, and an uncharacterized protein (n/a) and total amino acid similarity of various species in percentages compared with human (100%) is shown. Above the amino acid sequence of 1 PF4 monomer (70 amino acids, primary structure without signal sequence, UniProt P02776), the crystal structure of a PF4 tetramer (quaternary structure, PDB code 1f9q) is depicted from the front (left) and from the top view (right). Lysine residues of the C-terminus (K61, K62, K65, and K66) are highlighted in green; arginine (R20, R22, and R49), histidine (H23), and other lysine residues (K46) are highlighted in blue. The pictures of the crystal structure of PF4 were made using PyMOL Molecular Graphics System Version 1.3 Schrödinger LLC.
Amino acids of PF4 important for heparin binding are largely conserved among species. Alignment of the heparin binding region (R20, R22, H23, K46, R49, K61, K62, K65, and K66) of PF4, IL-8, and an uncharacterized protein (n/a) and total amino acid similarity of various species in percentages compared with human (100%) is shown. Above the amino acid sequence of 1 PF4 monomer (70 amino acids, primary structure without signal sequence, UniProt P02776), the crystal structure of a PF4 tetramer (quaternary structure, PDB code 1f9q) is depicted from the front (left) and from the top view (right). Lysine residues of the C-terminus (K61, K62, K65, and K66) are highlighted in green; arginine (R20, R22, and R49), histidine (H23), and other lysine residues (K46) are highlighted in blue. The pictures of the crystal structure of PF4 were made using PyMOL Molecular Graphics System Version 1.3 Schrödinger LLC.
Discussion
In this study, we identify the negatively charged phosphate groups in lipid A as the major binding sites for PF4 on Gram-negative bacteria and further demonstrate that PF4 bound to lipid A exposes epitopes on PF4 that can be recognized by human anti-PF4/heparin antibodies. The core- and O-polysaccharide structures of LPS appear to be dispensable as they rather protect from PF4 binding, most likely because of steric hindrance of the lipid A moiety.
The lipid A part of LPS is highly conserved among Gram-negative bacteria.30 Similarly, the PF4 residues that mediate PF4-binding to polyanions are conserved throughout vertebrate species, as shown by our comparative analyses of PF4 and PF4 analogues (Figure 6). This is in accordance with our hypothesis that PF4 may label bacteria for destruction by the immune system. By exposing the same neoepitope after binding to a variety of bacteria, PF4 allows an antibody with one specificity to opsonize a large variety of bacterial species.5 We further speculate that this antibacterial defense mechanism may have developed at an early stage of evolution.
Recently, Sachais et al showed, with a monoclonal antibody (KKO), which recognizes PF4/heparin complexes, that these antibodies are able to induce clustering of PF4, even in the absence of polyanions.31 In the present study, we show that PF4 binding to bacteria can be reduced depending on the composition of the polysaccharides of LPS. Some bacteria bind PF4 only weakly and could thereby escape the PF4/polyanion antibody-mediated defense mechanism. In this regard, it could be advantageous for the infected host if high-affinity anti-PF4/polyanion antibodies recognize PF4, even on pathogens that bind PF4 only weakly and then induce clustering of PF4 to augment binding of additional antibodies. This would help to opsonize also bacteria, which bind PF4 less efficiently (eg, by altering their LPS polysaccharide chain and to resist infection).
We identified lipid A of LPS as the binding site for PF4 by exploiting different LPS mutants of S enterica sv Typhimurium, which differ in their sizes of the saccharide portion, including O-chain and core-oligosaccharides. Consecutive shortening of the LPS oligosaccharide increased the binding activity of PF4 to the LPS mutants. The mutant S enterica sv Typhimurium SL1102 (hldE543), with only 2 Kdo residues attached to lipid A, displayed highest PF4 binding capacity. To test whether the lipid A part of LPS is sufficient for PF4 binding, we performed PF4 binding studies using the waaA mutant E coli KPM121. We had to change bacteria species, as E coli KPM121 is the only bacterial strain that is viable despite predominantly expressing only the lipid A precursor lipid IVA.17 This strain showed strong PF4 binding activity, suggesting that Gram-negative bacteria sequester PF4 via lipid A of LPS. To further corroborate the validity of this experiment, we also assessed the E coli K-12 wild-type strain BW30270 and the E coli K-12 ΔwaaC mutant KPM53 for their ability to bind PF4. As the results obtained with these strains were in good agreement with those obtained with S enterica sv Typhimurium, we conclude that lipid A, as the most conserved part of LPS, is the main binding site for PF4 on Gram-negative bacteria.
Besides LPS, proteins are components of the outermost surface of Gram-negative bacteria. As these proteins can also be negatively charged,15 they may potentially contain additional binding sites for PF4. Our data on proteolytic treatment of E coli strains BW30270, KPM53, and KPM121 suggest that bacterial surface proteins do not have a major impact on binding of PF4. However, a contribution of proteins to PF4 binding cannot be excluded because we did not test whether the proteolytic treatment eliminated all surface-exposed proteins.
We further ruled out the possibility that expression of capsular polysaccharide may result in altered PF4 binding to the bacterial cells used in this study as the E coli K-12 strains do not express a capsule or the M-antigen (colanic acid) under the growth conditions used.32,33 Likewise, S enterica sv Typhimurium LT2 does not produce a capsule (K. E. Sanderson, Salmonella Genetic Stock Center, Calgary, AB, e-mail, May 11, 2012). Whether expression of a capsule affects PF4 binding needs to be investigated in future studies.
In confirmatory experiments assessing the interaction of PF4 with lipid A, we found that PF4 bound in a dose-dependent manner to immobilized lipid A but not to Kdo alone or immobilized Kdo2-lipid A or LPS. In contrast, when Kdo2-lipid A or LPS was added in excess to the fluid phase, both were able to inhibit PF4 binding to bacteria (Figure 4C). We assume that this discrepancy was probably the result of a steric hindrance. Kdo2-lipid A and LPS can move freely in the fluid phase allowing the interaction of the lipid A part with PF4, whereas this is not possible for Kdo2-lipid A or LPS immobilized on a solid phase.
Because of the strong positive charge of PF4 and the inhibitory effect of polymyxin B, the 2 negatively charged phosphate groups of lipid A are very likely responsible for the interaction with PF4. Indeed, MPLA had a lower PF4 binding capacity compared with lipid A (which is bisphosphorylated; Figure 4A) and showed a reduced capacity to inhibit PF4 binding to E coli KPM121 compared with lipid A (Figure 4C).
The lipid A moiety is the endotoxic moiety of LPS responsible for inducing endotoxic shock.34 Thus, one role of PF4 might also be to protect from endotoxic shock, as has been shown for β2-glycoprotein I.35 Lipid A also binds antimicrobial peptides, such as the bactericidal/permeability-increasing protein (BPI)36,37 and the cyclic lipopeptide polymyxin B, which interacts with the phosphate groups of lipid A.38–42 In line with our other findings, BPI and polymyxin B inhibited PF4 binding to isolated lipid A (Figure 3B), further confirming that PF4 interacts with lipid A.
After identifying lipid A as the PF4 binding site, we asked the question whether this PF4-lipid A interaction also induces PF4/heparin-like epitopes to which anti-PF4/heparin antibodies can bind. Using PF4-coated E coli K-12 ΔwaaA (KPM121) and ΔwaaC (KPM53), we affinity purified anti-PF4/heparin antibodies from human serum by an adsorption-elution technique (Figure 5A). These antibodies did not only bind specifically to immobilized PF4/heparin complexes as tested by ELISA, but also mediated platelet activation in a functional assay for clinically relevant anti-PF4/heparin antibodies. This proves that lipid A and its biosynthetic precursor lipid IVA are capable to induce neoepitopes on PF4 recognized by anti-PF4/heparin antibodies. Moreover, we show that opsonization of bacteria expressing only lipid A but lacking LPS is enhanced when they were pretreated with PF4 and anti-PF4/heparin antibodies (Figure 5B).
We identified positively charged amino acids in the heparin binding site of PF4 (or the related chemokine IL-8, which also binds heparin43 and has been reported to induce the production of antibodies in HIT patients44,45 ), to be highly conserved in all vertebrates, including fish species. The interaction of PF4 and lipid A is inhibited by heparin (supplemental Figures 1 and 2, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), suggesting that the amino acids known to contribute to heparin binding are also involved in binding to lipid A. This would be also compatible with our hypothesis of an ancient role of PF4 in an antibacterial host defense mechanism. On the other hand, one might consider as possible consequence of this “primitive mechanism of defense” that the highly variable O-antigens of LPS have significantly contributed to an efficient escape mechanism of the bacteria during evolution to evade the humoral response of the host.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank R. W. Woodard (University of Michigan, Ann Arbor, MI) for providing E coli KPM121; K. E. Sanderson (Salmonella Genetic Stock Center, Calgary, AB) for the S enterica sv Typhimurium strains; U. Dobrindt (University of Münster, Münster, Germany) for providing E coli anti-Ag43 antibodies; and N. Gisch (Forschungszentrum Borstel, Borstel, Germany) for helpful discussions about LPS chemical structures.
K.K. was supported by the Bundesministerium für Bildung und Forschung, Zentrum für Innovationskompetenz ZIK HIKE Förderkennzeichen (BMBF FKZ 03Z2CN12). This work was supported by the Deutsche Forschungsgemeinschaft (grants HA 3125/2-1 and 4-2, SFB/TRR34 project C10; S.H.).
Authorship
Contribution: K.K. designed and performed the bacterial experiments and ELISA and heparin-induced platelet activation tests, analyzed and interpreted the data, helped with the amino acid sequence survey, and wrote the manuscript; C.W. designed and performed the bacterial experiments, interpreted the data, helped with the amino acid sequence survey, and wrote the manuscript; S.B. performed the amino acid sequence survey; U.Z. purified and provided lipid A and Kdo2-lipid A and revised the manuscript; U.M. constructed the E coli LPS mutants and revised the manuscript; A.G. reviewed the results and revised the manuscript; S.H. developed the concept, reviewed the results, and revised the manuscript; and all authors contributed to the study design and the data evaluation and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sven Hammerschmidt, Ernst-Moritz-Arndt-Universität Greifswald, Interfakultäres Institut für Genetik und Funktionelle Genomforschung, Genetik der Mikroorganismen, Friedrich-Ludwig-Jahn-Strasse 15a, D-17487 Greifswald, Germany; e-mail: sven.hammerschmidt@uni-greifswald.de; and Andreas Greinacher, Ernst-Moritz-Arndt-Universität Greifswald, Institut für Immunologie und Transfusionsmedizin, Sauerbruchstraße, D-17475 Greifswald, Germany; e-mail: greinach@uni-greifswald.de.
References
Author notes
K.K. and C.W. contributed equally to this study.
A.G. and S.H. share senior authorship of this study.

![Figure 5. PF4-coated E coli LPS mutants are recognized by anti-PF4/heparin IgG from sera of patients with HIT. (A) PF4-coated E coli LPS mutants ΔwaaC (KPM53) or ΔwaaA (KPM121) were incubated with sera of 3 patients with HIT known to contain anti-PF4/heparin antibodies, and bound IgG was affinity purified. Symbols represent reactivities of IgG antibodies eluted from PF4-coated E coli KPM53 (squares) or KPM121 (triangles); means are presented as horizontal lines. Binding of the purified antibodies to PF4/heparin complexes (first column) was inhibited by excess heparin (100 IU/mL unfractionated heparin [UFH], column 2), which disrupts PF4/heparin complexes. Antibodies did not react with PF4 alone (column 3). PF4-untreated bacteria served as control for unspecific binding of the antibodies to bacteria alone (column 4). (B) E coli K12 wild-type strain BW30270 and its isogenic ΔwaaA mutant KPM121 were labeled with FITC and preincubated with PF4 and additionally with heat-inactivated human serum containing anti-PF4/heparin IgG (preadsorbed with bacteria alone). After preincubation, the bacteria were subjected to phagocytosis. The figure shows the MFI multiplied by the percentage of FITC-positive polymorphonuclear leukocytes (PMNs) as a measure for bacterial phagocytosis. Data are mean ± SD of 4 different sera.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/120/16/10.1182_blood-2012-06-434985/4/m_zh89991298070005.jpeg?Expires=1768084270&Signature=OHtAoSQp5CGKSb8lXH-0J1S5Sike-PDXEGBq1L403Ij24zJ~HKoZBi51tnyGPkUif4TQg1JrDLa9donGUfULTPIlvoMH-KRwMKKGnIpeYPr6SI6ZrCSB9QcNZv8v-Pk91POo96Bgw~C1Pma5ejZRbgmsWTasgt9xsBnCYbK-iBSQbuW02B2Y4TycnUqa28MdaKwernExEFW5wqTSW~9fZ8i7zX07Z5ySx~SR1y4JKqrzdJoRv7kKembyu6NELkMczeE8VIAI5LxTW5q2oQuQERBXTCviFZmXK1~2l4pJcajMDBzuZkgP~x9ijHRWe6zj41xMnxorX~e56pxKEQHltw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 5. PF4-coated E coli LPS mutants are recognized by anti-PF4/heparin IgG from sera of patients with HIT. (A) PF4-coated E coli LPS mutants ΔwaaC (KPM53) or ΔwaaA (KPM121) were incubated with sera of 3 patients with HIT known to contain anti-PF4/heparin antibodies, and bound IgG was affinity purified. Symbols represent reactivities of IgG antibodies eluted from PF4-coated E coli KPM53 (squares) or KPM121 (triangles); means are presented as horizontal lines. Binding of the purified antibodies to PF4/heparin complexes (first column) was inhibited by excess heparin (100 IU/mL unfractionated heparin [UFH], column 2), which disrupts PF4/heparin complexes. Antibodies did not react with PF4 alone (column 3). PF4-untreated bacteria served as control for unspecific binding of the antibodies to bacteria alone (column 4). (B) E coli K12 wild-type strain BW30270 and its isogenic ΔwaaA mutant KPM121 were labeled with FITC and preincubated with PF4 and additionally with heat-inactivated human serum containing anti-PF4/heparin IgG (preadsorbed with bacteria alone). After preincubation, the bacteria were subjected to phagocytosis. The figure shows the MFI multiplied by the percentage of FITC-positive polymorphonuclear leukocytes (PMNs) as a measure for bacterial phagocytosis. Data are mean ± SD of 4 different sera.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/120/16/10.1182_blood-2012-06-434985/4/m_zh89991298070005.jpeg?Expires=1768084271&Signature=OE7Xj4D0Ew-6ya2Kde9N~hhrqTGMOvE47VoNygG6qk0zx5PV65j9o5edOUsYLdSKS-gUAk8czsNY6uZBZym~jInUReb9p3EGoHIJJDw1L6nhdIxA57aFzrk7oDWqy7cjgaJ0Y3jORbnCWA5DbQdJ4c5vxMSIS-KbNYBE5J1I6HzluwI2VjtGAyUAJlJyeMA3P0p~yTkGk0KlBckOaK7DtKUz~HYRh4X~SM7VBJ~LqrJUdOjaqYa3xLyn-CnPsDayKPDTSnCC6pn0O2NL-mVS3F6rAQLxjgvGYWicJlkekn3SmvxBU~2OEUaihPdmegRJ1xqRmH-jvTg6GsS0FbYrBQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)