Abstract
Proviral integration site for Moloney murine leukemia virus (Pim) kinases are serine/threonine/tyrosine kinases and oncoproteins that promote tumor progression. Three isoforms of Pim kinases have been identified and are known to phosphorylate numerous substrates, with regulatory functions in transcription, translation, cell cycle, and survival pathways. These kinases are involved in production, proliferation, and survival of normal B cells and are overexpressed in B-cell malignancies such as mantle cell lymphoma (MCL). SGI-1776 is a small mol-ecule and Pim kinase inhibitor with selectivity for Pim-1. We hypothesize that Pim kinase function can be inhibited by SGI-1776 in MCL and that inhibition of phosphorylation of downstream substrates will disrupt transcriptional, translational, and cell cycle processes and promote cell death. SGI-1776 treatment in 4 MCL cell lines resulted in apoptosis induction. Phosphorylation of transcription (c-Myc) and translation targets (4E-BP1), tested in Jeko-1 and Mino, was declined. Consistent with these data, Mcl-1 and cyclin D1 protein levels were decreased. Importantly, similar to cell line data, MCL primary cells but not normal cells showed similar inhibition of substrate phosphorylation and cytotoxicity from SGI-1776 treatment. Genetic knockdown of Pim-1/Pim-2 affected similar proteins in MCL cell lines. Collectively these data demonstrate Pim kinases as therapeutic targets in MCL.
Introduction
Mantle cell lymphoma (MCL) is an aggressive lymphoma characterized by overexpression of cyclin D1 caused by t(11:14)(q13;q32).1 Although current therapeutic approaches provide a good response rate (> 90%) and progression-free survival (∼ 2.5 years), there is no effective cure for this disease.2,3 Hence, identification of novel targets and their inhibition are needed in MCL.
Proviral integration site for Moloney murine leukemia virus (Pim) kinases are oncoproteins that promote tumor progression.4 To date, 3 Pim kinases have been identified. Pim-1, -2, and -3 are highly conserved serine/threonine/tyrosine kinases that are important for normal B-lymphocyte development5 and that are overexpressed in B-cell malignancies, such as chronic lymphocytic leukemia (CLL)6 and MCL.7-9 In addition, Pim-1 and Pim-2 have been found to be highly expressed in other hematologic malignancies10 as well as in solid tumors, such as prostate cancer.11 c-Myc, also a Pim kinase substrate, has been observed to coexpress with Pim kinase in B-cell malignancies.12 In addition, elevated Pim-1 expression in MCL has been reported to induce the p53 pathway and correlate with increased expression of MDM2.13 Furthermore, Pim-1 expression is known to be highly associated with poor outcome in MCL patients.7 These observations suggest that Pim kinases could be potential therapeutic targets in MCL.
Pim kinase genes are early responders to growth factors and cytokines.14 These kinases are highly conserved throughout evolution, yet Pim-1, -2, and -3 triple-knockout mice are viable and fertile, revealing the dispensability of these proteins in crucial physiologic developmental processes.5 Pim kinases phosphorylate several substrates, including c-Myc and Histone H3 (H3) that drive the transcription process.12,15 Pim-1 phosphorylation of Histone H3 at Ser10 has been reported to be a necessary event for c-Myc–driven transcription.15 Pim-1 phosphorylates c-Myc at Ser62 to stabilize this protein.16 Notably, both Pim-1 and Pim-2 work synergistically with c-Myc, as confirmed by double-knockout studies in mice.5 The translation regulator eukaryotic elongation factor 4E-BP1 is also a substrate of Pim kinases. Pim kinases phosphorylate the priming sites Thr37/46, allowing for the hyperphosphorylation of 4E-BP1, including Ser65 phosphorylation by Pim-2, causing it to dissociate from eukaryotic initiation factor 4.17,18 Dissociation of eukaryotic initiation factor 4 contributes to the activation of cap-dependent translation.18,19 In addition, Pim-1 and Pim-2 phosphorylate Bcl-2–associated death promoter (Bad) at Ser112; this phosphorylation disrupts binding of Bad to the antiapoptotic protein B-cell lymphoma-extra large (Bcl-XL) and allows Bad to bind scaffold protein 14-3-3 to sequester its proapoptotic function, thereby activating a cell survival pathway.20,21 Furthermore, cell cycle proteins such as CDKN1B (or p27) and cell division cycle 25A/C are phosphorylated by Pim kinases and are involved in promoting proliferation.22-24 Pim kinases phosphorylate p27 (Kip1) at Thr157/198, thereby inducing binding of p27 to 14-3-3, and causing p27 to be exported from nucleus and degraded by proteasomes.24 There are several other known Pim kinase substrates such as transcriptional regulator Myb, runt-related transcription factors 1 and 3, cyclin-dependent kinase inhibitor 1 (p21), signaling transducer and suppressor of cytokine signaling 1 and 3, drug-resistant mediator ATP-binding cassette subfamily G member 2, and also p65 (REL-A) as part of NF-κB pathway.25
SGI-1776, an imidazo[1,2-b] pyridazine compound, was developed based on the known crystal structure of Pim-1, high-throughput screening, and lead optimization technique. This compound specifically binds to the ATP binding site of Pim-1 and competes with ATP binding. SGI-1776 is a potent inhibitor of all 3 Pim kinases; the IC50 values for Pim-1, -2, and -3 are 7, 363, and 69nM, respectively.6 The Kinase Profiler Assay (EMD Millipore) indicated that SGI-1776 is highly selective for Pim kinases and only inhibits Flt3 and TrkA, with nanomolar inhibitory concentrations.6 In preclinical models of human acute myeloid leukemia and CLL and prostate cancer, the efficacy of SGI-1776 has been demonstrated, either as a single agent or in combination with other chemotherapeutic agents, such as cytarabine in acute myeloid leukemia.6,10,26,27
As a novel therapeutic target, the exact pathophysiologic roles of Pim kinases in MCL are not clearly understood. Using 4 established MCL cell lines, as well as MCL primary tumor samples, we investigated the cytotoxicity and mechanism of action of SGI-1776 (a pharmacologic inhibitor of Pim kinase) and PIM small interfering RNAs (siRNAs; genetic knockdown of Pim kinases) in MCL. Our investigation establishes utility of Pim kinase inhibition for treatment of MCL and suggests that primary targets were transcription and translation.
Methods
Cell lines
The MCL cell lines JeKo-1, Mino, Granta 519, and SP-53 were provided by Dr Hesham Amin (MD Anderson Cancer Center). These cell lines express high levels of MCL signature protein markers, including cyclin D1, c-Myc, Mcl-1, Bcl-2, and Bcl-XL.28 JeKo-1, Mino, and SP-53 were maintained in RPMI 1640 medium (Corning Cellgro for JeKo-1 and SP-53, American Type Culture Collection for Mino) supplemented with 10% (JeKo-1) or 20% (Mino and SP-53) fetal bovine serum (FBS; Invitrogen). Granta 519 was maintained in Dulbecco modified Eagle medium with high glucose (Cellgro) and 20% FBS. The cells were tested routinely for Mycoplasma infection using a MycoTect kit (Invitrogen) and were authenticated using an AmpF/STR Identification kit (Applied Biosystems).
Drug
SGI-1776 was provided by SuperGen (now Astex Pharmaceuticals), and the powder was dissolved in dimethylsulfoxide (DMSO) and stored at −20°C.
Patient samples and lymphocyte isolation
Samples were obtained after informed consent from patients and healthy donors (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) through MD Anderson Cancer Center institutional review board–approved protocols and in accordance with the Declaration of Helsinki. Fresh apheresis or whole blood was collected from patients or healthy donors into heparinized tubes and diluted in phosphate-buffered saline. Mononuclear cells (PBMCs) were isolated using the standard Ficoll-Hypaque (Invitrogen) density gradient centrifugation as described previously.6 The isolated PBMCs were washed twice and then resuspended in RPMI 1640 medium (with 10% FBS) and maintained at 107 cells/mL. The concentrations were determined by a Coulter channelyzer (Beckman Coulter).
Immunohistochemistry
Immunohistochemical analysis was performed using formalin-fixed, paraffin-embedded tissue sections. We used heat-induced antigen retrieval (Dako target retrieval pH6, Dako North America), and a manual procedure was followed.29 Pim-1, Pim-2, and Pim-3 (all rabbit; Abcam) were used. Detection was performed with biotinylated secondary antibody, horseradish peroxidase, and 3,3′-diaminobenzidine as a chromogen (Abcam). Slides were counterstained with hematoxylin. Appropriate negative and positive control slides were run in parallel. Photographs were taken using an Olympus U-TV0 camera (Olympus Corporation) mounted on an Olympus BX41 microscope. Hematoxylin staining image was obtained at 100× magnification (objective Plan N 10×) while Pim kinases staining images were obtained at 400× magnification (objective Plan N 40×). Images were processed using CellSens software (Olympus).
Apoptosis and cell cycle assays
Cells were treated with DMSO or with SGI-1776. The annexin V and propidium iodide (PI) positivity for apoptosis and PI positivity for cell cycle (105 cells) were measured using an FACSCalibur flow cytometer (BD Biosciences) as described previously.6
RNA synthesis assays
Cells were incubated with DMSO or SGI-1776 and then incubated with [5,6-3H]uridine (1.0 mCi/mL; Moravek Biochemicals) for the last 30 minutes. Incorporated radioactivity was measured as described previously.6
Protein expression analysis using immunoblots
Untreated or treated cells were lysed, and their protein contents were measured as described previously.6 Total protein aliquots (30-50 μg) were loaded to gels and transferred to nitrocellulose membranes (GE Osmonics Labstore) and then probed with various antibodies as described previously.6 Primary antibodies list is provided in supplemental Table 2. Infrared-labeled secondary antibodies also were probed and visualized using an Odyssey infrared imager (LI-COR Biosciences), as described previously.6
Gene expression assays
Total RNA from MCL cells was isolated using the RNAeasy Mini kit (QIAGEN) and quantified using the NanoDrop ND1000 spectrophotometer (Thermo Fisher Scientific). Gene expression was measured with an ABI prism 7900 Sequence Detection System (Applied Biosystems) using a 1-step TaqMan RT-PCR (master mix was purchased from Applied Biosystems). TaqMan primers and probes for PIM1 (Hs01065498_m1), PIM2 (Hs00179139_m1), PIM3 (Hs00420511_g1), MCL1 (HS00172036_m1), and 18S (4333760) were purchased from Applied Biosystems. Samples were assayed in triplicate, and relative gene expression levels were normalized to endogenous control 18S.
siRNA and transfection
PIM1 and PIM2 genes were knocked down individually or in combination with human siRNA for PIM1 (S10527) and PIM2 (S21749; Ambion/Invitrogen). siRNA transfection in MCL cells was performed using Amaxa Nuceofector I device (Lonza Group) paired with Amaxa Cell Line Nucleofector kit V (Lonza Group). Negative control siRNA #1(AM4611, Ambion/Invitrogen) was used and siGLO Red Transfection Indicator (Dharmacon RNA Technologies/Thermo Fisher Scientific) was used to measure transfection efficiency.
Data and statistical analyses
All plots were created using GraphPad Prism Version 5 software, and statistical analyses were performed using the same software (GraphPad). Most data were analyzed by the Student paired, 2-tailed t test. Phosphorylated and total protein levels detected via immunoblot analysis were quantified using this software and were normalized to control. When protein phosphorylation was detected, a ratio of phosphorylated to total protein level was calculated. When only total proteins were detected, a ratio of protein to GAPDH level was determined. Vehicle DMSO was used as control.
Results
Induction of apoptosis by SGI-1776 treatment in MCL cell lines
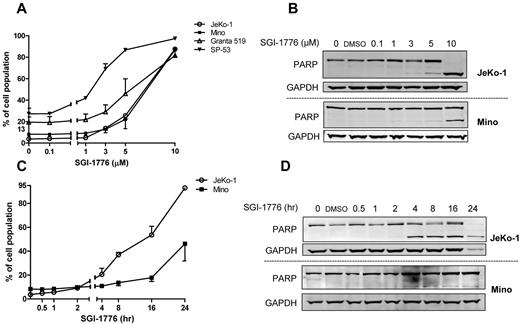
To evaluate whether SGI-1776 induces cytotoxicity, all 4 MCL cell lines were treated with DMSO or SGI-1776. Because up to 95% of the drug is bound to human plasma protein and to a lesser extent in FBS, micromolar concentrations (0.1, 1, 3, 5, and 10μM) of SGI-1776 were used; such concentrations will lead to the nanomolar levels of free drug.6 At 24 hours, there was a dose-dependent death in all 4 cell lines. All cells showed an average of 1% to 15% Annexin V positivity at 1μM, 5% to 40% at 3μM, 15% to 60% at 5μM, and 60% to 80% at 10μM SGI-1776 (Figure 1A). Consistent with these data, in both JeKo-1 and Mino, cleaved poly(ADP-ribose) polymerase (PARP) band was visible at 5μM SGI-1776, and this was stronger in JeKo-1 than in Mino cells (Figure 1B).
Induction of apoptosis by SGI-1776 treatment in MCL cell lines. (A) Dose-dependent induction of SGI-1776–mediated annexin V and PI positivity in MCL cells. MCL cell lines (JeKo-1, Mino, Granta 519, and SP-53) were treated with DMSO alone or with 0.1, 1, 3, 5, or 10μM SGI-1776 for 24 hours, stained with annexin V and PI, and analyzed by flow cytometry. (B) Apoptosis in JeKo-1 and Mino was confirmed by PARP cleavage assay. MCL cells were treated with SGI-1776 for 24 hours in the concentrations mentioned in panel A for immunoblots. (C) Time-dependent induction of SGI-1776–mediated annexin V/PI–positive MCL cells. JeKo-1 and Mino cells were treated with DMSO alone or with 10μM SGI-1776 for 0.5, 1, 2, 4, 8, 16, or 24 hours, stained with annexin V and PI, and analyzed by flow cytometry. (D) Apoptosis in JeKo-1 and Mino was confirmed by PARP cleavage assay. MCL cells were treated with 10μM SGI-1776 for time points mentioned above for immunoblots. All experiments were performed in triplicate.
Induction of apoptosis by SGI-1776 treatment in MCL cell lines. (A) Dose-dependent induction of SGI-1776–mediated annexin V and PI positivity in MCL cells. MCL cell lines (JeKo-1, Mino, Granta 519, and SP-53) were treated with DMSO alone or with 0.1, 1, 3, 5, or 10μM SGI-1776 for 24 hours, stained with annexin V and PI, and analyzed by flow cytometry. (B) Apoptosis in JeKo-1 and Mino was confirmed by PARP cleavage assay. MCL cells were treated with SGI-1776 for 24 hours in the concentrations mentioned in panel A for immunoblots. (C) Time-dependent induction of SGI-1776–mediated annexin V/PI–positive MCL cells. JeKo-1 and Mino cells were treated with DMSO alone or with 10μM SGI-1776 for 0.5, 1, 2, 4, 8, 16, or 24 hours, stained with annexin V and PI, and analyzed by flow cytometry. (D) Apoptosis in JeKo-1 and Mino was confirmed by PARP cleavage assay. MCL cells were treated with 10μM SGI-1776 for time points mentioned above for immunoblots. All experiments were performed in triplicate.
JeKo-1 and Mino cells also were treated with 10μM of SGI-1776 for various times. Cell death in JeKo-1 was 5%, 17%, 33%, 50%, and 89% at 2, 4, 8, 16, and 24 hours, respectively, whereas in Mino it was 2%, 3%, 5%, 10%, and 38% at these time points (Figure 1C). Cleaved PARP was visible at 4 and 8 hours in JeKo-1 and Mino, respectively (Figure 1D).
Expression of Pim kinases in cell lines and primary patient tissues
Expression of Pim-1, Pim-2, and Pim-3 in MCL cell lines was determined by immunoblot analysis (Figure 2A). Pim-1 and Pim-2 have isoforms of varying sizes detected as multiple bands; Pim-3 was observed mainly as 1 band. Although Pim kinase expression varied, both cell lines expressed these proteins.
Expression Pim kinase protein in cell lines and lymphoma patient tissue samples. (A) Pim-1, -2, and -3 expressions in JeKo-1 and Mino cells. Cells were either untreated or treated with DMSO, and immunoblots were prepared for Pim protein expressions. Note: multiple bands in Pim-1 and Pim-2 are expected because of multiple translation initiation sites.25 (B) Histologic section of case of MCL stained with hematoxylin and eosin, and immunohistochemical staining of MCL patient tissue samples, stained with rabbit anti–Pim-1, -2, and -3 antibodies and counterstained with hematoxylin.
Expression Pim kinase protein in cell lines and lymphoma patient tissue samples. (A) Pim-1, -2, and -3 expressions in JeKo-1 and Mino cells. Cells were either untreated or treated with DMSO, and immunoblots were prepared for Pim protein expressions. Note: multiple bands in Pim-1 and Pim-2 are expected because of multiple translation initiation sites.25 (B) Histologic section of case of MCL stained with hematoxylin and eosin, and immunohistochemical staining of MCL patient tissue samples, stained with rabbit anti–Pim-1, -2, and -3 antibodies and counterstained with hematoxylin.
A tissue array was built containing duplicate samples of 12 different MCL patient specimens (Figure 2B). Reactivity for Pim-1, Pim-2, and Pim-3 was detected in 11 (92%) cases (supplemental Table 3). Reactivity was uniform throughout individual tumor samples, but the intensity and patterns of expression differed between tumors. Expression of Pim-1 was faint to moderate and mostly nuclear and cytoplasmic, Pim-2 was strong and mostly cytoplasmic, and Pim-3 was very faint and mostly nuclear (Figure 2B).
Effect of SGI-1776 treatment on Pim kinase targets
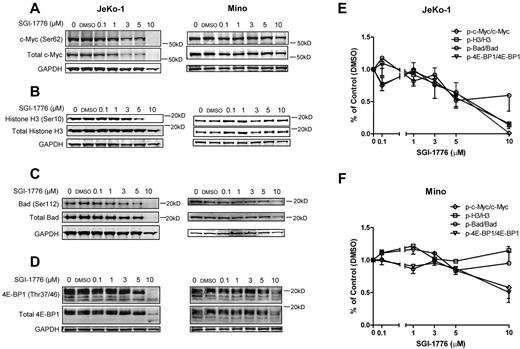
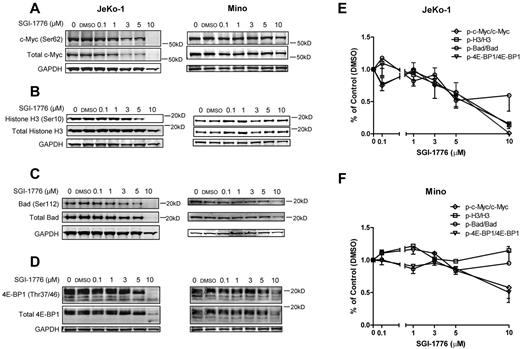
To investigate the mechanisms of action of Pim kinase inhibition, we evaluated the effect of SGI-1776 on the Pim kinase targets in JeKo-1 and Mino cell lines. In JeKo-1 cells, phosphorylation of c-Myc at Ser62 and total c-Myc, as well as phosphorylation of Histone H3 at Ser10, was decreased after treatment with SGI-1776 in a dose-dependent manner. However, in Mino cells, phospho-Histone H3 (Ser10) was not reduced after SGI-1776, suggesting that an alternative or redundant pathway of regulation might be involved (Figure 3A-B).
Effects of SGI-1776 treatment on Pim kinases targets. Effect of SGI-1776 on phospho–c-Myc (Ser62) and total c-Myc (A) phospho-Histone H3 (Ser10) and total Histone H3 (B), phospho-Bad (Ser112) and total Bad (C), and phospho–4E-BP1(Thr37/46) and total 4E-BP1 (D) protein expressions. JeKo-1 and Mino cell were treated with 0.1, 1, 3, 5, or 10μM SGI-1776 for 24 hours, and then cell lysates were analyzed via immunoblot. Quantitation of immunoblots with phosphorylation targets (c-Myc, Histone H3, Bad, and 4E-BP1) is shown in panel E for JeKo-1 cells and in panel F for Mino cells. Phospho-to-total protein ratios were calculated, and data are presented as means of 3 independent experiments ± SEM.
Effects of SGI-1776 treatment on Pim kinases targets. Effect of SGI-1776 on phospho–c-Myc (Ser62) and total c-Myc (A) phospho-Histone H3 (Ser10) and total Histone H3 (B), phospho-Bad (Ser112) and total Bad (C), and phospho–4E-BP1(Thr37/46) and total 4E-BP1 (D) protein expressions. JeKo-1 and Mino cell were treated with 0.1, 1, 3, 5, or 10μM SGI-1776 for 24 hours, and then cell lysates were analyzed via immunoblot. Quantitation of immunoblots with phosphorylation targets (c-Myc, Histone H3, Bad, and 4E-BP1) is shown in panel E for JeKo-1 cells and in panel F for Mino cells. Phospho-to-total protein ratios were calculated, and data are presented as means of 3 independent experiments ± SEM.
The level of phospho-Bad (Ser112) compared with total protein expression was measured (Figure 3C). In JeKo-1 cells, there was a decrease in phospho-Bad (Ser112) at concentrations > 1μM. Notably, total Bad protein levels also decreased at 3 and 5μM. Mino cells also showed decreased phospho- and total Bad protein levels beginning at the 3μM, and a more dramatic decrease was observed at 10μM.
The phosphorylation status of 4E-BP1 in both JeKo-1 and Mino cells was detected and the multiple bands confirmed the hyper- and multiple phosphorylation status of 4E-BP1 (Figure 3D). The phospho–4E-BP1 (Thr37/46) levels decreased after dose-dependent treatments of SGI-1776, as indicated by a decrease in the number of phosphorylation bands visible at 3μM in JeKo-1 and 5μM in Mino.
Effect of SGI-1776 treatment on global transcription and Mcl-1 expression
c-Myc and 4E-BP1 were among the substrates that were affected in both JeKo-1 and Mino cell lines. Hence, we hypothesized that RNA synthesis would be affected and short-lived mRNA and that protein expressions would decline. The global RNA synthesis levels in JeKo-1 cells were decreased to 75%, 57%, and 28% of DMSO-only–treated cells at 3, 5, and 10μM of SGI-1776 (Figure 4A). In Mino cells, there was a slight increase in global RNA synthesis at lower concentrations (up to 1μM), which declined at 3μM. At 10μM, there was a 40% decrease in RNA synthesis (Figure 4A). Granta 519 cells showed a similar pattern as JeKo-1, with a more dramatic decrease. But in SP-53 cells, a gradual reduction was observed. In time-dependent experiments, total RNA synthesis in JeKo-1 and Mino cells were reduced to 75% to 85% of control within 30 minutes, decreased to 50% to 60% after 1 hour, and then was further reduced to ∼ 40% at 24 hours (Figure 4B). Because RNA synthesis was inhibited in all 4 cell lines, the action of SGI-1776 on Mcl-1 mRNA and protein expressions in a dose- (Figure 4C) and time-dependent (Figure 4D) manner was evaluated. MCL1 mRNA levels in JeKo-1 cells decreased at 1μM and reached 35% to 38% at 3 and 5μM. Similarly, in Mino cells, although MCL1 mRNA declined in dose- and time-dependent manner; MCL1 mRNA level at 5μM was comparable to 10μM (data not shown). In JeKo-1 cells, there was a rapid decrease in mRNA expression for the first 4 hours and then expression plateaued. In Mino cells, mRNA expression levels were decreased gradually. At 16 and 24 hours, both cell lines had similar levels of MCL1 mRNA remaining.
Reduction of global RNA synthesis and Mcl-1 mRNA and protein expression in MCL cell lines by SGI-1776. (A) Dose-dependent effects of SGI-1776 treatment on global transcription level in JeKo-1, Mino, Granta 519, and SP-53 cells. Cells were treated with DMSO alone or with 0.1, 1, 3, 5, or 10μM SGI-1776 for 24 hours and then incubated with uridine for 30 minutes, and radioactive incorporation was measured via scintillation counter. (B) Time-dependent effects of SGI-1776 treatment on global transcription level in JeKo-1 and Mino cells. Cells were treated with DMSO alone or with 10μM SGI-1776 for 0.5, 1, 2, 4, 8, 16, or 24 hours and then prepared for uridine incorporation as described in panel A. Dose-dependent (C) and time-dependent (D) effects of SGI-1776 on MCL1 mRNA expressions in JeKo-1 and Mino cells. Cells were treated with SGI-1776 in dose-dependent or time-dependent manner as mentioned in Panels A and B, and RNA was isolated and analyzed using real-time RT-PCR. Impact of SGI-1776 in dose-dependent (E) and time-dependent (F) treatments on Mcl-1 protein expression in JeKo-1 and Mino cells. Mcl-1 protein expression was measured by immunoblots in both dose- and time-dependent SGI-1776 treatments (see panels A and B) of JeKo-1 and Mino cells. Quantitation of immunoblot with dose-dependent SGI-1776 treatment in JeKo-1 (G) and Mino (H) cells. Mcl-1-to-GAPDH ratio was calculated, and data are presented as means of 3 independent experiments ± SEM.
Reduction of global RNA synthesis and Mcl-1 mRNA and protein expression in MCL cell lines by SGI-1776. (A) Dose-dependent effects of SGI-1776 treatment on global transcription level in JeKo-1, Mino, Granta 519, and SP-53 cells. Cells were treated with DMSO alone or with 0.1, 1, 3, 5, or 10μM SGI-1776 for 24 hours and then incubated with uridine for 30 minutes, and radioactive incorporation was measured via scintillation counter. (B) Time-dependent effects of SGI-1776 treatment on global transcription level in JeKo-1 and Mino cells. Cells were treated with DMSO alone or with 10μM SGI-1776 for 0.5, 1, 2, 4, 8, 16, or 24 hours and then prepared for uridine incorporation as described in panel A. Dose-dependent (C) and time-dependent (D) effects of SGI-1776 on MCL1 mRNA expressions in JeKo-1 and Mino cells. Cells were treated with SGI-1776 in dose-dependent or time-dependent manner as mentioned in Panels A and B, and RNA was isolated and analyzed using real-time RT-PCR. Impact of SGI-1776 in dose-dependent (E) and time-dependent (F) treatments on Mcl-1 protein expression in JeKo-1 and Mino cells. Mcl-1 protein expression was measured by immunoblots in both dose- and time-dependent SGI-1776 treatments (see panels A and B) of JeKo-1 and Mino cells. Quantitation of immunoblot with dose-dependent SGI-1776 treatment in JeKo-1 (G) and Mino (H) cells. Mcl-1-to-GAPDH ratio was calculated, and data are presented as means of 3 independent experiments ± SEM.
Consistent with the changes in the Mcl-1 transcript levels, Mcl-1 protein levels showed a clear dose- and time-dependent decrease in JeKo-1 cells. However, Mcl-1 levels in Mino decreased only at a higher dose (10μM; Figure 4E-F). The immunoblots assays were quantitated (Figure 4G-H). In contrast to Mcl-1, other antiapoptotic proteins such as Bcl-2, Bcl-XL, and XIAP were not decreased by SGI-1776 treatment (supplemental Figure 1). These antiapoptotic proteins are not known to be Pim kinase targets.30,31
Similar to the Mcl-1 protein, another fast turnover protein, cyclin D1, also was decreased in dose- and time-dependent manner in Jeko-1, but in contrast, increased in Mino (supplemental Figure 2). To determine whether the discrepancy in cyclin D1 data were because of modulation in cell cycle profile, G1, S, and G2/M populations were quantitated in both cell lines. In Mino, but not in JeKo-1, there was an increase in G1 population (supplemental Figure 3).
Effect of PIM1 and PIM2 siRNA treatments on Pim kinase targets and Mcl-1
To confirm the selective effect of SGI-1776 on Pim kinase targets, we specifically knocked down expression of PIM1, PIM2, or both using siRNAs. For each siRNA, 150nM was used, and they were incubated with MCL cells for 24 and 48 hours. siGLO transfection indicator also was used, and it was confirmed by FACS analysis that there was high transfection efficiency in both cell lines (> 70%; data not shown).
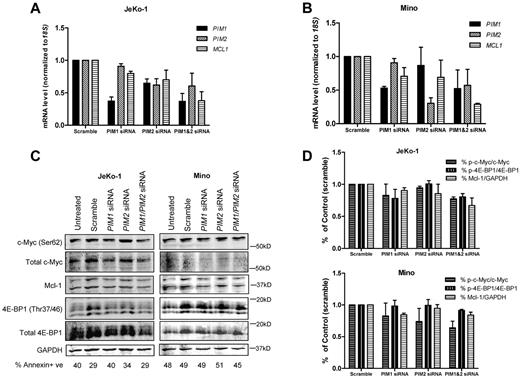
In JeKo-1 cells, PIM1 siRNA treatment alone and in combination reduced PIM1 expression to 36% of scramble, whereas PIM2 siRNA reduced the PIM2 transcript to 60% of scramble; MCL1 mRNA levels were reduced to 80% of scramble when treated with PIM1 siRNA alone and further reduced to 70% and 37% when treated with PIM2 or both PIM1 and PIM2 siRNAs, respectively (Figure 5A). Meanwhile, in Mino cells, single PIM1 and PIM2 siRNA treatments reduced respective mRNAs to 50% and 35% of scramble; but when used in combination, levels were reduced to 62% and 67% of scramble. In addition, both siRNA alone caused MCL1 transcript levels to reduce to 86% of scramble, and levels were further declined to 29% when both siRNAs were used (Figure 5B). When treated with siRNA for 48 hours, in both JeKo-1 and Mino cells, similar trend occurred, but the decrease in MCL1 was most pronounced when both siRNA were used together (supplemental Figure 4).
Effect of PIM1, PIM2 siRNAs on PIM1, PIM2 and MCL1 mRNA and Pim kinase targets in MCL cell lines. Effects of PIM1, PIM2 siRNAs and their combination on PIM1, PIM2 and MCL1 mRNA expressions in JeKo-1 (A) and Mino (B) cells. Cells were treated with 150nM scramble, siGLO, PIM1, PIM2 siRNA as well as PIM1 and PIM2 siRNA combination (150nM for each) for 24 hours. Then, RNA was extracted and analyzed using RT-PCR. (C) Effects of PIM1, PIM2 siRNA and their combination on total and phospho–c-Myc (Ser62), total and phospho–4E-BP1 (Thr37/46), as well as Mcl-1 protein expression were measured by immunoblots. (D) Quantitation of immunoblots in cells with siRNA treatments. Phospho- and total protein ratios or Mcl-1-to-GAPDH ratios were calculated, and data are presented as means of 3 independent experiments ± SEM. Percentage of cell death is marked under each lane of immunoblot measured by annexin V and PI positivity.
Effect of PIM1, PIM2 siRNAs on PIM1, PIM2 and MCL1 mRNA and Pim kinase targets in MCL cell lines. Effects of PIM1, PIM2 siRNAs and their combination on PIM1, PIM2 and MCL1 mRNA expressions in JeKo-1 (A) and Mino (B) cells. Cells were treated with 150nM scramble, siGLO, PIM1, PIM2 siRNA as well as PIM1 and PIM2 siRNA combination (150nM for each) for 24 hours. Then, RNA was extracted and analyzed using RT-PCR. (C) Effects of PIM1, PIM2 siRNA and their combination on total and phospho–c-Myc (Ser62), total and phospho–4E-BP1 (Thr37/46), as well as Mcl-1 protein expression were measured by immunoblots. (D) Quantitation of immunoblots in cells with siRNA treatments. Phospho- and total protein ratios or Mcl-1-to-GAPDH ratios were calculated, and data are presented as means of 3 independent experiments ± SEM. Percentage of cell death is marked under each lane of immunoblot measured by annexin V and PI positivity.
Next, we analyzed effects of PIM1and PIM2 siRNA on Pim kinase targets c-Myc and 4E-BP1 as well as Mcl-1 protein levels (Figure 5C). Quantitation of the immunoblots was shown as ratios of phosphorylated to total protein in c-Myc and 4E-BP1, as well as Mcl-1 protein to GAPDH levels in JeKo-1 and Mino (Figure 5D). In JeKo-1, PIM1 siRNA alone seemed to decrease the phosphorylation levels of c-Myc Ser62 and 4E-BP1 Thr37/46, but not Mcl-1. PIM2 siRNA alone did not affect the 2 phosphorylation targets of Pim kinase but reduced Mcl-1 protein level. PIM1 and PIM2 siRNA combination reduced the expression of both the phosphorylated proteins and Mcl-1 levels. In Mino cells, PIM1and PIM2 siRNA treatments alone both reduced c-Myc Ser62 phosphorylation but not 4E-BP1 Thr37/46. Only PIM1 not PIM2 siRNA treatment decreased Mcl-1 protein level. When PIM1and PIM2 siRNA were combined, c-Myc Ser62 phosphorylation and Mcl-1 proteins were decreased.
Effect of SGI-1776 treatment in samples from MCL patients and healthy donors
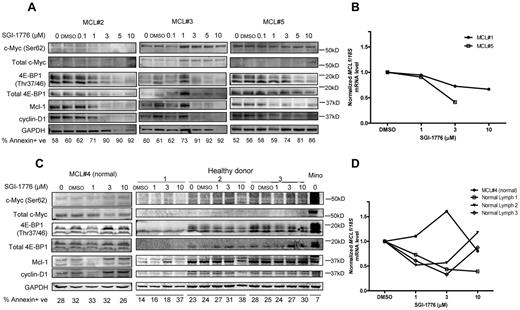
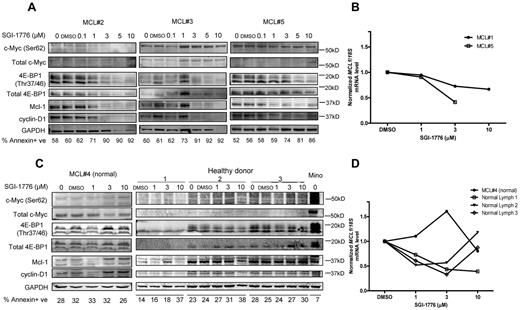
To validate the mechanisms of action of Pim kinase inhibition in MCL primary cells, we evaluated the effect of SGI-1776 on Pim kinase targets and short-lived mRNA and proteins. The 2 Pim kinase phosphorylation targets c-Myc and 4E-BP1 were selected because they were consistently affected by SGI-1776 treatment in MCL cell lines. Results in MCL #2 and #5 indicated clear dose-dependent decrease of phospho–c-Myc (Ser62) levels, but not in MCL #3, whereas total c-Myc levels were not strongly detected in these primary cells therefore results are not conclusive. In all 3 samples, both phospho- (Thr37/46) and total 4E-BP1 levels were decreased with SGI-1776 treatment. Both Mcl-1 and cyclin D1 levels showed significant decrease at low levels of SGI-1776 (1-3 μM), and their expression continued to decrease with higher concentrations of drug treatment (Figure 6A).
Effects of SGI-1776 on MCL malignant lymphocytes and normal PBMCs. Effects of SGI-1776 on phospho–c-Myc (Ser62) and total c-Myc, Mcl-1, cyclin D1 and phospho–4E-BP1 (Thr37/46), and total 4E-BP1 protein expressions in MCL malignant lymphocytes (A) and normal PBMCs from MCL patients and apparently healthy donors (C). Primary patient cells were treated with 0.1, 1, 3, 5, or 10μM, and normal PBMCs were treated with 1, 3, or 10μM SGI-1776 for 24 hours, and then cell lysates were analyzed via immunoblot. Untreated Mino cells were positive control. Apoptosis levels measured by annexin V and PI positivity is marked under each lane of the immunoblot in each primary sample. Effects of SGI-1776 on MCL1 mRNA expression levels in primary MCL malignant lymphocytes (B) and normal PBMCs from MCL patients and healthy donors (D). Primary PBMCs were treated with SGI-1776 using concentrations mentioned in this legend; RNA was isolated and analyzed using real-time RT-PCR.
Effects of SGI-1776 on MCL malignant lymphocytes and normal PBMCs. Effects of SGI-1776 on phospho–c-Myc (Ser62) and total c-Myc, Mcl-1, cyclin D1 and phospho–4E-BP1 (Thr37/46), and total 4E-BP1 protein expressions in MCL malignant lymphocytes (A) and normal PBMCs from MCL patients and apparently healthy donors (C). Primary patient cells were treated with 0.1, 1, 3, 5, or 10μM, and normal PBMCs were treated with 1, 3, or 10μM SGI-1776 for 24 hours, and then cell lysates were analyzed via immunoblot. Untreated Mino cells were positive control. Apoptosis levels measured by annexin V and PI positivity is marked under each lane of the immunoblot in each primary sample. Effects of SGI-1776 on MCL1 mRNA expression levels in primary MCL malignant lymphocytes (B) and normal PBMCs from MCL patients and healthy donors (D). Primary PBMCs were treated with SGI-1776 using concentrations mentioned in this legend; RNA was isolated and analyzed using real-time RT-PCR.
Consistent with the effect of SGI-1776, these primary cells showed an increase in annexin-positive cells (Figure 6A). In all 3 samples, there was increase of cell death by at least 20% at 3μM SGI-1776, and > 30% of cell death was observed at 5 and 10μM. In MCL #5, we measured the effect of SGI-1776 on global RNA synthesis by uridine incorporation, and there was a very clear dose-dependent decrease of RNA synthesis starting at 1μM (data not shown).
To determine whether the effect of SGI-1776 on Mcl-1 was also at mRNA levels, transcript levels were measured in 2 MCL primary samples, MCL #1 and MCL #5 (Figure 6B). Both samples showed a dose-dependent decrease of MCL1 transcript levels. At 3μM, MCL1 mRNA level was reduced to 73% and 41% of control for MCL #1 and MCL #5, respectively; and at 10μM, the mRNA level was further reduced to 67% for MCL #1.
One patient with MCL, MCL #4, had localized disease with normal circulating mononuclear cells in peripheral blood. In addition, we obtained whole blood samples from 3 healthy donors. Phosphorylation targets and short-lived proteins Mcl-1 and cyclin D1 in these normal PBMCs were analyzed (Figure 6C). Untreated Mino cells were used as positive control. In contrast to the MCL malignant cells, phosphorylation targets or short-lived proteins did not show consistent decrease with SGI-1776 treatment in normal PBMCs. For the MCL1 transcript levels, in the same samples there was a decrease of the mRNA but not protein (Figure 6D). Also in contrast to malignant cells, apoptosis was not induced in normal PBMCs after SGI-1776 treatment.
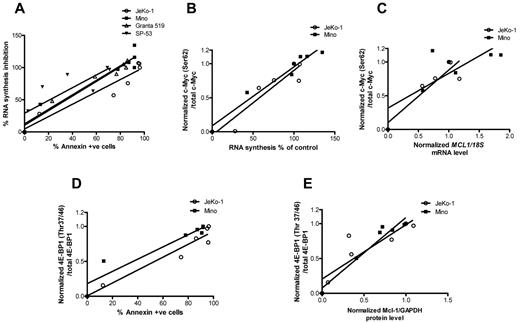
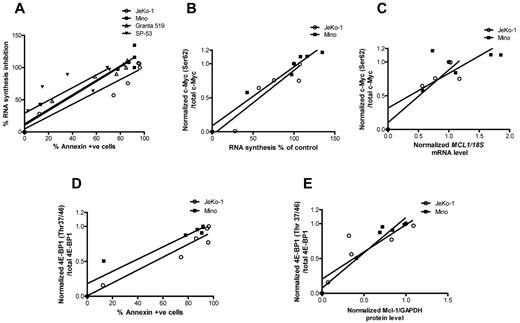
Association between transcription and translation inhibition and cell death
To identify parameters associated with SGI-1776–induced cell death, we correlated transcription and protein translation with cell death. At first, we tried to establish the association between inhibition of global RNA synthesis and apoptosis induced by SGI-1776 treatments in all 4 MCL cell lines (Figure 7A). The linear regression value was > 0.9 for 3 cell lines and 0.68 for SP-53. Collectively for all 4 cell lines, RNA synthesis decrease was directly associated with increase of apoptosis levels (r2 = 0.8, P < .0001). Because c-Myc was consistently affected in MCL cell lines, we evaluated the associations among transcription inhibition and SGI-1776–induced changes of c-Myc phosphorylation. A strong association (Figure 7B) was observed between c-Myc phosphorylation at Ser62 and RNA synthesis (r2 values for either JeKo-1 or Mino, or both cell lines combined were > 0.9; P < .02 for all). c-Myc phosphorylation status also was directly associated with a decline in MCL1 mRNA levels (r2 = 0.8 in JeKo-1, 0.6 in Mino and 0.7 in 2 cell lines together; P < .03 for all; Figure 7C). Ratios of phospho–4E-BP1 (Thr37/46) to total 4E-BP1 protein were correlated with the apoptosis levels in JeKo-1 and Mino (Figure 7D). Linear regression values for each cell line and 2 cell lines combined were ≥ 0.9 (P < .01). Consistent with these data, Mcl-1 protein levels were strongly associated with decline in 4E-BP1 phosphorylation (r2 > 0.8 for each cell line, P = .09 for JeKo-1, and < 0.004 for Mino; Figure 7E).
Association between transcription and translation inhibition and cell death. (A) Global RNA synthesis inhibition versus apoptosis levels and its linear regression. RNA synthesis was measured in percentages and apoptosis was defined by annexin V positivity. Values were taken from dose-dependent SGI-1776 treatments in JeKo-1, Mino, SP-53, and Granta-519 cell lines. In experiments performed in JeKo-1 and Mino after dose-dependent SGI-1776 treatments for 24 hours, the following plots were generated: c-Myc (Ser62) phosphorylation versus global RNA synthesis, where c-Myc (Ser62) levels were normalized to total c-Myc to obtain the ratios (B); c-Myc (Ser62) phosphorylation versus MCL1 mRNA expressions, where c-Myc (Ser62) levels were normalized to total c-Myc and MCL1 mRNA was normalized to 18S rRNA controls to obtain the ratios (C); inhibition of 4E-BP1 (Thr37/46) phosphorylation versus apoptosis, where 4E-BP1 (Thr37/46) phosphorylation levels were normalized to total 4E-BP1 to obtain the ratios (D); and 4E-BP1 (Thr37/46) phosphorylation versus Mcl-1 protein expression, where phospho–4E-BP1 (Thr37/46) was normalized to total 4E-BP1 protein and Mcl-1 protein was normalized to GAPDH levels (E). Linear regression was performed, and r2 values and P values were calculated using the GraphPad Prism Version 5 software.
Association between transcription and translation inhibition and cell death. (A) Global RNA synthesis inhibition versus apoptosis levels and its linear regression. RNA synthesis was measured in percentages and apoptosis was defined by annexin V positivity. Values were taken from dose-dependent SGI-1776 treatments in JeKo-1, Mino, SP-53, and Granta-519 cell lines. In experiments performed in JeKo-1 and Mino after dose-dependent SGI-1776 treatments for 24 hours, the following plots were generated: c-Myc (Ser62) phosphorylation versus global RNA synthesis, where c-Myc (Ser62) levels were normalized to total c-Myc to obtain the ratios (B); c-Myc (Ser62) phosphorylation versus MCL1 mRNA expressions, where c-Myc (Ser62) levels were normalized to total c-Myc and MCL1 mRNA was normalized to 18S rRNA controls to obtain the ratios (C); inhibition of 4E-BP1 (Thr37/46) phosphorylation versus apoptosis, where 4E-BP1 (Thr37/46) phosphorylation levels were normalized to total 4E-BP1 to obtain the ratios (D); and 4E-BP1 (Thr37/46) phosphorylation versus Mcl-1 protein expression, where phospho–4E-BP1 (Thr37/46) was normalized to total 4E-BP1 protein and Mcl-1 protein was normalized to GAPDH levels (E). Linear regression was performed, and r2 values and P values were calculated using the GraphPad Prism Version 5 software.
Discussion
Our results indicate that Pim kinases are expressed in MCL cell lines as well as in primary MCL cells (Figure 2) and are potential drug targets, because inhibition of these kinases caused cytotoxicity in MCL cells in vitro (Figure 1). This biologic response is related to inhibition of phosphorylation of the transcription regulator c-Myc and translational controller 4E-BP1 (Figures 3, 5, and 6), attenuation of global RNA synthesis, and short-lived Mcl-1 and cyclin D1 expression in cell lines and primary MCL cells (Figures 4 and 6; supplemental Figure 2).
In cell lines, as well as MCL primary samples, c-Myc (Ser62) phosphorylation was consistently reduced after SGI-1776 treatment (Figures 3A and 6A). Notably, levels of cell death are inversely correlated with c-Myc (Ser62) phosphorylation levels in MCL cell lines (Figure 7) and primary samples. Accordingly, reduction of c-Myc phosphorylation is strongly associated with inhibition of global RNA synthesis and reduction of MCL1 mRNA levels (Figure 7B-C). This observation suggests that c-Myc–driven transcription is an important regulator for Pim kinases in this disease context. Phosphorylation of c-Myc at Ser62 stabilizes c-Myc protein by prolonging its half-life from minutes to hours, and both Pim-1 and Pim-2 phosphorylate this target site.16,32 Our results are in accord with these findings and for a strong and direct relationship between the c-Myc phosphorylation status and either inhibition of RNA synthesis or levels of MCL1 mRNA (Figure 7B-C).
To cooperate with c-Myc–driven transcription, phosphorylation of Histone H3 (Ser10) by Pim-1 has been demonstrated to be a critical step.15 Our data suggest a dose-dependent decline in H3Ser10 phosphorylation with SGI-1776 treatment in the JeKo-1 but not in Mino (Figure 3B). We speculate that the H3Ser10 phosphorylation was either cell type specific, or the altered experimental conditions such as serum starvation would have affected the intracellular localization patterns of Pim kinases.15 Nonetheless, in our model system, the role of H3 phosphorylation in Myc/Max mediated transcription activation may not be as important as direct phosphorylation of c-Myc and c-Myc–driven transcription.
4E-BP1 phosphorylation at its priming site Thr37/46, a Pim-1 target, was consistently reduced in both cell lines and primary samples after SGI-1776 treatment. After this priming event, Pim-2 can further phosphorylate 4E-BP1 at Ser65, and hyperphosphorylated 4E-BP1 dissociates from eukaryotic initiation factor 4 to activate cap-dependent translational.17,18,33 Early response genes such as cyclin D1, c-Myc, Mcl-1, and Pim kinases require cap-dependent protein translation.31,34,35 Hence, impact on 4E-BP1 phosphorylation may contribute to the expression levels of these 4 short-lived proteins, all of which play essential roles in MCL.31,34,35 In accord with these reports, there was strong and significant association between changes in phosphorylation of 4E-BP1 and either Mcl-1 protein levels or cell death (Figure 7D-E). In hematologic malignancies, cap-dependent translation is regulated independently by Pim-1 and Pim-2 kinases, particularly Pim-2, as well as the PI3K/Akt/mTOR pathway, and it has been shown that inhibition of cap-dependent translation contributes substantially to therapy-induced cell death in these models.18,33,35,36 Given that Pim kinases, and especially Pim-2, are expressed at high levels, and mTOR pathway is constitutively active in MCL,33 cap-dependent translation may be a potential therapeutic target for treating MCL patients, and that Pim kinase and AKT/mammalian target of rapamycin (mTOR) inhibitors could be used in combination.35
Mcl-1 mRNA and protein are expressed at high level in MCL cell lines and primary samples (Figures 4C-H and 6A-B). As an important antiapoptotic protein in the Bcl-2 family, Mcl-1 has been reported to be overexpressed in many B-cell malignancies, and both the gene and protein products are short lived, making them sensitive to drug-induced changes.37,38 Because Pim kinases target both transcription and translation, and c-Myc and PI3K/Akt pathways up-regulate Mcl-1, it can be indirectly regulated by Pim kinases. Mcl-1 has been known to be stabilized by Pim kinases via c-Myc pathways.6,38 After SGI-1776 treatment, Mcl-1 mRNA and protein expression were reduced in both cell lines (Figure 4C-H). Genetic knowdown of PIM1 and PIM2 also resulted in MCL1 mRNA decrease (Figure 5A-B), further establishing Pim-mediated action of SGI-1776. In contrast with Mcl-1, other antiapoptotic proteins, such as Bcl-2 and Bcl-XL, were not affected by SGI-1776 treatment (supplemental Figure 1). These results imply that Mcl-1 is a prominent antiapoptotic protein in the Bcl-2 family that is regulated by Pim kinases, and down-regulation of Mcl-1 by SGI-1776 may contribute to the cell death observed in MCL cell lines and primary samples (Figures 1 and 6A).
In addition to Mcl-1, overexpression of another rapid-turnover protein, cyclin D1, has been identified as a hallmark of the disease39,40 and is subjected to transcriptional regulation via multiple factors, including STAT3 and STAT5.41,42 Because STAT3 is also a target of Pim kinases, SGI-1776 may have triggered down-regulation of cyclin D1 gene expression and thereafter protein expression.43 JeKo-1 and MCL primary samples express high levels of cyclin D1, and this expression was readily reduced after SGI-1776 treatment (Figure 6A; supplemental Figure 2). As an important oncogene in MCL, cyclin D1 degradation likely contributes to therapy-induced cell death in MCL.
Results from the Pim kinase phosphorylation targets showed cell line–specific effects with SGI-1776 treatment; however, extensive cell death was observed in all 4 MCL cell lines and primary patient samples (Figures 1A and 6A). We propose that c-Myc–driven transcription and cap-dependent translation are likely to be the main survival factors regulated by Pim kinases; and as a result, SGI-1776–induced degradation of short-lived oncogenic proteins such as Mcl-1 and cyclin D1 may have expedited this process of cell death.16,35,38,42 Although these actions were observed in cell lines and MCL primary tumor cells, normal PBMCs from these patients did not show such attenuation in Mcl-1 after treatment with SGI-1776; accordingly, there was no induction of apoptosis in these healthy cells (Figure 6).
Although our data suggest a role of c-Myc in response to SGI-1776 in 4 MCL cell lines and primary MCL cells (Figures 3A,E-F and 6A), additional factors, not investigated in the present study, may play a role in drug-induced cytotoxicity. Notably, growth factors such as B-cell receptor/B-lymphocyte stimulator, other B-cell kinases such as spleen tyrosine kinase, or transcription factors such as NF-κB and/or PI3K/Akt-regulated pathways, all of which were found to be activated in MCL and are known to positively regulate cyclin D1 and Mcl-1, may be involved.44-47 Pim kinases, particularly Pim-2, are known to be downstream of B-lymphocyte stimulator and NF-κB, and are both regulator and target of STAT3.48-50 All of these pathways interconnect, and it is reasonable to postulate potential overlap and interactions between Pim kinases and these growth or transcription factors.
In summary, our investigation demonstrates that Pim kinases are expressed in MCL cell lines as well as primary cells from MCL patients. These multisubstrate kinases are potential drug targets in MCL. Because of these multimodality actions, Pim kinases inhibitors could be attractive targets for combinations. In fact, preliminary data with bendamustine (an approved agent for MCL) suggested additive or synergistic cell kill in MCL cell lines (data not shown). Among various signaling pathways downstream of Pim kinase inhibition, transcription and translation seem to be predominantly affected in MCL cell lines as well as primary samples. Finally, decline in the protein levels of Mcl-1 and cyclin D1 seems to be a primary correlate with cell death. Importantly, none of these actions of SGI-1776 were observed in normal PB mononuclear cells, providing a therapeutic index for this kinase inhibition.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Rakesh Sharma, Sanat Dave, and Yuling Chen for obtaining and processing patient samples.
This work is supported by Lymphoma Specialized Program of Research Excellence (SPORE) grant CA136411 (V.G., L.J.M., S.S.N.) and a CLL Alliance/Global Research Foundation grant (V.G.). The primary tumor samples were provided by The University of Texas MD Anderson Cancer Center Lymphoma SPORE Biospecimens Core and Tissue Bank, supported by Cancer Center support grant CA16672.
National Institutes of Health
Authorship
Contribution: Q.Y. performed experiments, analyzed data, and wrote the manuscript; L.S.C. designed research project, analyzed data, and reviewed the manuscript; V.G. supervised research project, analyzed data, obtained funding, and reviewed manuscript; L.J.M. and R.N.M. performed and analyzed immunohistochemistry studies and reviewed the manuscript; and S.S.N. identified patients and provided primary MCL cells.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Varsha Gandhi, Department of Experimental Therapeutics, The University of Texas MD Anderson Cancer Center, Unit 1950, PO Box 301429, Houston, TX 77230-1429; e-mail: vgandhi@mdanderson.org.