Abstract
Chronic lymphocytic leukemia is characterized by relapse after treatment and chemotherapy resistance. Similarly, in other malignancies leukemia cells accumulate mutations during growth, forming heterogeneous cell populations that are subject to Darwinian selection and may respond differentially to treatment. There is therefore a clinical need to monitor changes in the subclonal composition of cancers during disease progression. Here, we use whole-genome sequencing to track subclonal heterogeneity in 3 chronic lymphocytic leukemia patients subjected to repeated cycles of therapy. We reveal different somatic mutation profiles in each patient and use these to establish probable hierarchical patterns of subclonal evolution, to identify subclones that decline or expand over time, and to detect founder mutations. We show that clonal evolution patterns are heterogeneous in individual patients. We conclude that genome sequencing is a powerful and sensitive approach to monitor disease progression repeatedly at the molecular level. If applied to future clinical trials, this approach might eventually influence treatment strategies as a tool to individualize and direct cancer treatment.
Introduction
Despite significant progress in the management of lymphomas and leukemias, relapse remains the major cause of death. Increased use of expensive targeted therapies and toxic chemotherapies (especially in the elderly) confronts us with an urgent need to improve response prediction for all cancer patients to reduce side effects and costs from ineffective treatment. Current diagnostic approaches to treatment selection, response monitoring, and relapse prediction are limited to single genes and apply only to a minority of hematologic cancers. This is at odds with modern concepts of tumor propagation and maintenance, which propose that every cell in an individual cancer is characterized by a combination of mutation events that comprise tumorigenic (driver) mutations, passive (passenger) mutations, and possibly predisposing germ-line risk variants. Cancer cells propagate and diversify during tumor growth, resulting in a heterogeneous population of genotypically and phenotypically distinct subclones that are related in a hierarchical lineage. As the composition of the local environment changes, for example as a consequence of drug treatment, tumor cell populations adapt and evolve by Darwinian selection.1-3
Whole-genome sequencing (WGS) of a single tumor sample can be used to generate a comprehensive catalog of variants that provides a snapshot of the cell population en masse at a particular time point.2,4-6 However, over time and with continued evolution of the cancer, this snapshot becomes progressively less representative of the disease. Recent reports have described whole-tumor genomes from single patients or cohorts of individuals mostly at single time points and irrespective of treatment.7-10 This approach has enabled identification of mutations representative and in some cases highly predictive of histologic cancer type, outcome, and/or treatment response.11-15 Comparison of sequence data from primary and metastatic tumor samples, or from multiple locations within a tumor, reveals major differences in the somatic mutation profiles within an individual, illustrating the dynamic nature of tumor evolution.16-18 Recently, 2 time point analyses of relapsed19 and secondary20 acute myeloid leukemia have also demonstrated clonal evolution at a molecular level.
We elected to study subclonal evolution in B-cell chronic lymphocytic leukemia (CLL). CLL is characterized by immunodeficiency, autoimmunity, a chronically relapsing course, and the development of chemotherapy resistance, making it an ideal model to study tumor progression. Using WGS analysis, we tracked molecular changes in pretreatment, posttreatment, and relapse samples in 3 patients. We defined cellular subpopulations on the basis of somatic mutation profiles and revealed changes within the tumor clonal architecture over time as patients were subjected to multiple rounds of treatment. We describe for the first time the heterogeneous patterns of clonal evolution in patients with IgHV unmutated CLL throughout the lifetime of their disease. This proof-of-principle study enabled us to evaluate how large-scale sequence information might be used in future clinical trials to evaluate response and to target therapies more effectively for patients suffering from CLL and perhaps other cancers.
Methods
Samples
Informed consent from CLL patients was obtained in line with the Declaration of Helsinki and in line with the Oxford IRB ethics 09/H0606/5. DNA and RNA were extracted from peripheral blood CLL lymphocytes and control DNA from buccal smear samples.
WGS
Application of sequencing by synthesis (SBS) to human WGS was previously described.21 Cluster formation and sequencing to an average depth of 40× using were carried out the Illumina TruSeq Version 3 Cluster and SBS kits, respectively. Paired-end sequence reads (100 bp) were generated using HiSeq 2000.
Somatic mutation calling and analysis
The Illumina CASAVA Version 1.8 pipeline was used for quality control, alignment to human GRCh37.1 reference and variant calling for single genome analysis. Identification of somatic mutations was performed with prediction software that uses a joint Bayesian model combining analysis of the tumor and normal genomes. Annotation of mutation consequence was performed using Ensembl Variant Effect Predictor on Ensembl database release e62.22 Mutant allele frequencies (AFs) and consecutive AF differences were calculated at each tumor stage for base substitutions. Mutations were clustered into groups showing similar mutation AF profiles using a k-means algorithm.
Targeted deep amplicon sequencing
Selected somatic substitution sites in protein-coding genes were amplified from genomic DNA by 2-step PCR. Partial adapter sequences were added as 5′ extensions to target-specific primers and used as priming sites in the second PCR step to complete the adapter sequences required for cluster generation and SBS. Amplicons were sequenced using the Genome Analyzer IIx to an average depth of 100 000×.
See supplemental Methods for further details and for a list of all primer sequences (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Results
Somatic mutation detection at 5 time points in each patient's disease progression
We selected 3 patients with CLL who received multiple different treatments sequentially for a period of up to 7 years (supplemental Table 1 and supplemental Methods). We took peripheral blood samples at 5 specific time points during disease progression together with 1 matched buccal swab per patient (supplemental Table 2), and performed WGS and mutation analysis (supplemental Tables 3-5). Genome-wide somatic mutations were in the range of 1744 to 2829 substitutions and 204 to 385 insertions/deletions per sample (supplemental Tables 4-5). There was a clear bias toward C > T / G > A substitutions (supplemental Figure 1) as seen previously in other cancers.2,5 C > T substitutions have been previously linked to UV light exposure in skin cancers23 and recently to specific sequence signatures (eg, TpCpX) in breast cancer.24
Between 14 and 22 mutations per sample are predicted to alter protein-coding sequences (supplemental Tables 4-8). WGS analysis confirmed copy number aberrations (CNAs) seen in a previous array-based analysis and revealed additional CNAs (supplemental Figure 2, supplemental Table 9). CLL003 had 3 large CNAs: del11q23.2 and del13q14.1 remained unchanged over time and were detected at all time points); loss/gain of 8p/8q was first seen at first relapse (time point b) in a small subclone that subsequently expanded. CLL077 developed a deletion of chromosome 6q first observed before ofatumumab treatment (time point d). CLL006 had trisomy 12 at all time points (data not shown).
Mutation frequency profiles differ between patients and change over time
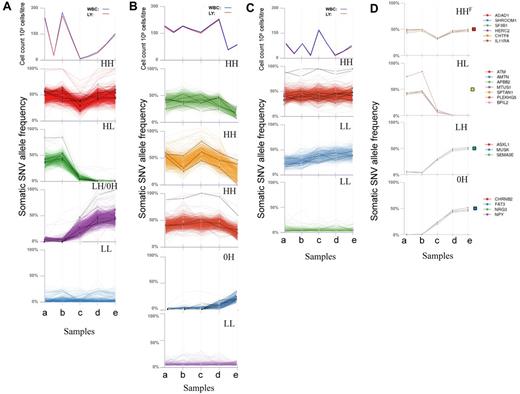
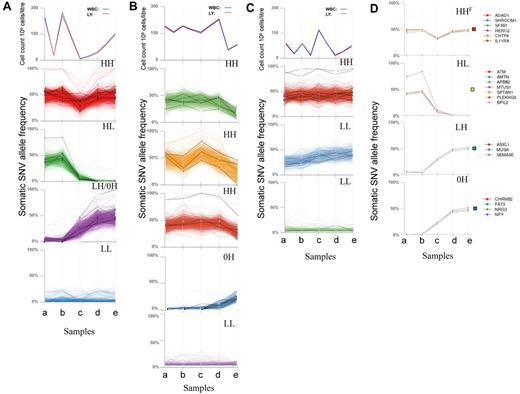
We determined allele frequencies of all somatic single nucleotide variants (SNVs) at each disease stage, established a profile for each mutation during disease progression and grouped similar mutation profiles together. This revealed changes in mutation profiles over time and clear differences between patients (Figure 1A-C). The most dynamic profiles were seen in CLL003. Mutation profiles of CLL077 were relatively stable initially and then underwent a change at later stages. Finally, CLL006 mutation profiles remained relatively stable throughout.
Genome-wide clustering reveals changes in mutation profiles. (A-C) Grouping of somatic mutation profiles for all single nucleotide variants (SNVs). Absolute white blood cell (WBC) and lymphocyte (LY) counts are shown at the top of each figure. The bottom panels show genome-wide SNV frequencies plotted against the 5 time points. Mutation profiles for coding genes are shown as black lines. (A) CLL003; a indicates before chlorambucil; b, before fludarabine, cyclophosphamide, rituximab; c, immediately after 6 cycles of fludarabine, cyclophosphamide, rituximab; d, before ofatumumab; and e, after ofatumumab. Coding mutations: red plot SLC9A11, NLRP3, SF3B1, ADAD1, IL11RA, TRIM58, HERC2, RPGRIP1, MUC16, SHROOM1; green plot ATM, PLEKHG5, NFATC1, FCGBP, BPIL2, AMTN, MTUS1, SPTAN1; purple plot SLITRK4, SEMA3E, ASXL1, MUSK, NPY, CHRNB2, ZNF534, FAT3; and blue plot: noncoding mutations only. (B) CLL077; a indicates before chlorambucil; b, before fludarabine, cyclophosphamide; c, immediately after 4 cycles of fludarabine, cyclophosphamide; d, before ofatumumab; and e, relapse 9 months after ofatumumab. Coding mutations: green plot: OCA2, SLC12A1, PLA2G16, DAZAP1, EXOC6B, LRRC16A; orange plot: NAMPTL, BCL2L13, GHDC; red plot: SAMHD1, IRF2BP2, GPR158; blue plot: MAP2K1, ZFHX4, HMCN1, DDX1, KLHDC2, NOD1, ZNF566, COL24A1; and purple plot: noncoding mutations only. (C) CLL006; a indicates before fludarabine, cyclophosphamide; b, before Rituximab; c, before Ofatmumab; d, immediately after Ofatumumab; and e, relapse 12 months after Ofatumumab. Coding mutations: red plot: MED12, KLHL4, CNOT7, SLK1, U2AF1, C3orf43, PILRB, ARHGAP29, KIAA0182, MAP4, TMPRSS9; blue plot: PCLO, IRF4, LRRC37B, KIAA0319L; and green plot: RBPJ. (D) Mutation profiles based on deep sequencing in patient CLL003. Colored boxes to the right of each plot indicate mutation profile type (HHF indicates red box, HL, yellow box, LH, green box, and 0H, blue box). See supplemental Table 5.
Genome-wide clustering reveals changes in mutation profiles. (A-C) Grouping of somatic mutation profiles for all single nucleotide variants (SNVs). Absolute white blood cell (WBC) and lymphocyte (LY) counts are shown at the top of each figure. The bottom panels show genome-wide SNV frequencies plotted against the 5 time points. Mutation profiles for coding genes are shown as black lines. (A) CLL003; a indicates before chlorambucil; b, before fludarabine, cyclophosphamide, rituximab; c, immediately after 6 cycles of fludarabine, cyclophosphamide, rituximab; d, before ofatumumab; and e, after ofatumumab. Coding mutations: red plot SLC9A11, NLRP3, SF3B1, ADAD1, IL11RA, TRIM58, HERC2, RPGRIP1, MUC16, SHROOM1; green plot ATM, PLEKHG5, NFATC1, FCGBP, BPIL2, AMTN, MTUS1, SPTAN1; purple plot SLITRK4, SEMA3E, ASXL1, MUSK, NPY, CHRNB2, ZNF534, FAT3; and blue plot: noncoding mutations only. (B) CLL077; a indicates before chlorambucil; b, before fludarabine, cyclophosphamide; c, immediately after 4 cycles of fludarabine, cyclophosphamide; d, before ofatumumab; and e, relapse 9 months after ofatumumab. Coding mutations: green plot: OCA2, SLC12A1, PLA2G16, DAZAP1, EXOC6B, LRRC16A; orange plot: NAMPTL, BCL2L13, GHDC; red plot: SAMHD1, IRF2BP2, GPR158; blue plot: MAP2K1, ZFHX4, HMCN1, DDX1, KLHDC2, NOD1, ZNF566, COL24A1; and purple plot: noncoding mutations only. (C) CLL006; a indicates before fludarabine, cyclophosphamide; b, before Rituximab; c, before Ofatmumab; d, immediately after Ofatumumab; and e, relapse 12 months after Ofatumumab. Coding mutations: red plot: MED12, KLHL4, CNOT7, SLK1, U2AF1, C3orf43, PILRB, ARHGAP29, KIAA0182, MAP4, TMPRSS9; blue plot: PCLO, IRF4, LRRC37B, KIAA0319L; and green plot: RBPJ. (D) Mutation profiles based on deep sequencing in patient CLL003. Colored boxes to the right of each plot indicate mutation profile type (HHF indicates red box, HL, yellow box, LH, green box, and 0H, blue box). See supplemental Table 5.
To extend the sensitivity of the study we selected specific somatic mutations from each profile, focusing on those predicted to alter protein structure. We performed targeted deep sequencing to an average depth of 100 000× to quantify the mutation frequency at each stage to high accuracy and to observe low levels of somatic mutations (down to ∼ 0.5%) previously undetected by WGS (Figure 1D, supplemental Figure 3, supplemental Tables 5-7). All mutations selected were confirmed by deep sequencing (see supplemental Methods). The quantitative analysis revealed a striking similarity in frequency profiles for different mutations in the same group (Figure 1D, supplemental Figure 3). Considering the deep sequence and WGS data we defined 5 mutation profiles: (1) high (H) frequency at initial diagnosis and later (HH); (2) high at diagnosis then low (L) or disappearing after treatment (HL); (3) initially at low frequency but then increasing (LH); (4) undetectable by deep sequencing at diagnosis (0H); and (5) present at low frequency throughout (LL). All 5 profiles are evident in CLL003, but only HH, 0H, and LL profiles are present in CLL077, whereas CLL006 is characterized exclusively by HH and LL profiles (Figure 1, supplemental Figure 3).
Defining leukemia architecture and founder mutations
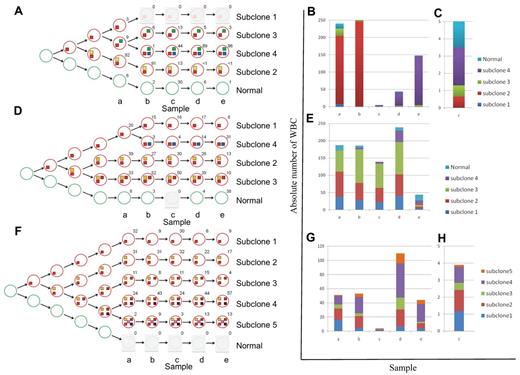
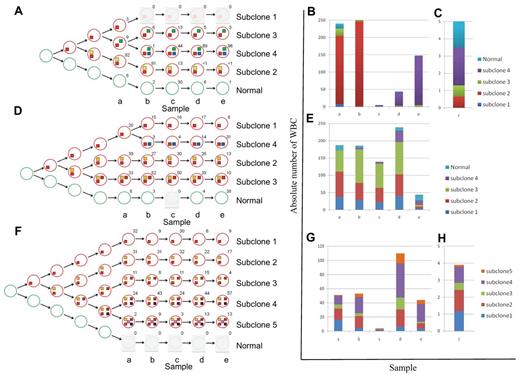
We used the deep-sequencing data to define tumor subclones and to infer an evolving and branching cellular hierarchy of tumor cells for the 3 patients (Figure 2). This analysis enabled us to define a founder subclone in each patient who was genetically characterized by mutations present in all tumor cells at all time points (mutation profile HHF in Figures 1D and 2, and supplemental Figure 3). Mutations of this type should include the initial drivers of tumorigenesis, as well as passenger mutations that were fixed in the originating tumor cell. Additional subclone diversity was because of other mutations (HL, LH, 0H, and HH profiles in Figures 1D and 2, and supplemental Figure 3) that arose on the background of the founder mutations.
Schematic presentation of the changes in subclonal architecture over time. (A,D,F) Schematic representation of the subclonal hierarchy for patients CLL003 (A), CLL077 (D), and CLL006 (F). Tumor subclones (red circles) are mapped to each stage and extrapolated back to the origin. The number beside each circle shows the percentage of cells calculated using mutant allele frequencies. Colored boxes denote mutation profile groups (Figure 1D, supplemental Figure 3). (B-C,E,G-H) Graphic illustration of absolute cell numbers for each subclone at all stages for patients CLL003 (B-C), CLL077 (E), and CLL006 (G-H). Plots are expanded for stage c of patient CLL003 (C) and stage c of patient CLL006 (H).
Schematic presentation of the changes in subclonal architecture over time. (A,D,F) Schematic representation of the subclonal hierarchy for patients CLL003 (A), CLL077 (D), and CLL006 (F). Tumor subclones (red circles) are mapped to each stage and extrapolated back to the origin. The number beside each circle shows the percentage of cells calculated using mutant allele frequencies. Colored boxes denote mutation profile groups (Figure 1D, supplemental Figure 3). (B-C,E,G-H) Graphic illustration of absolute cell numbers for each subclone at all stages for patients CLL003 (B-C), CLL077 (E), and CLL006 (G-H). Plots are expanded for stage c of patient CLL003 (C) and stage c of patient CLL006 (H).
The list of mutated genes was unique to each patient (supplemental Tables 6-8). However, each carried 1 or more candidate driver mutations based on recurrence in CLL25-30 or other cancers31 (supplemental Table 10). Importantly, a single somatic founder mutation in each patient affected a gene recurrently mutated in CLL (CLL003: SF3B1; CLL077: SAMHD1; CLL006: MED12). Further 5 to 10 nonrecurrent mutations were fixed within the founder clone and could include both driver and passenger events. By contrast, ATM, PLEKHG5, and IRF4 mutations, although recurrent in CLL, were clearly secondary events, as they were not observed in all tumor cells and because their allele frequency reduced during treatment.
Patterns of clonal evolution are heterogeneous in CLL
Next, we explored how the patterns of subclonal evolution differed between the 3 patients. Using conventional prognostic markers, all 3 patients belonged to an intermediate risk group (IgVH unmutated, no TP53 abnormalities, no genomic complexity32-36 ; supplemental Table 1) and were treated with similar combinations of alkylating agents, purine analogues, and immunotherapy.
CLL003 showed dramatic shifts in subclonal composition over time. At diagnosis, 82% of cells from CLL003 carried a nonsense mutation in ATM and had lost the other copy of the gene as the result of an 11q deletion (“subclone 2” in Figure 2A-C). Subclone 2 expanded at first relapse and accounted for more than 90% of the tumor cells. The patient subsequently achieved a minimal residual disease (MRD) positive complete remission after fludarabine/cyclophosphamide/rituximab (FCR; time point c; supplemental Figure 4). This coincided with a dramatic contraction of subclone 2. However, another subclone defined by mutations in genes known to be mutated in malignancies (FAT3, NPY, NRG3, ASXL1, MUSK, SEMA3E)31 was detected in a large fraction of this remission sample and became dominant at later stages (“subclone 4” in Figure 2A-C). ASXL1, MUSK, and SEMA3E mutations were detected at low level by WGS in a sample 2 years previously and before the patient ever received treatment.
By contrast to patient CLL003, the major subclones of CLL077 remained initially largely unchanged (time points a-c), consistent with the clinical picture of refractory but stable disease. As with CLL003, clinical disease progression in CLL077 coincided with expansion of a subclone (“subclone 4” in Figure 2D-E) containing a mutation in a cancer gene (MAP2K1c.171G > T, supplemental Figure 5). MAP2K1 mutations are rare events in lung cancer,37 and MAP2K1 (Mek-1) inhibitors have entered phase 3 clinical trials for solid tumors. However, we did not find any MAP2K1 mutations by targeted sequencing of 90 patients as a follow-up in this study (data not shown). The MAP2K1c.171G > T mutation causes increased phosphorylation of ERK1/2 in transfection experiments using heterologous cells.37 We showed that phosphorylated ERK1/2 increased over time in lymphocytes of patient CLL077 and that this mirrored the expansion of the subclone containing the mutation (supplemental Figure 6). Although this does not prove the pathogenic mechanism, it implies that the mutated MAP2K1 gene product was active in this patient at these time points. At relapse before death subclone 4 was predominant in CLL077 (time point e). Importantly, the presence of the MAP2K1 change was detected at low levels by WGS in the pre-Ofatumumab sample taken 9 months earlier (time point d).
Clonal evolution in CLL006 was characterized by the absence of expanding or emerging subclones. With every treatment, absolute lymphocyte cell numbers decreased dramatically. The same subclones re-emerged at relapse albeit in different proportions. At later stages (time points d-e), subclones containing IRF4 mutations (4 and 5) outcompeted all other cells.
The findings for patient CLL006 demonstrate that relapse does not always coincide with expansion of subclones containing new or rare mutations. Instead, it could be because of cell-extrinsic factors, such as the pharmacokinetic properties of monoclonal antibodies resulting in incomplete penetration of lymphoid organs and subsequent redistribution of residual leukemic cells into the periphery.
Discussion
Cancers initiate from a single cell with one or more founder mutations and acquire additional mutations, some of which may give rise to resistance or confer sensitivity to treatment. We demonstrate that WGS provides an abundance of mutated sites whose allele frequency profiles can be grouped to reveal the probable evolving subclonal hierarchy of leukemia. In addition, WGS provides information on somatic mutations in noncoding regions, whose significance for cancer currently remains to be determined.
In all 3 patients, we identified founder events that could be future targets for curative therapy in CLL. Further, in all patients WGS defined the genetic composition of subclones that later became dominant even before initiation of relapse treatment. In some cases these changes to the molecular phenotype of the tumor became apparent months or years ahead of an obvious clinical phenotype, thus offering the possibility for earlier targeted and sequential treatment selection directed against these subclones, which are characterized by the presence of nonrecurrent or low-recurrence mutations. However, in practice this approach might only shift the balance of the different subclones but not affect the ultimate outcome for the patient.
We show that genome-wide tracking of somatic mutation profiles over time reveals heterogeneous patterns of clonal evolution in CLL. All 3 patients had multiple subclones before treatment. With the exception of subclone 2 in CLL003, which became undetectable and never recurred, these subclones persisted through later disease stages. This is different from ALL1 and AML models of relapse where either the dominant (model 1) or a single minor subclone (model 2) give rise to relapse,19 and is more similar to the pattern observed in MDS/AML progression.20 Relapse in 2 of the CLL patients is also characterized by emergence and expansion of subclones that were not present at diagnosis.
We identify both dynamic/rapid and stable/gradual shifts in the interclonal balance. The clinical and prognostic significance of these subclonal shifts remains to be established. For example, we have seen that a relatively stable molecular phenotype in patient CLL006 correlates with slow disease progression and a good response to repeated treatment, whereas emerging or increasing subclones in the other 2 patients correlate with resistance to different treatments and death. It could be that in patients CLL003 and CLL077 the chemotherapy itself selected for chemo-resistant subclones and/or induced new mutations causing resistance through DNA damage. These conferred a survival advantage and led to expansion of resistant subclones. By contrast, this did not occur in CLL006 who was treated almost exclusively with antibodies. Going forward, the potential clinical utility of our analysis approach will need to be evaluated systematically within clinical trials in larger cohorts of patients. Depending on the outcome of these studies, longitudinal WGS studies may eventually provide a means to individualize treatment.38
Challenges remain to genome-wide sequencing being applied within clinical trials. These include simplifying and standardizing the currently complex analysis methods, and improving turnaround time, costs, and interpretation of clinically actionable information. Clinical implementation also depends on the availability of sequential biopsies, access to integrated phenotype-genotype databases and effective therapeutics.
Given progress in all these areas, we anticipate that genome-wide sequencing will become an effective approach to monitor disease progression systematically and also prospectively, and that it will direct future clinical trials and therapeutic decisions. Its successful implementation could fundamentally change our strategy for treatment selection and monitoring and provide the tool for delivering more successful and cost-effective healthcare with better outcomes for individual patients.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Helen Northen for performing experimental work.
This work was supported by the Oxford Partnership Comprehensive Biomedical Research Center with funding from the Department of Health's National Institute of Health Research (NIHR) Biomedical Research Centre funding scheme. The views expressed in this publication are those of the authors and not necessarily those of the Department of Health.
Authorship
Contribution: A.S., S.M.F., M.R., and D.B. designed, analyzed, and interpreted experiments; J.B. performed bioinformatics; S.H., A.A., A.B., R.C., R.G., S.H., I.K., Z.K., S.L., D.M., L.M., T.M., and A.T. performed experiments; A.S. wrote the first draft of the paper; and A.S., J.T., M.R., and D.B. wrote the final draft of the paper.
Conflict-of-interest disclosure: J.B., S.B., A.A., R.G., I.K., Z.K., S.L., D.M., L.M., M.R., and D.B. are employees of Illumina Inc, a public company that develops and markets systems for genetic analysis. The remaining authors declare no competing financial interests.
Correspondence: Anna Schuh, Oxford NIHR Biomedical Research Centre, Molecular Diagnostic Laboratory, Level 4 John Radcliffe Site, University of Oxford, Oxford OX3 9DS, United Kingdom; e-mail: anna.schuh@ndcls.ox.ac.uk.