Key Points
Naturally occurring oncogenic GATA1 mutants with internal deletions contribute to transient abnormal myelopoiesis in Down syndrome.
Abstract
Children with Down syndrome have an increased incidence of transient abnormal myelopoiesis (TAM) and acute megakaryoblastic leukemia. The majority of these cases harbor somatic mutations in the GATA1 gene, which results in the loss of full-length GATA1. Only a truncated isoform of GATA1 that lacks the N-terminal 83 amino acids (GATA1-S) remains. We found through genetic studies of 106 patients with TAM that internally deleted GATA1 proteins (GATA1-IDs) lacking amino acid residues 77-119 or 74-88 (created by splicing mutations) contributed to the genesis of TAM in 6 patients. Analyses of GATA1-deficient embryonic megakaryocytic progenitors revealed that the GATA1 function in growth restriction was disrupted in GATA1-IDs. In contrast, GATA1-S promoted megakaryocyte proliferation more profoundly than that induced by GATA1 deficiency. These results indicate that the internally deleted regions play important roles in megakaryocyte proliferation and that perturbation of this mechanism is involved in the pathogenesis of TAM.
Introduction
Children with Down syndrome (DS) are known to have a high risk of developing transient abnormal myelopoiesis (TAM) and subsequent acute megakaryoblastic leukemia (DS-AMKL).1-4 Blast cells in the majority of patients with TAM and DS-AMKL have mutations in the second exon of the GATA1 gene.5,6 The mutations turn off the production of full-length GATA1. Instead, N-terminally truncated GATA1 protein (GATA1-S) was translated from the second methionine at codon 84, which is identical to the truncated GATA1 isoform found in the healthy human.7 In contrast, only a few patients with AMKL have been reported to harbor 21-disomy blasts with the GATA1 mutation.8,9 Therefore, GATA1-S is believed to be a prerequisite for the pathogenesis of TAM and DS-AMKL in children with DS, and unrestricted proliferation of megakaryocytic progenitors in DS-AMKL is thought to be provoked by a mechanism involving GATA1-S. However, the molecular mechanism of how GATA1-S contributes to the genesis of TAM and DS-AMKL remains elusive.
GATA1 regulates the proliferation of immature megakaryocytic progenitors. Indeed, active proliferation of immature megakaryocytic progenitors derived from GATA1-deficient mouse embryos is restricted by introduction of wild-type GATA1, but not by GATA-S.10 GATA1-deficient mice rescued with transgenic expression of GATA1-S (or GATA1-ΔNT) are found to exhibit hyper-megakaryopoiesis in a limited embryonic and postnatal period, resembling the phenotype in human TAM cases.11 In contrast, another report indicates that targeting mice expressing GATA1 protein with a deletion of 64 N-terminal amino acids, but retaining the 65th to 83rd amino acid residues intact, has demonstrated that the embryos display a transient megakaryocytic phenotype only during the early embryonic stage, not in the late-embryonic and postnatal stages.12 We surmise that this difference simply may be a result of missing the region corresponding to the 65th to 83rd amino acids.
Here, we have identified novel GATA1 mutants with internal deletions (IDs) of either amino acid residues 77-119 or 74-88 (GATA1-IDs) in 6 patients. We found that the GATA1-IDs lost their activity in the regulation of megakaryocyte growth. These results demonstrate that disruption of ID regions is implicated in the pathogenesis of TAM.
Study design
This study was approved by the Ethics Committee of the Hirosaki University Graduate School of Medicine. All animal experiments were approved by the Institutional Animal Experiment Committee of Tohoku University. All clinical samples were obtained with informed consent from the parents of all patients with TAM in accordance with the Declaration of Helsinki. Additional information can be found in the supplemental text on the Blood website.
Results and discussion
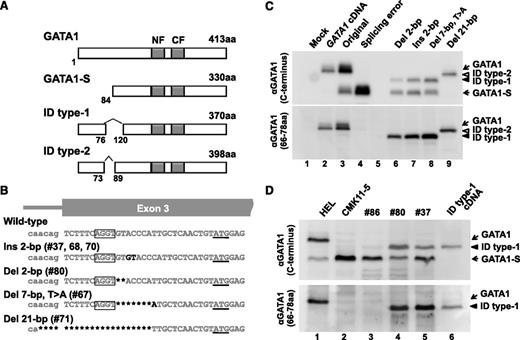
Between 2003 and 2010, we screened GATA1 mutations by direct sequencing, using cDNAs prepared from TAM blasts provided by 106 patients with DS on request from referring hospitals. Acquired GATA1 mutations were detected in 99 (93.4%) patients (supplemental Table 1). The majority of the mutations resulted in the GATA1-S mutant protein, which lacks the entire N-terminal transactivation domain. Importantly, we found new mutations harboring IDs of 43 and 15 amino acids in 5 patients (patients 37, 67, 68, 70, and 80) and in 1 patient (patient 71), respectively. We refer to these mutants as GATA1-ID type 1 and GATA1-ID type 2, respectively (Figure 1A). Clinical features in patients with TAM who have GATA1-ID mutations were shown in supplemental Table 2. All of these patients showed high white blood cell counts in the peripheral blood, which is known to be a risk factor for early death.13
GATA1 mutant proteins with internal deletions. (A) A schema of mutant GATA1 proteins observed in patients with TAM. The amino acid sequence of GATA1-ID proteins was deduced from the sequence of GATA1 cDNA obtained from patients with TAM. Dark boxes indicate N-finger (NF) and C-finger (CF) domains. ID indicates internal deletion. (B) Somatic mutations of the GATA1 gene found in ID type 1 and type 2 patients. Missing, inserted, or substituted nucleotides are highlighted with dark color. A second translation initiation codon located in the third exon is underlined. The AGGT sequence functioning as an alternative splice donor site in mutant GATA1 genes of ID type 1 patients is circled. Note that a mutant GATA1 gene found in TAM patient 71f (ID type 2) lost a splice acceptor site in exon 3 because of the 21-nucleotide deletion. (C) Expression of GATA1 proteins in cells transfected with minigenes using anti-GATA1 antibodies recognizing the C terminus (upper) and residues between the 66th and 78th amino acids (lower) of the GATA1 protein. GATA1-ID proteins are recognized by the antibody against amino acid residues 66-78 of GATA1, whereas GATA1-S is not (lanes 6-9). Cells transfected with mock pcDNA3.1 (lane 1), pcDNA3.1-GATA1 cDNA (lane 2), original minigene (lane 3), and GATA1 minigene harboring a splicing error mutant in the 3′ boundary of intron 113 (lane 4) are used as positive and negative controls for GATA1 and GATA1-S, respectively. (D) GATA1 ID type 1 protein and GATA1-S are detected in the TAM blast cells from patients 80 (lane 4) and 37 (lane 5), whereas only GATA1-S is expressed in the blast cells from patient 86 harboring a conventional type of GATA1 gene mutation in TAM cases (lane 3). Note that relatively abundant GATA1-S is recognized in patient 37 because of the intermixing of genetically distinct clone of cells expressing only GATA1-S (supplemental Table 1). Human erythroleukemia cells (HEL, lane 1) were used as a control for GATA1 and GATA1-S. DS-AMKL cells (CMK11-5, lane 2) and BHK-21 cells transfected with cDNA encoding GATA1 ID type 1 protein (lane 6) were used as controls for GATA1-S and GATA1 ID type 1, respectively.
GATA1 mutant proteins with internal deletions. (A) A schema of mutant GATA1 proteins observed in patients with TAM. The amino acid sequence of GATA1-ID proteins was deduced from the sequence of GATA1 cDNA obtained from patients with TAM. Dark boxes indicate N-finger (NF) and C-finger (CF) domains. ID indicates internal deletion. (B) Somatic mutations of the GATA1 gene found in ID type 1 and type 2 patients. Missing, inserted, or substituted nucleotides are highlighted with dark color. A second translation initiation codon located in the third exon is underlined. The AGGT sequence functioning as an alternative splice donor site in mutant GATA1 genes of ID type 1 patients is circled. Note that a mutant GATA1 gene found in TAM patient 71f (ID type 2) lost a splice acceptor site in exon 3 because of the 21-nucleotide deletion. (C) Expression of GATA1 proteins in cells transfected with minigenes using anti-GATA1 antibodies recognizing the C terminus (upper) and residues between the 66th and 78th amino acids (lower) of the GATA1 protein. GATA1-ID proteins are recognized by the antibody against amino acid residues 66-78 of GATA1, whereas GATA1-S is not (lanes 6-9). Cells transfected with mock pcDNA3.1 (lane 1), pcDNA3.1-GATA1 cDNA (lane 2), original minigene (lane 3), and GATA1 minigene harboring a splicing error mutant in the 3′ boundary of intron 113 (lane 4) are used as positive and negative controls for GATA1 and GATA1-S, respectively. (D) GATA1 ID type 1 protein and GATA1-S are detected in the TAM blast cells from patients 80 (lane 4) and 37 (lane 5), whereas only GATA1-S is expressed in the blast cells from patient 86 harboring a conventional type of GATA1 gene mutation in TAM cases (lane 3). Note that relatively abundant GATA1-S is recognized in patient 37 because of the intermixing of genetically distinct clone of cells expressing only GATA1-S (supplemental Table 1). Human erythroleukemia cells (HEL, lane 1) were used as a control for GATA1 and GATA1-S. DS-AMKL cells (CMK11-5, lane 2) and BHK-21 cells transfected with cDNA encoding GATA1 ID type 1 protein (lane 6) were used as controls for GATA1-S and GATA1 ID type 1, respectively.
We determined the genomic DNA sequences of these cases. As shown in Figure 1B, the mutations in GATA1-ID type 1 were located in a site immediately 3′ of the consensus motif for a splice donor site AGGT14 (Ins 2-bp in patients 37, 68, and 70; Del 2-bp in patient 80; and Del 7-bp T>A in patient 67), whereas 21 bp containing a splice acceptor site in front of exon 3 was deleted in GATA1-ID type 2 (Del 21-bp). To verify the transcripts achieved through the putative splice donor site created by mutations in GATA1-ID type 1, we introduced identified mutations into GATA1 minigene expression vectors13 and transduced them into hamster fibroblast cell line BHK-21. We found 3 variant transcripts in the cases of GATA1-ID type 1 mutations (supplementary Figure 1A-B): a full-length transcript with deletion or insertion of nucleotides [Ex-2 (+) (PTC)], a short transcript lacking exon 2 by alternative splice variant skipping of exon 2 for GATA1-S [Ex-2 (−)], and an aberrant transcript in which 129 nucleotides were spliced out from exon 3 (Del 129-bp). In contrast, 2 disparate transcripts with deletions of 45 or 137 nucleotides were created by mutation in GATA1-ID type 2, using alternative acceptor sites in exon 3.
To examine whether the GATA1-ID proteins were produced from the mutant alleles, we performed immunoblotting analysis with 2 distinct antibodies recognizing the C terminus and amino acids 66-78, respectively. We detected GATA1-ID type 1 protein in addition to GATA1-S in the cells transfected with the minigenes harboring Ins 2-bp, Del 2-bp, or Del 7-bp T>A mutations, whereas only GATA1-ID type 2 protein was expressed on transfection of the minigene with a Del 21-bp mutation (Figure 1C). Consistent with the minigene results, a significant amount of GATA1-ID type 1 protein and GATA1-S had accumulated in patients 80 and 37, whereas only GATA1-S was detected in the TAM blasts of patient 86, who had only a short transcript skipping exon 2 because of a point mutation in the exon 2–intron 2 boundary (Figure 1D). Thus, splicing errors were occurred in GATA1-ID type 1 and type 2 patients, leading to the production of GATA1-ID proteins.
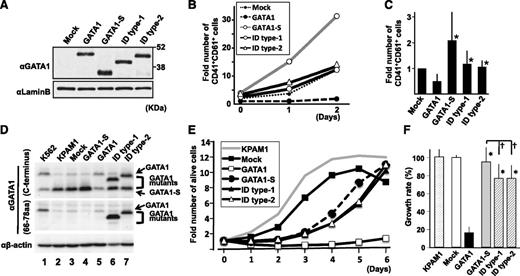
We next examined how GATA1-ID proteins affect the proliferation of embryonic megakaryocytic progenitors. We retrovirally transduced GATA1-S and GATA1-ID mutants into lineage-negative cells derived from megakaryocyte-specific Gata1-deficient (Gata1ΔneoΔHS) embryos15 and induced differentiation toward the megakaryocytic lineage. The number of CD41+CD61+ megakaryocytes was significantly higher in cases transduced with GATA1-ID proteins than with wild-type GATA1, despite almost equivalent expression levels of GATA1 proteins (Figure 2A-C). GATA1-S-transduced cells unexpectedly acquired a hyperproliferative potential compared with mock cells, probably because of an unknown function that resides in the GATA1 N-terminal region (Figure 2B-C).
GATA1 ID proteins showed restricted antiproliferative activity. (A) Expression of GATA1 and GATA1 mutant proteins in cultured megakaryocytes at day 0, using an antibody against the C terminus of GATA1. The amount of protein loaded was quantified using an anti-Lamin B antibody on the same membrane. (B) Time-course change in the number of CD41+CD61+ cells. The value in the mock case at day 0 is set to 1. The result is representative of 4 independent experiments. (C) Comparison of the number of CD41+CD61+ cells at day 2. The value in the mock case is set to 1 in every experiment. The mean values and standard deviations from 4 independent experiments are presented. Asterisks indicate a significant difference compared with wild-type GATA1 (P < .05). (D) Immunoblot analysis of ectopic expression of GATA1 proteins in KPAM1 cells using anti-GATA1 antibodies against C terminus (upper) and residues between amino acids 66 and 78 (middle). The loading volume was quantified using anti-β-actin antibody (lower). (E) Growth curves of KPAM1 cells after ectopic expression of GATA1 proteins. Average values obtained from 6 wells are shown. The value at day zero is set to 1 for each. The growth curve of the original KPAM1 cells was analyzed as a control. Representative data from 3 independent experiments are shown. (F) Relative growth rate of KPAM1 cells at 5 days after ectopic expression of GATA1 mutant proteins. The average value of growth rate in the mock case is set to 100% in every experiment. The mean values and standard deviations from 18 wells obtained in 3 independent experiments (6 wells in each) are presented. Asterisks and daggers indicate significant differences compared with wild-type GATA1 and GATA-S, respectively (P < .01).
GATA1 ID proteins showed restricted antiproliferative activity. (A) Expression of GATA1 and GATA1 mutant proteins in cultured megakaryocytes at day 0, using an antibody against the C terminus of GATA1. The amount of protein loaded was quantified using an anti-Lamin B antibody on the same membrane. (B) Time-course change in the number of CD41+CD61+ cells. The value in the mock case at day 0 is set to 1. The result is representative of 4 independent experiments. (C) Comparison of the number of CD41+CD61+ cells at day 2. The value in the mock case is set to 1 in every experiment. The mean values and standard deviations from 4 independent experiments are presented. Asterisks indicate a significant difference compared with wild-type GATA1 (P < .05). (D) Immunoblot analysis of ectopic expression of GATA1 proteins in KPAM1 cells using anti-GATA1 antibodies against C terminus (upper) and residues between amino acids 66 and 78 (middle). The loading volume was quantified using anti-β-actin antibody (lower). (E) Growth curves of KPAM1 cells after ectopic expression of GATA1 proteins. Average values obtained from 6 wells are shown. The value at day zero is set to 1 for each. The growth curve of the original KPAM1 cells was analyzed as a control. Representative data from 3 independent experiments are shown. (F) Relative growth rate of KPAM1 cells at 5 days after ectopic expression of GATA1 mutant proteins. The average value of growth rate in the mock case is set to 100% in every experiment. The mean values and standard deviations from 18 wells obtained in 3 independent experiments (6 wells in each) are presented. Asterisks and daggers indicate significant differences compared with wild-type GATA1 and GATA-S, respectively (P < .01).
We next analyzed cell proliferation using the DS-AMKL cell line KPAM1, in which GATA1-S was predominantly expressed with a very low level of full-length GATA1 (Figure 2D).16 On transduction with full-length GATA1 retrovirus, proliferation of KPAM1 cells was markedly reduced. In contrast, GATA1-ID type 1 and type 2 moderately restricted the proliferation of KPAM1 cells, but the restriction activity was significantly stronger than that of GATA1-S (Figure 2E-F). These results thus demonstrate that the ID regions indeed contribute to the regulation of AMKL cell proliferation.
Our newly identified GATA1-ID mutants have highlighted a much narrower set of sequences responsible for the pathogenesis of TAM than has previously been suggested by the loss of the N-terminal sequence, as in GATA1-S. The missing region identified by the GATA1-ID proteins contains a consensus motif (LxCxE, amino acids 81-85) essential for the interaction with pRb,17 which is also lost in GATA1-S. Interaction with hypophosphorylated pRb-E2F complex has been reported to be important for GATA1 to support the normal proliferation and differentiation of erythroid progenitors.17 Consistent with this notion, GATA1-S failed to repress E2F activation, which was followed by activation of mTOR signaling in the GATA1-S fetal megakaryocytes and DS-AMKL cells.18 Because the protein levels of cyclin D1 and p27Kip are reciprocally regulated by the mTOR pathway, and thereby cause pRb to be phosphorylated,19 cell-cycle progression in response to the mTOR pathway may be potentiated by the enfeebled function of LxCxE motif of GATA1-S. Thus, we are one step closer to a molecular understanding of GATA1-related leukemias.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (R.S., T.T., M.Y., and E.I.), sciences research grants from the Ministry of Health, Labour and Welfare of Japan (E.I.), the Asahi Glass Foundation (R.S.), the Mitsubishi Foundation (R.S. and M.Y.) and the Takeda Foundation (M.Y.).
Authorship
Contribution: T.T., R.K., E.K., H. Kaneko, R.W., and K.T. contributed to the experiments; T.T., M.S., R.S., M.Y., and E.I. contributed to the study design, funding, project conception, and manuscript writing; and H. Kanegane, M.M., M.E., T.M., S.A., and Y.H. contributed to the clinical sample collection and phenotype analyses.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Masayuki Yamamoto, Department of Medical Biochemistry, Tohoku University Graduate School of Medicine, 2-1 Seiryo-cho, Aoba-ku, Sendai 980-8575, Japan; e-mail: masiyamamoto@med.tohoku.ac.jp; and Etsuro Ito, Department of Pediatrics, Hirosaki University Graduate School of Medicine, 5 Zaifu-cho, Hirosaki, 036-8562, Japan; e-mail: eturou@cc.hirosaki-u.ac.jp.