Key Points
Quantitative clonal analysis demonstrates directional changes in contributions of stem cells to blood.
The pool of aged hematopoietic stem cells is comprised of many, but small clones, while young stem cells are less numerous, but more potent.
Abstract
The number of hematopoietic stem cells (HSCs) that contributes to blood formation and the dynamics of their clonal contribution is a matter of ongoing discussion. Here, we use cellular barcoding combined with multiplex high-throughput sequencing to provide a quantitative and sensitive analysis of clonal behavior of hundreds of young and old HSCs. The majority of transplanted clones steadily contributes to hematopoiesis in the long-term, although clonal output in granulocytes, T cells, and B cells is substantially different. Contributions of individual clones to blood are dynamically changing; most of the clones either expand or decline with time. Finally, we demonstrate that the pool of old HSCs is composed of multiple small clones, whereas the young HSC pool is dominated by fewer, but larger, clones.
Introduction
Hematopoietic stem cells (HSCs) provide a paradigm of how stem cells maintain the function of an organ over the lifespan of an organism. Multiple views have been formulated on how HSCs contribute to hematopoiesis throughout life. The main hypotheses include (1) clonal succession, which argues that distinct HSCs contribute to hematopoiesis in a sequential order1-3 ; (2) clonal stability, which implies that HSC clones steadily contribute to hematopoiesis4 ; and (3) more recently, dynamic repetition,5 in which stem cells undergo a reversible switch between dormant and active self-renewal states, also affecting their contribution to differentiation.
These models need to be reconciled with recent data that demonstrate profound heterogeneity within the HSC pool. Single-cell transplantation studies argue that individual HSCs are very different in terms of their lineage differentiation potential,6,7 repopulation capacity,7,8 and self-renewal ability.8 Additional levels of HSC heterogeneity were shown to exist with respect to their cycling behavior2,3 and their homing and migration ability.9,10 Furthermore, proportions of HSCs with different characteristics change during aging.11-13 Aging results in an increase of the number of HSCs with a lymphoid-deficient differentiation program,6,11-13 impaired repopulation potential per stem cell,14,15 and decreased homing efficiencies.10,13 Given such a high level of heterogeneity, it is unclear how different HSCs coexist in a polyclonal environment and how many HSC clones are simultaneously active.16
Reliable clonal analysis depends on the method of clonal labeling and detection. Cellular barcoding was recently introduced for the analysis of clonal fluctuations in T cells17 and hematopoietic cell populations.18,19 These techniques are based on inserting a random DNA sequence tag into viral vectors. Upon transduction, the tag is stably integrated into the genome of a target cell and is inherited by its progeny.
Here, we used a cellular barcoding method of highly purified young and old HSCs in combination with multiplex parallel sequencing to perform detailed clonal tracking in the hematopoietic system. We characterized clone size, developmental potential, and homing ability in several hundreds of young and old HSC clones. Our data provide detailed insight into functional differences, and into the homing ability of young and old HSCs. We demonstrate that most transplanted clones consistently contribute to hematopoiesis, arguing in favor of clonal stability, however, there are directional changes (growth/decline) and age-dependencies in the clonal output of individual HSCs.
Materials and methods
Mice
C57BL/6 (B6) mice were purchased from Harlan Laboratories (Boxmeer, The Netherlands). C57BL/6.SJL and C57BL/6.SJLxC57BL/6 mice were bred in the Central Animal Facility of University Medical Centre Groningen (Groningen, The Netherlands). C57BL/6J-kitW-41J/kitW-41J (W41) mice were obtained from E. Dzierzak (Rotterdam, The Netherlands), and were crossed with B6.SJL to obtain CD45.1 W41, as previously described.13 All experiments were approved by the University of Groningen Animal Care Committee.
Barcoded vector libraries
Design and validation of the MIEV-based barcode library (total, 800 barcodes) has been previously described.18 The pGIPZ vector was purchased from Open Biosystems and modified in the following way: An IRES-puro-miR cloning site was cut with BsrgI-MluI restriction enzymes and was replaced with a short linker carrying ClaI, BamHI, SmaI restriction sites. A barcode of the following structure: GTACAAGTAAGGNNNACNNNGTNNNCGNNNTANNNCANNNTGNNNGACGGCCAGTGAC was cloned via BsrgI-BamHI sites as previously described.18 Although a modification was introduced in the design of the barcode backbone of 2 libraries, preparation of the libraries and retrieval of the barcodes were exactly the same. The pGIPZ-based library (450 barcodes) (supplemental Figure 1) was used in the calibration experiment (Figure 1) for establishing the technical limitations of the method, whereas the MIEV library was used in all other experiments.
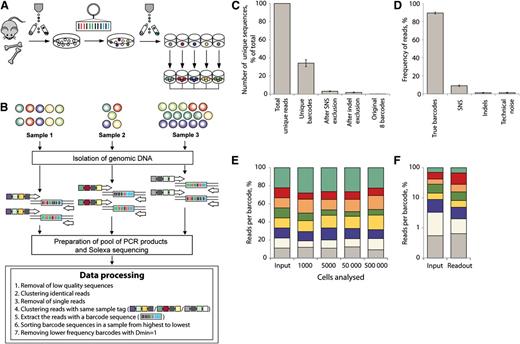
Combining cellular barcoding with multiplex deep sequencing – setup and method validation. (A) Individually barcoded LSK48−150+ cells were monoclonally expanded in liquid culture. (B) Different numbers of cells from expanded barcode cultures were mixed to generate samples with different ratios of barcodes and different total cell numbers. After isolation of genomic DNA, individual samples were amplified with primers bearing multiplex tags. Pooled PCR products were analyzed on Illumina HiSeq 2000 (Illumina, Inc., San Diego, CA). Steps of data processing and noise filtering are described. Dmin refers to minimal distance or nucleotide difference between 2 barcodes. (C) Number of unique sequencing reads (sequences different at any sequence position from all other reads) that remain after removal of noise calculated based on calibration samples. (D) Calculated reads frequencies (proportions of total number of reads in a multiplexed sample) related to true barcodes and various sources of noise. (E) Distribution of barcode frequencies in calibration samples with different cell content (1000 to 500 000). The original ratio of mixed barcodes was 1:1:1:1:1:1:1:2 (top barcode, green). Each color represents a distinct barcode. (F) Barcode analysis in a sample with highly unequal barcode composition (1:5:10:10:25:25:50:55). Barcodes comprising 0.55% of the total mix (gray) could be quantitatively detected.
Combining cellular barcoding with multiplex deep sequencing – setup and method validation. (A) Individually barcoded LSK48−150+ cells were monoclonally expanded in liquid culture. (B) Different numbers of cells from expanded barcode cultures were mixed to generate samples with different ratios of barcodes and different total cell numbers. After isolation of genomic DNA, individual samples were amplified with primers bearing multiplex tags. Pooled PCR products were analyzed on Illumina HiSeq 2000 (Illumina, Inc., San Diego, CA). Steps of data processing and noise filtering are described. Dmin refers to minimal distance or nucleotide difference between 2 barcodes. (C) Number of unique sequencing reads (sequences different at any sequence position from all other reads) that remain after removal of noise calculated based on calibration samples. (D) Calculated reads frequencies (proportions of total number of reads in a multiplexed sample) related to true barcodes and various sources of noise. (E) Distribution of barcode frequencies in calibration samples with different cell content (1000 to 500 000). The original ratio of mixed barcodes was 1:1:1:1:1:1:1:2 (top barcode, green). Each color represents a distinct barcode. (F) Barcode analysis in a sample with highly unequal barcode composition (1:5:10:10:25:25:50:55). Barcodes comprising 0.55% of the total mix (gray) could be quantitatively detected.
Purification and transduction of HSCs and progenitor cells
Bone marrow cells were isolated from bones of donor mice and stained with a cocktail of antibodies against Sca1, c-Kit, CD150, CD48, and lineage markers (Ter119, CD11b, CD3ε, B220, and Gr1) (BioLegend, San Diego, CA). Sorted lineage− Sca1+ c-Kit+ CD150+ CD48− (LSK48−150+) cells were pre-stimulated in Stemspan medium (STEMCELL Technologies, Vancouver, BC, Canada) supplemented with 300 ng/mL of stem cell factor (SCF), 1 ng/mL Flt3 ligand (both Amgen, Thousand Oaks, CA), and 20 ng/mL interleukin-11 (IL-11) (R&D Systems, Minneapolis, MN) for 24 hours. Production of MIEV supernatant was described earlier.18 For production of pGIPZ supernatant, 293T human embryonic kidney cells were transfected with pCMV, VSV-G, and pGIPZ plasmids as previously described.20 After 24 hours, the medium was changed to Stemspan (STEMCELL Technologies). Supernatant containing lentiviral particles was harvested after 12 hours and stored at −80°C. Retroviral and lentiviral transduction was performed overnight in viral supernatant supplemented with SCF, FLT3-ligand, and IL-11 in retronectin-coated plates (Takara Bio, Otsu, Shiga, Japan) in the presence of 2 µg/mL of polybrene (Sigma-Aldrich, St. Louis, MO).
Monoclonal liquid cultures of barcoded cells
At 20 to 22 hours post-transduction, green fluorescent protein-positive (GP+) cells were single-cell sorted in 96-well round-bottom plates in Stemspan medium supplemented with 10% fetal calf serum, 300 ng/mL of SCF, 1 ng/mL Flt3 ligand, and 20 ng/mL IL-11. For vector copy number analysis, cells were harvested once the colony reached the size of approximately 30 000 cells. For generation of calibration samples, monoclonal colonies were passaged to 12-well plate and expanded until a culture reached several million cells.
Hematopoietic cell transplantation and blood analysis
For limiting dilution transplantation of GFP+ cells, the cells were sorted 20 to 22 hours post-transduction. Doses of 10, 50, 70, 700, 800, and 1600 GFP+ cells were transplanted into 6 to 10 W41 recipient animals irradiated with 3.5 Gy. For transplantations of unfractionated transduced cells donor cells were transplanted into lethally (9.5 Gy) irradiated B6 mice, simultaneously with 2 × 106 radioprotective cells of either W41 origin or from previously transplanted B6 mice. For limiting dilution studies of unfractionated cells, the doses were 100, 500, and 17 360 cells per mouse, and the transduction efficiency was 20%. There were 6 to 8 mice per dose transplanted. Because not all transduced cells express GFP at the moment of transplantation, gene transfer efficiencies were determined at 3 to 5 days post-transduction in a small aliquot of cells left in the culture. Blood samples were stained with antibodies against Gr1, B220 and CD3ε. Antibodies against CD45.1 and CD45.2 were used to discriminate between donor and recipient cells. Granulocytes (Gr1+ side scatterhigh cells), B cells (B220+), and T cells (CD3+) were sorted. For estimation of HSC frequencies, we used extreme limiting dilution analysis software (ELDA Software; http://bioinf.wehi.edu.au/software/elda/).21
Barcode analysis in LSK48-150+ cells after transplantation
LSK48-150+ cell purification from transplant recipients was performed as previously described, with addition of antibodies against CD45.2 and CD45.1 for young and old HSC discrimination. Viable GFP+ lineage- Sca1+ c-Kit+ CD48− CD150+ donor-derived cells were individually sorted to generate monoclonal cultures as previously described. In some cases, the origin (old or young donor) of a clone was established after monoclonal expansion and detection of CD45 polymorphisms was performed using PCR and restriction analysis.22
Barcode detection in monoclonal cultures
Genomic DNA was extracted from LSK48−150+ cell-derived colonies by the use of the REDExtract-N-Amp Tissue PCR Kit (Sigma-Aldrich). Barcode sequences were amplified with primers against MIEV vector sequences and sequenced as previously described.18 Sanger sequencing was used here because the majority of monoclonal colonies contained only 1 or 2 barcodes.
Design of multiplexing tags for deep sequencing
For the present study, we generated a list of more than 300 individually tagged primers against the barcode vector sequence (supplemental Table 1, as shown in supplemental Methods). The structure of multiplex primers ensured at least 3 nucleotide differences between different tags to allow unambiguous sample identification post-sequencing. In addition, such differences allow correction of single nucleotide substitution errors, if necessary.23
Preparation of batches for multiplex sequencing
Extraction of genomic DNA was performed as previously described.18 Individual samples for each deep sequencing run were amplified with assigned multiplexing primers. Using DreamTaq Green mastermix (Fermentas, Burlington, ON, Canada), 35-cycle amplification was performed. Each sample was amplified in duplicate. Polymerase chain reaction (PCR) products were purified with the use of the High Pure PCR Cleanup Micro Kit (Roche, Basel, Switzerland), and they were pooled. Before sequencing, pools of products were phosphorylated with polynucleotide kinase (Fermentas, Burlington, ON, Canada) (30 minutes at 37°C, inactivation for 10 minutes at 75°C) and an additional round of purification was performed using High Pure PCR Cleanup Micro Kit. Sequencing was performed using an Illumina HiSeq2000 sequencer in the sequencing facility of University Medical Centre Groningen or at BaseClear Group (Leiden, The Netherlands).
Data processing and noise removal
A detailed protocol is described in the supplemental methods. In short, custom-written scripts were applied to FASTQ sequence data to remove low quality reads and sequences occurring only once. Samples were retrieved on the basis of exact match to sample tag and adjacent primer sequence. Each sequencing batch comprised up to 211 samples; 103 to 106 reads per sample were retrieved. Barcode sequences were retrieved and cross-compared for linear similarity. In barcode pairs differing by a single nucleotide, the lower frequency barcode was removed.
Results
Cellular barcoding combined with high-throughput sequencing permits sensitive quantitative clonal analysis
Before clonal analysis of biological samples can be reliably carried out in vivo, it was important to test the sensitivity and accuracy of our barcoding method. Therefore, defined mixtures of clonally expanded barcoded cells were analyzed in several dilutions (Figure 1). Purified lineage-negative LSK48−150+ cells were transduced with a barcoded vector library and transgene-positive cells were single-cell sorted and cultured to generate large uniquely barcoded colonies (Figure 1A). Expanded cells were mixed in different ratios and samples ranging from 1000 to 500 000 cells were compared (Figure 1B). Individual DNA samples were amplified with primers containing a unique multiplex tag and pooled multiplexed PCR products were sequenced (Figure 1B). After removal of low quality and single reads from each sample (filtering protocol is described in supplemental methods), we retrieved >100 000 reads per sample. The initial mixture of clones contained 8 barcodes that were identified in ∼88.5% of all sequencing reads per sample (Figure 1D). The remaining 11.5% of reads were constituted by 3578 to 5359 unique barcode sequences per sample and must be explained by sequencing/PCR errors and technical noise. We observed that 96.5% of these unique barcode sequences occurred due to single nucleotide substitutions, derived from the true barcodes, and a further 1.7% were due to single nucleotide insertion/deletion mutations of the true barcodes. Remaining sequences unrelated to true barcodes (technical noise) had low frequencies and cumulatively constituted ∼1% of all reads (Figure 1D). The proportions of reads and unique barcode sequences related to different sources of noise are shown in Figure 1C-D. We developed a filtering protocol for noise removal (described in supplemental Methods) and applied it for all other data sets described as follows.
Barcodes could be reliably quantified in as few as 1000 cells (Figure 1E) and clones that represent only 0.55% of the starting population could successfully be detected (Figure 1F). We estimated that the relative measurement error of barcode detection was on average 22% (Figure 1E; supplemental Methods). Thus, if the real contribution of a barcode is 10%, values for clonal contribution between 8% and 12% can be expected.
Concordance between estimated numbers of transplanted HSCs and barcode count in granulocytes, but not lymphocytes
To test whether this method is accurate and sensitive enough to follow clonal contributions in vivo, barcode readout was combined with limiting dilution analysis, currently the gold standard for HSCs quantification. This allowed us to assess whether the estimated numbers of barcoded clones agreed with numbers of HSCs predicted from limiting dilution. Furthermore, it allowed us to establish an upper limit of expected clones in recipients transplanted with a high HSC dose. To this end, we transplanted different numbers of non-transduced freshly isolated or transduced barcoded LSK48−150+ cells into irradiated recipients and measured donor and GFP+ chimerism (Figure 2A). Reconstituted mice were defined by at least 1% donor chimerism (for naive cells) or 1% GFP chimerism (for transduced cells) in granulocytes 16 weeks or longer post-transplantation. The HSC frequency equaled 1 in 9 (95% confidence interval [CI] 1 in 5 to 16.5) for freshly isolated LSK48-150+ cells and decreased to 1 in 134 (95% CI 1 in 53.9 to 334) after transduction. When GFP+ cells were resorted prior to transplantation, the frequency of functional long-term repopulating stem cells further decreased to 1 in 243 (95% CI 1 in 132 to 448) (Figure 2B). It should be noted that at 20 to 22 hours post-transduction, when cells were resorted, not all of the transduced cells already expressed GFP, and the final gene transfer efficiency was established in cell aliquot 3 to 5 days after transduction. The frequency of functionally defined HSCs is higher among cells with delayed GFP expression (cells that express GFP later than 20 to 22 hours after gene transfer) (supplemental Figure 2). This explains the lower HSC prevalence in the sorted cells (Figure 2B).
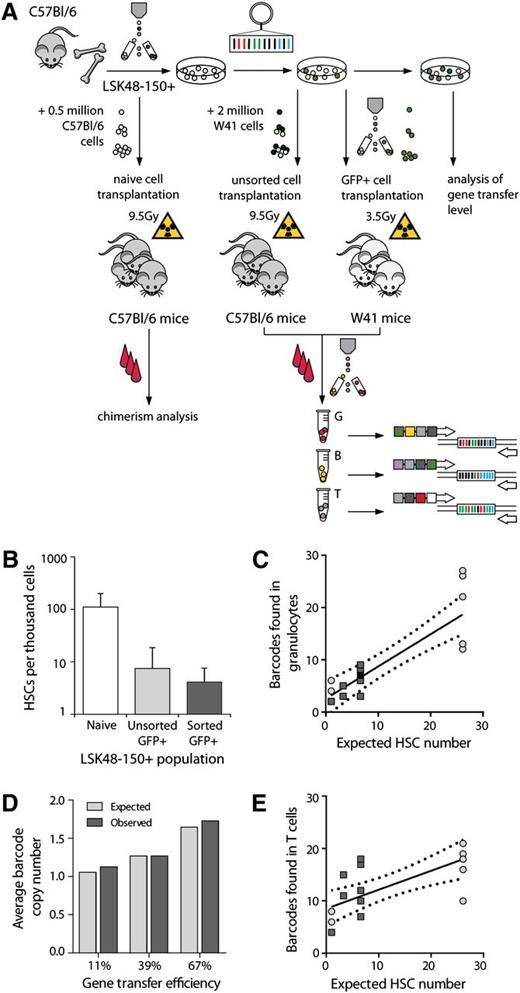
Relationship between the number of transplanted HSCs and detected barcodes. (A) Overview of limiting dilution experiments. LSK48−150+ cells were purified by cell sorting. For establishing the frequency of functional HSCs in this population, naïve cells were transplanted into irradiated recipients. Alternatively, LSK48−150+ cells were transduced with barcoded viruses and different doses of transduced cells were transplanted into irradiated hosts, either without (middle arrow) or with (right arrow) selection for GFP+. Mouse strains, competitors, and irradiation regimen used in every experiment are indicated. Congenic donor and recipient B6 animals were used to allow donor and recipient cell discrimination. Granulocytes (G), B- and T-lymphocytes (indicated as B and T, respectively) were isolated for further barcode analysis at regular time points after transplant. (B) Assessment of HSCs frequencies by limiting dilution analysis in the naive LSK48−150+ population, in transduced nonsorted and in sorted GFP+ cells. (C) Relationship between the expected number of transplanted HSCs and the number of barcodes, as detected in granulocytes. Each dot represents an individual mouse. Light gray circles indicate data generated using nonsorted cells and dark gray squares reflect experiments with sorted cells. In one mouse, data on granulocytes were not available for week 20, so week 28 data are shown instead with a black square. The best-fit line (line equation Y = 0.61X + 2.58) and 95% confidence intervals are plotted. Note that the 95% confidence interval includes X/Y intercept. (D) Observed and expected vector copy number per transduced cell at different transduction efficiencies. The average number of barcodes in 15 to 22 colonies is depicted as a function of gene transduction efficiency. (E) Same as shown in (C), but now data are shown for T-cell clones. Equation for best-fit line was Y = 0.36X + 8.5.
Relationship between the number of transplanted HSCs and detected barcodes. (A) Overview of limiting dilution experiments. LSK48−150+ cells were purified by cell sorting. For establishing the frequency of functional HSCs in this population, naïve cells were transplanted into irradiated recipients. Alternatively, LSK48−150+ cells were transduced with barcoded viruses and different doses of transduced cells were transplanted into irradiated hosts, either without (middle arrow) or with (right arrow) selection for GFP+. Mouse strains, competitors, and irradiation regimen used in every experiment are indicated. Congenic donor and recipient B6 animals were used to allow donor and recipient cell discrimination. Granulocytes (G), B- and T-lymphocytes (indicated as B and T, respectively) were isolated for further barcode analysis at regular time points after transplant. (B) Assessment of HSCs frequencies by limiting dilution analysis in the naive LSK48−150+ population, in transduced nonsorted and in sorted GFP+ cells. (C) Relationship between the expected number of transplanted HSCs and the number of barcodes, as detected in granulocytes. Each dot represents an individual mouse. Light gray circles indicate data generated using nonsorted cells and dark gray squares reflect experiments with sorted cells. In one mouse, data on granulocytes were not available for week 20, so week 28 data are shown instead with a black square. The best-fit line (line equation Y = 0.61X + 2.58) and 95% confidence intervals are plotted. Note that the 95% confidence interval includes X/Y intercept. (D) Observed and expected vector copy number per transduced cell at different transduction efficiencies. The average number of barcodes in 15 to 22 colonies is depicted as a function of gene transduction efficiency. (E) Same as shown in (C), but now data are shown for T-cell clones. Equation for best-fit line was Y = 0.36X + 8.5.
To determine how many hematopoietic clones are active, we analyzed the barcode composition in short-lived granulocytes, and long-lived B and T lymphocytes at 18 to 20 weeks post-transplantation (Figure 2A) in 17 mice from cohorts of mice transplanted with sorted and unfractionated barcoded cells. A conservative threshold for barcode identification of 0.5% of total barcode reads was used. In each mouse, the range of barcodes varied from 2 to 27 in granulocytes, 4 to 21 in T cells, and 6 to 33 in B cells. We compared the number of barcodes detected in mice transplanted with different doses of transduced cells with HSC estimates from limiting dilution analysis (Figure 2C-E). In granulocytes, the number of detected barcodes linearly increased with transplanted HSC dose (Figure 2C), indicating consistency between the 2 methods. However, this linearity was less evident in the case of T cells (Figure 2E) and B cells (supplemental Figure 3). Although 95% confidence intervals for data of granulocytes intercepts the X/Y coordinate, this is not the case for T cells. These 17 reconstituted mice were used for time-course clonal tracking experiments further described.
Correction of clonal counts for multiple vector integrations
Integration of multiple vectors in a single cell can lead to an overestimation of stem cell counts. To correct for this, we compared the occurrence of multiple barcodes integrating in single HSCs at different transduction efficiencies (11%, 39%, and 67% GFP+ cells). Transduced LSK48−150+ cells were monoclonally expanded to large colonies. Sanger sequencing was used to analyze the number of barcodes in 15 to 22 of such colonies for each group. Between 1 and 6 barcodes per clone were detected. The distributions of vector copy numbers per HSC followed a Poisson distribution (supplemental Figure 4), and therefore could be considered a random event.24 The average number of vector insertions per cell varied from 1.1 at 11% gene transfer to 1.7 at 67% (Figure 2D). To calculate the number of active hematopoietic clones in blood, we divided the numbers of barcodes detected in blood by average barcode copy number at the respective transduction efficiency (supplemental Figure 4). For clonal analysis, these observations implied that the proportion of transduced HSCs with multiple integrated vectors may vary from 13% (at 11% GFP+) to 40% (at 67% GFP+).
Clonal composition of granulocytes, T cells, and B cells is stable over time, but these lineages are maintained by different subsets of primitive cells
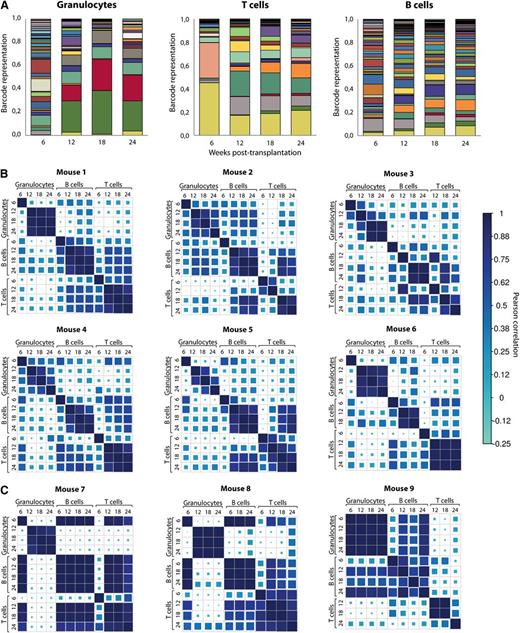
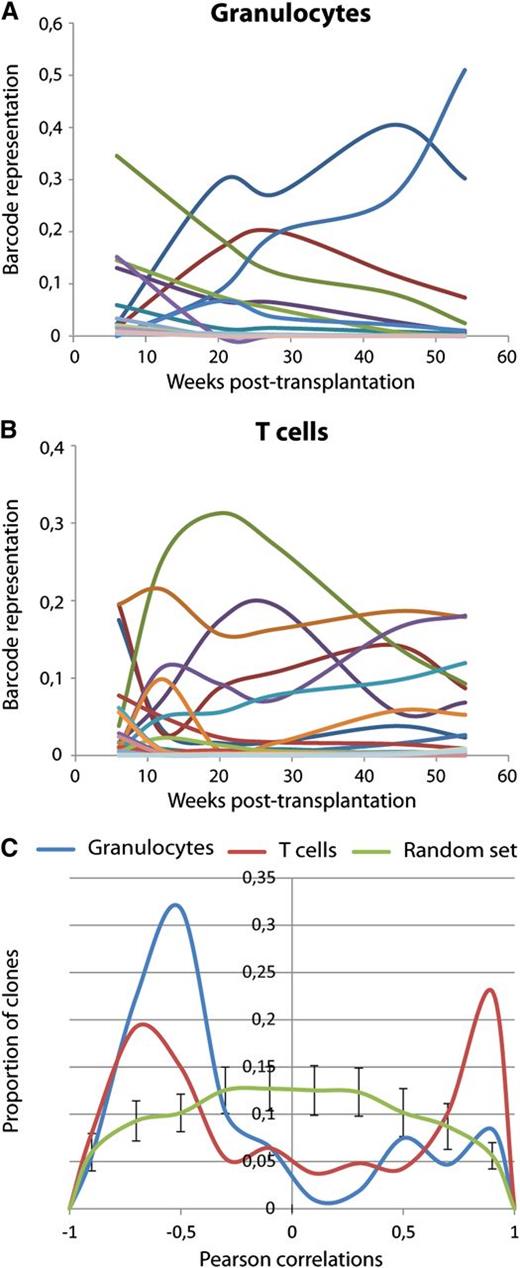
Next, we studied whether the clonal composition of blood cells of the previously described mice showed any signs of variation with time in a cohort of 9 mice transplanted with unfractionated barcoded cells. As an illustrative example, the barcode composition of granulocytes, T and B cells of one of these mice is shown in Figure 3A. Barcode contributions were compared among different time points and among the 3 blood lineages. To do so, we used Pearson correlation as a measure of the similarity for all data points. Results of analysis in 9 mice are shown in Figure 3. As expected, we observed that initial time points after transplantation are substantially different from later time points for the same lineage. At this time point, short-lived progenitors will contribute to blood cell development. Starting from week 12, clonal composition within each lineage showed very good correlation, demonstrating that at 12 weeks after transplant peripheral blood cell counts already reflect HSC engraftment. Interestingly, when we analyzed mice transplanted with HSCs at limiting dilution, barcodes found in granulocytes, B cells, and T cells were consistently different (Figure 3C). This provides a cautionary tale when interpreting limiting dilution experiments as truly clonal.
Clonal dynamics in mice transplanted with barcoded cells. (A) Barcode composition of granulocytes, T- and B lymphocytes in one of the mice transplanted with ∼26 barcoded HSCs. Different colors reflect different barcodes. This and other mice shown here originate from a cohort of mice transplanted with nonsorted transduced LSK48−150+ cells (these mice are identified with light gray circles as shown in Figure 2). (B) Pearson correlations between barcode compositions of 3 cell types at 4 time points from 6 mice transplanted with ∼26 HSCs. Note good correlations between samples collected from 12 to 24 weeks within each of the cell types. Mouse 1 corresponds to the data shown in (A). (C) The same analysis as in panel (B) was performed for 3 mice transplanted with 100 barcoded cells. At these doses, Poisson distribution predicts that more than two-thirds of the positive animals are transplanted with a single HSC.
Clonal dynamics in mice transplanted with barcoded cells. (A) Barcode composition of granulocytes, T- and B lymphocytes in one of the mice transplanted with ∼26 barcoded HSCs. Different colors reflect different barcodes. This and other mice shown here originate from a cohort of mice transplanted with nonsorted transduced LSK48−150+ cells (these mice are identified with light gray circles as shown in Figure 2). (B) Pearson correlations between barcode compositions of 3 cell types at 4 time points from 6 mice transplanted with ∼26 HSCs. Note good correlations between samples collected from 12 to 24 weeks within each of the cell types. Mouse 1 corresponds to the data shown in (A). (C) The same analysis as in panel (B) was performed for 3 mice transplanted with 100 barcoded cells. At these doses, Poisson distribution predicts that more than two-thirds of the positive animals are transplanted with a single HSC.
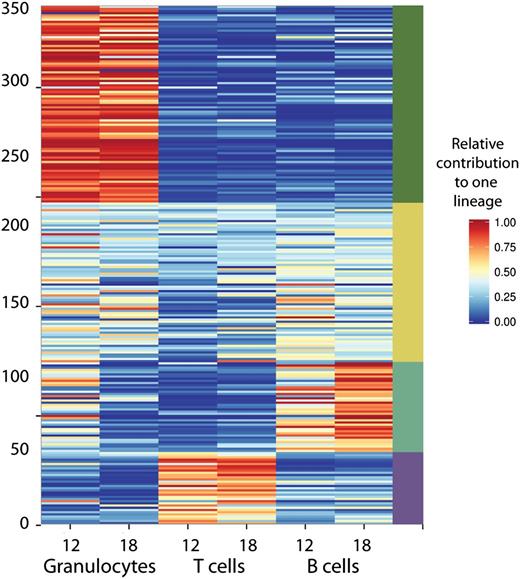
The barcode compositions between the lineages were only moderately correlated, indicating differences in clonal composition in each of the lineages. We followed the behavior of 350 clones found in 6 polyclonally repopulated mice. Contributions of all clones to at least 1 of these three lineages is shown in Figure 4. Importantly, although we transduced highly purified cells that have robust multilineage potential, B and T lymphocyte populations were frequently supported by different clones.
Relative lineage contributions of 350 barcoded clones found in 6 polyclonally-repopulated mice. Only clones that contributed at least 0.5% to one of the lineages 12 to 24 weeks post-transplant are shown. To assess relative contributions to granulocytes (and other lineages), the barcode representation in granulocytes was divided by total barcode representation (granulocytes + T cells + B cells). Clonal fluctuations within these mice are shown in Figure 3B.
Relative lineage contributions of 350 barcoded clones found in 6 polyclonally-repopulated mice. Only clones that contributed at least 0.5% to one of the lineages 12 to 24 weeks post-transplant are shown. To assess relative contributions to granulocytes (and other lineages), the barcode representation in granulocytes was divided by total barcode representation (granulocytes + T cells + B cells). Clonal fluctuations within these mice are shown in Figure 3B.
Additionally, to establish the correlation between barcode composition of HSCs and mature blood cells after transplantation, we analyzed 4 hematopoietic cells types (ie, blood granulocytes, T and B cells, and bone marrow LSK48−150+ cells) in a cohort of 3 mice (supplemental Figure 5). LSK48−150+ cells were cultured in cytokine-supplemented medium to generate colonies of approximately 30 000 cells. Monoclonal expansion served 2 purposes. First, it allowed us to confirm functionality of phenotypically defined HSCs, as it assesses the high proliferative potential of these cells. Second, the large number of cells allowed us to perform robust barcode analysis of each colony. Correlation analysis revealed that clonal spectrum of bone marrow and blood compartments were highly variable, ranging from high overlap between LSK48−150+ and blood to medium and low correlation (supplemental Figure 5).
Majority of clones in granulocytes and T cells are either expanding or declining in time
As long as 6 months after transplantation, the same clones were persistently detected in the same cell types. Although this may be considered clonal stability, this was only qualitative, meaning that no major clonal changes occurred between 2 consecutive time points among all mice and cell lineages. Quantitatively, however, many clones showed systematic changes in their contribution. A cohort of 7 mice, representing 228 barcoded clones, was followed for a period of up to 1 year after transplant. We used this cohort to analyze time trends. Clonal kinetics in one of these mice is shown in Figure 5A-B. Because the number of data points was limited (5 to 6 time points), we restricted our dynamic analysis only to linear trends in time (positive or negative) using Pearson correlation. In this analysis, a positive correlation indicates consistent clonal growth and a negative correlation detects gradual decline, whereas the absence of correlation implies fluctuation of clone size around a certain value without a defined direction. We summarized the values of Pearson correlation for 107 individual clones in granulocytes and 187 clones in T cells (Figure 5C). The data were compared with a random model, thus allowing establishing thresholds for significant nonrandom dynamic behavior. As shown in Figure 5C, in randomly simulated data, most clones do not follow a time trend. Experimental data, on the other hand, show a clear bimodal distribution of correlation values, indicating that most clones undergo systematic change in their clone size. In granulocytes, most of the clones were declining in time, whereas in T cells, both growing and declining populations were seen. Although it is possible that clonal fluctuations will differ from 1 experiment to the other, it may depend on the number of clones that contributes to hematopoiesis, and it may also reflect the presence of progenitor cells in the transplanted population; it is apparent that a large fraction of clones will systematically change in time. Therefore, barcode tracking over multiple time points is necessary for establishing the direction of clonal dynamics.
Clonal dynamics in long-term hematopoiesis. (A-B) Barcode fluctuations from 6 to 54 weeks after transplant are shown for one mouse in granulocytes (A) and T lymphocytes (B). Different colors represent different barcodes. (C) Pearson correlations of clonal sizes with time trend for clones detected at 0.5% or higher frequency at any of the time points in the respective lineage. Positive correlation indicates that the clone is consistently growing, and negative correlation reflects decline. Proportions of T cells (red) and granulocytes (blue) are plotted. The green line shows randomly expected correlations for 200 clones simulated 20 times (average values and standard deviations for 20 simulations are shown). Fluctuations in granulocytes reflect behavior of 107 barcodes and in T cells of 187 barcodes. Mice used for this analysis were transplanted with sorted barcoded cells (that were identified in Figure 2 with dark squares).
Clonal dynamics in long-term hematopoiesis. (A-B) Barcode fluctuations from 6 to 54 weeks after transplant are shown for one mouse in granulocytes (A) and T lymphocytes (B). Different colors represent different barcodes. (C) Pearson correlations of clonal sizes with time trend for clones detected at 0.5% or higher frequency at any of the time points in the respective lineage. Positive correlation indicates that the clone is consistently growing, and negative correlation reflects decline. Proportions of T cells (red) and granulocytes (blue) are plotted. The green line shows randomly expected correlations for 200 clones simulated 20 times (average values and standard deviations for 20 simulations are shown). Fluctuations in granulocytes reflect behavior of 107 barcodes and in T cells of 187 barcodes. Mice used for this analysis were transplanted with sorted barcoded cells (that were identified in Figure 2 with dark squares).
Clonal tracking of young and old HSCs
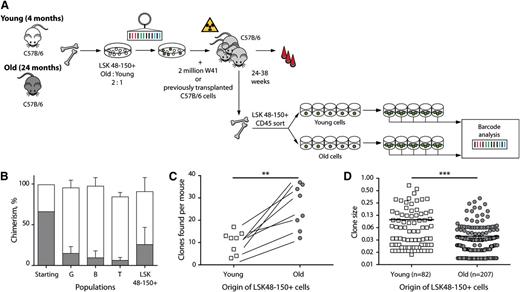
Finally, we asked how the composition of HSC pool changes with aging. To this end, we cotransplanted barcoded LSK48−150+ cells isolated from young (4 months) and old (24 months) donors into 8 recipients. Studies of unmanipulated old and young HSCs have indicated an ∼twofold reduced functional activity of old cells.13 To compensate for the expected decrease of functional activity in old HSCs, we initiated the experiment with LSK48−150+ cells from young and old mice cotransplanted in a 1:2 ratio (Figure 6A). At 24 to 38 weeks post-transplantation, we analyzed blood chimerism and barcode composition of engrafted LSK48−150+ bone marrow cells. Although twofold more of the old LSK48−150+ cells were transplanted, chimerism in the peripheral blood was predominantly derived from young HSCs (Figure 6B). Old HSCs contributed the least to lymphoid populations (Figure 6B). Interestingly, whereas old HSCs were functionally inferior in producing peripheral cells, the frequency of old LSK48−150+ cells in the bone marrow was substantially higher than predicted from blood values, indicating hampered differentiating activity of aged HSCs (Figure 6B). The number and size distributions of old HSC clones was subsequently determined. We sorted single transduced LSK48−150+ HSCs of old and young origin from the 8 transplanted recipients into cytokine-supplemented medium and grew colonies. Between 38 and 146 single cell colonies per mouse were successfully expanded and barcode sequences were retrieved and analyzed. Each animal contained between 15 and 53 uniquely barcoded HSC clones. Contrary to our expectations,13 in each mouse, the number of unique old LSK48−150+ clones exceeded the number of young clones (Figure 6C) in the 2:1 ratio of originally mixed HSCs. These results argue against the previously suggested decreased homing potential of old HSCs. However, old clones were significantly smaller than their young counterparts, resulting in lower overall contributions to the LSK48−150+ pool and to blood cell production (Figure 6D). We did not observe enrichment for clones carrying multiple inserts or for larger clone size, both in young and old HSCs (supplemental Figure 6).
Analysis of HSC pool in mice transplanted with old and young cells. (A) Shows experimental setup of LSK48−150+ cells that were purified from CD45 congenic young (4 months) and old (24 months) donor mice, mixed in 1:2 ratio and transduced with the barcoded vector library. There were 20 500 transduced cells transplanted simultaneously with W41 or previously transplanted B6 cells into 2 cohorts of 4 lethally irradiated B6 mice. There were 6 mice sacrificed 6 months post-transplantation, and 2 additional mice sacrificed at 8 months after transplantation. GFP+ LSK48−150+ cells of young and old origin were single-cell sorted in 96-well plates and expanded in liquid culture in presence of cytokines for subsequent barcode analysis. (B) Contribution from young (white bars) and old (gray bars) to different cell populations before (starting) and after transplantation. (C) Number of uniquely barcoded clones detected in expanded colonies of young and old LSK48−150+ cells. Lines connect data points derived from the same mice. The number of clones detected within the old compartment was significantly higher than the number of clones within the young population (P = .0011, paired two-sided t test). (D) Contribution of individual young and old LSK48−150+ HSCs to the stem cell compartment. Horizontal lines indicate mean values. Young LSK48−150+ cells produced larger clones than old LSK48−150+ cells (P < .0001, two-tailed Mann Whitney nonparametric U test).
Analysis of HSC pool in mice transplanted with old and young cells. (A) Shows experimental setup of LSK48−150+ cells that were purified from CD45 congenic young (4 months) and old (24 months) donor mice, mixed in 1:2 ratio and transduced with the barcoded vector library. There were 20 500 transduced cells transplanted simultaneously with W41 or previously transplanted B6 cells into 2 cohorts of 4 lethally irradiated B6 mice. There were 6 mice sacrificed 6 months post-transplantation, and 2 additional mice sacrificed at 8 months after transplantation. GFP+ LSK48−150+ cells of young and old origin were single-cell sorted in 96-well plates and expanded in liquid culture in presence of cytokines for subsequent barcode analysis. (B) Contribution from young (white bars) and old (gray bars) to different cell populations before (starting) and after transplantation. (C) Number of uniquely barcoded clones detected in expanded colonies of young and old LSK48−150+ cells. Lines connect data points derived from the same mice. The number of clones detected within the old compartment was significantly higher than the number of clones within the young population (P = .0011, paired two-sided t test). (D) Contribution of individual young and old LSK48−150+ HSCs to the stem cell compartment. Horizontal lines indicate mean values. Young LSK48−150+ cells produced larger clones than old LSK48−150+ cells (P < .0001, two-tailed Mann Whitney nonparametric U test).
Discussion
Detailed investigations of HSC clonality in the hematopoietic system require reliable heritable marking of HSCs to trace their progeny in time (reviewed in Bystrykh et al16 ). In this article, a barcoding method has been used to provide quantitative and dynamic tracking of individual HSCs regarding their lineage contribution and pool composition upon aging.
We show here that cellular barcoding reliably measures the number of clones contributing to hematopoiesis. The observed concordance between barcoding and limiting dilution in granulocytes indicates that most of HSC clones contribute to hematopoiesis. However, neither method can exclude the presence of quiescent stem cells. The notion of dormant HSCs is supported by the detection of clones that were prominently contributing to LSK48−150+, but not to mature blood cells (supplemental Figure 5). Additional hematopoietic stress, such as secondary transplantation or longer observation time might be necessary to activate these cells.
This study confirmed the view that blood sampling at early time points post-transplantation is not representative for long-term repopulating cells.25 However, the clonal make-up of blood starting 3 months post-transplantation was consistent for all time points for all individual lineages. At the same time, tracking clones for longer time periods was essential for understanding the dynamics of clonal fluctuations. Our data agree best with the theory of clonal stability of hematopoiesis. However, the relevance of other models cannot be excluded, because the definition of clones varies and clonal behavior is heterogeneous. For instance, clonal fluctuation between active and quiescent states will be unnoticed if only a fraction of 1 barcoded clone will undergo those changes. We also cannot exclude that progeny of an originally barcoded HSC does not participate in hematopoiesis sequentially on the basis of their division history, as proposed by the theory of clonal succession.1
Although most differentiated cells originated from a common precursor, the number of uniquely barcoded clones in myeloid and lymphoid cells substantially varied. Interestingly, also the clonal repertoire of B and T cells was only moderately correlated, demonstrating that HSC clones do not only differentially contribute to the myeloid and lymphoid lineages, but similar bias was observed within the lymphoid lineage. In the future, barcode analysis of progenitor populations can help to identify the stages of differentiation in which such commitment occurs.
Unexpectedly, in mice transplanted with a cell dose close to limiting dilution, lymphoid, and myeloid populations were supported by different groups of barcodes, suggesting more hematopoietic clones than predicted (Figure 3C).
Previous studies we conducted, and others,13,14 suggested an ∼twofold decrease in functionality of old purified stem cells. This was mostly attributed to a defect in homing of aged HSCs.10,13 In concordance with these data, we observed that transplantation of twice as many old than young LSK48−150+ cells resulted in lower chimerism of old LSK48−150+ cells in recipient animals (Figure 6). However, barcode analysis of individual LSK48−150+ cells showed that the initial 2:1 relation between the number of old and young clones was preserved (Figure 6), although old clones were smaller compared with clones from young HSCs. This argues against the previously described homing defect of old HSCs.10,13 In addition, the output of old HSCs to mature blood lineages was drastically lower than that of young cells. In classical competitive repopulation assays, this would result in a lower frequency of old HSC, because the contribution of many old HSCs would remain below the detection threshold.
Previously, it was suggested that transduction of hematopoietic cells leads to clonal dominance26 and preferential survival of clones with multiple insertions.27 Observations made in our study did not confirm these data. First, although we detected abundant clones contributing more than 20% of a respective cell population, the number of barcoded clones found >3 months post-transplantation correlated well with the expected HSC frequency, arguing against clonal selection (Figure 2). The presence of such large clones is likely reflecting intrinsic differences in repopulating capacity of HSCs. Secondly, we observed neither preferential survival nor larger clone size in HSC with multiple insertions among ≃150 young and old HSCs 6-months post-transplantation (supplemental Figure 6).
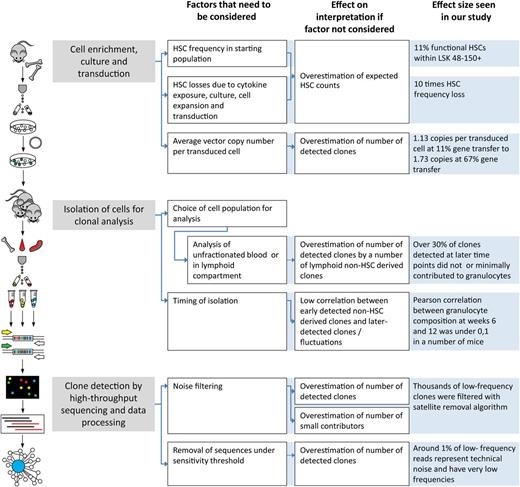
Recently, we discussed methodologic constraints that can lead to misinterpretation of clonal tracking studies.16 Limited resolution, insufficient quantification, and failure of noise detection can severely impact conclusions of the analysis. Here, we define principles of establishing cutoff values for minimizing noise. We demonstrate that this approach allows robust quantitative clonal tracking in clones present at frequencies of less than 1% of the transduced population. We believe that our current experimental design compares favorably to several related barcoding approaches, including our own, that had lower sensitivity,17 did not use high-throughput sequencing,18 or did not allow quantitative comparison of clones within the same animal.19 We briefly summarized several methodological steps that can influence clonal count analysis in Figure 7.
Factors that influence HSC clonal counts at different stages of analysis. Methodologic steps of planning, implementation of experiments, and barcode data analysis are shown. For each step, we indicate how experimental setup and approach may influence conclusions of analysis. The diagram aims to point out the factors that we found critical in the currently described experiments, but it is not exhaustive. For instance, the setup of transplantation experiments, number of transplanted cells, and irradiation regimens can also influence clonal counts.
Factors that influence HSC clonal counts at different stages of analysis. Methodologic steps of planning, implementation of experiments, and barcode data analysis are shown. For each step, we indicate how experimental setup and approach may influence conclusions of analysis. The diagram aims to point out the factors that we found critical in the currently described experiments, but it is not exhaustive. For instance, the setup of transplantation experiments, number of transplanted cells, and irradiation regimens can also influence clonal counts.
Although the setup of our experiments, using a conservative 0.5% detection threshold, would preclude detection of >200 (equally represented) barcodes, we would argue that an exact count of HSCs in complex polyclonal situations is neither possible nor relevant. First, there is the problem of discrimination between signal and noise at the very small clone sizes. We expect that some false positive and false negative clones will persist through any statistical filtering of noise. Second, the question of biologically relevant clone size has to be addressed. Large clones are most important, whereas small clones are biologically less relevant. We expect that some small contributors identified in our study would not satisfy the stem cell definition accepted in single-cell transplantation studies.
In conclusion, our data document heterogeneity in multiple aspects of HSC functioning and demonstrate how HSC clones comanifest themselves in polyclonally reconstituted recipient animals. Further studies will demonstrate how such HSC behavior can be influenced by hematopoietic stress or can contribute to the development of blood malignancies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank H. Moes, G. Mesander and R.-J. van der Lei for expert cell sorting assistance; E. Weersing, E. Wojtowicz, and L. Bosman for technical assistance; B. Dykstra and H. Schepers for valuable discussions, suggestions, and assistance in the laboratory; and P. van der Vlies and J. Bergsma for assistance with high-throughput sequencing.
This work was supported by grants from the Netherlands Organization for Scientific Research (G.d.H.) and TopTalent (E.V.), the National Roadmap for Large Scale Infrastructure (Mouse Clinic for Cancer and Aging), and the Netherlands Institute for Regenerative Medicine.
Authorship
Contribution: E.V., L.V.B., and G.d.H. designed research; E.V., M.J.C.B., M.R., and R.v.O. performed research; E.V., L.V.B., and E.Z. analyzed and interpreted data; and E.V., L.V.B., and G.d.H. wrote the manuscript with contributions from R.v.O.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Leonid V. Bystrykh, Laboratory of Ageing Biology and Stem Cells, European Research Institute for the Biology of Ageing, University Medical Centre Groningen, University of Groningen, Antonius Deusinglaan 1, Groningen 9713 AV, The Netherlands; e-mail: l.bystrykh@umcg.nl; and Gerald de Haan, Laboratory of Ageing Biology and Stem Cells, European Research Institute for the Biology of Ageing, University Medical Centre Groningen, University of Groningen, Antonius Deusinglaan 1, Groningen 9713 AV, The Netherlands; e-mail: g.de.haan@umcg.nl.