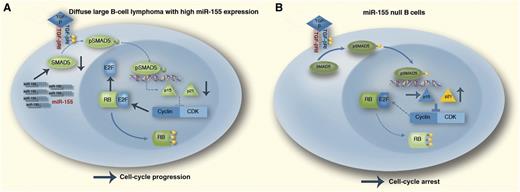
In this issue of Blood, Jiang and Aguiar1 present a novel mechanism in diffuse large B-cell lymphoma (DLBCL) cell lines by which microRNA (miR)-155 deregulate the crucial retinoblastoma protein (RB)/E2F cascade by repressing SMAD5. The resulting hyperphosphorylated RB is inactive and mediates unrestricted cell-cycle progression. Conversely, they demonstrate in mature B lymphocytes from miR-155 knockout (KO) mice elevated SMAD5 levels accompanied by a hypophosphorylated RB state and a more pronounced cell-cycle arrest. This might contribute to the reduced numbers of germinal center B cells and impaired T cell–dependent antibody response found in these mice.1
The TGF-β1–pRB axis in distinct miR-155 contexts. Outcome of the TGF-β1 activation of SMAD5 in DLBCL (A), which often expresses high levels of miR-155. In these malignant cells, miR-155 will markedly suppress SMAD5 expression, resulting in defective p15/p21 transcriptional induction and limited inhibition of the cyclin-dependent kinase (CDK)–cyclin complex–mediated phosphorylation of RB. The resulting hyperphosphorylated (inactive) RB allows tumor cells to progress through the cell cycle irrespective of prohibitive extracellular TGF-β signals instructing G0/G1 arrest. Mature B cells lacking miR-155 (B). In this instance, elevated SMAD5 expression sensitizes the cells to TGF-β1–mediated induction of p15/p21. The high expression of these CDK inhibitors blocks the CDK–cyclin complex and the hypophosphorylated (active) RB promotes cell-cycle arrest. This scenario may explain the defective germinal center B-cell development found in miR-155 null mice. In this diagram, stimulatory interactions are indicated with →; inhibitory interactions with ⊥. Professional illustration by Marie Dauenheimer.
The TGF-β1–pRB axis in distinct miR-155 contexts. Outcome of the TGF-β1 activation of SMAD5 in DLBCL (A), which often expresses high levels of miR-155. In these malignant cells, miR-155 will markedly suppress SMAD5 expression, resulting in defective p15/p21 transcriptional induction and limited inhibition of the cyclin-dependent kinase (CDK)–cyclin complex–mediated phosphorylation of RB. The resulting hyperphosphorylated (inactive) RB allows tumor cells to progress through the cell cycle irrespective of prohibitive extracellular TGF-β signals instructing G0/G1 arrest. Mature B cells lacking miR-155 (B). In this instance, elevated SMAD5 expression sensitizes the cells to TGF-β1–mediated induction of p15/p21. The high expression of these CDK inhibitors blocks the CDK–cyclin complex and the hypophosphorylated (active) RB promotes cell-cycle arrest. This scenario may explain the defective germinal center B-cell development found in miR-155 null mice. In this diagram, stimulatory interactions are indicated with →; inhibitory interactions with ⊥. Professional illustration by Marie Dauenheimer.
Despite recent advances in the understanding of the molecular pathogenesis of lymphomas, clinical parameters are still used to identify patients at risk receiving immunochemotherapy. A major step in deciphering molecular events in lymphomagenesis was the discovery of microRNAs (miRNA) and their causal involvement in multiple cancerous processes and tumor types. miRNA are small noncoding RNAs that posttranscriptionally regulate expression of target RNAs by reducing their stability. Mature miRNA are single-stranded molecules of 20-23 nucleotides in length that regulate gene expression in many physiological and pathological processes. In cancer, including lymphomas, miRNA dysregulation is a ubiquitous phenomenon; however, their exact functional role in lymphomagenesis has not been thoroughly investigated in many instances.2
Dysregulation of a single miR-155 in transgenic mice was the first report to prove that miRNA can cause cancer. Overexpression of miR-155 was sufficient to develop polyclonal lymphoid proliferation followed by frank B-cell malignancies.3 More recently, the underlying mechanisms for the transforming events have been described: the Src homology 2 domain-containing inositol-5-phophatase (SHIP) and the CCAAT enhancer–binding protein β (C/EBPβ) are 2 important direct targets of miR-155. By downregulating C/EBPβ and SHIP, the interleukin-6 pathway is liberated, which in turn generates a block in the B-cell differentiation and favors the accumulation of proliferating apoptosis-resistant pre-B lymphocytes.4 Another publication reported that the ectopic expression of miR-155 in hematopoietic stem cells by retroviral infection causes a myeloproliferative disorder, corroborating the profound oncogenic role of miR-155 in hematopoietic tissues.5 Subsequently, miR-155 has also been shown to play a key role in lymphocyte biology and function of the immune system. miR-155–deficient mice are immunocompromised and B cells generate reduced immunoglobulin levels following antibody treatment.6 Furthermore, total numbers of germinal center-B (GC-B) cells are also significantly reduced. Finally, higher expression of miR-155 in aggressive lymphomas is associated with an adverse outcome.7
The article by Jiang and Aguiar is a continuation of previous observations by the same group identifying the transcription factor SMAD5 as a direct target of miR-155 and defining a novel mechanism adopted by lymphoma cells to escape transforming growth factor-β (TGF-β) growth inhibitory effects.8 In their actual work, the authors precisely dissect the molecular events of miR-155–mediated disruption of the TGF-β–SMAD5 pathway in normal and malignant B lymphocytes. TGF-β exerts tumor suppressive effects in normal and malignant B lymphocytes predominantly through upregulation of the cyclin-dependent kinase inhibitors p15 (CDKN2B) and p21 (CDKN1A) and subsequent cell-cycle arrest. In stably expressed miR-155 DLBCL cell lines, the authors detected lower levels of SMAD5 and showed that stable expression of miR-155 could limit the TGF-β–induced G0/G1 arrest in a SMAD5-dependent fashion. Also, after TGF-β stimulation, they found increased RB phosphorylation and a significantly higher amount of free E2F1 in miR-155–expressing lymphoma cells compared with their empty vector control cells. As a consequence, the effects exerted by TGF-β exposure (ie, enhanced pRB-E2F1 complex formation and a hypophosphorylated RB state leading to cell-cycle arrest) are antagonized by miR-155. By an RNA interference (RNAi) strategy, the authors demonstrated that a specific SMAD5 knockdown following TGF-β signaling phenocopies miR-155 expression in DLBCL, underscoring the crucial role of this transcription factor in mediating TGF-β signals. Subsequently, in an RNAi experiment targeting the downstream mediators p15 and p21 in miR-155 overexpressing DLBCL cells exposed to TGF-β, p15 and p21 downregulation significantly reduced the TGF-β effects on RB phosphorylation. Together, these results support a model in which miR-155 attenuates the tumor suppressive properties of TGF-β by inhibiting SMAD5, resulting in a diminished transcriptional induction of p15/p21 and limited inhibition of the CDK-cyclin complex–mediated RB phosphorylation. The resulting hyperphosphorylated (inactive) RB allows tumor cells to progress through the cell cycle irrespective of prohibitive extracellular TGF-β signals (see figure). The authors also studied normal B-cell development by using mature B cells purified from the spleen of miR-155 wild-type (WT) or KO mice. They were able to demonstrate that the B cells from miR-155 null mice had higher expression of SMAD5. Upon TGF-β exposure, mature B cells from the miR-155 KO mice induced a more pronounced cell-cycle arrest compared with their WT counterpart. This higher sensitivity to TGF-β signals with a higher amount of resting KO cells might serve as explanatory model for the B-lymphocyte deficiency observed in miR-155 null mice (see figure).
In summary, these findings have important implications for DLBCL and normal B-cell development: miR-155 deregulation represents a new model to suppress RB in aggressive lymphoma cells leading to unrestrained cell-cycle progression (and poor outcome). Of interest, recent publications underscore the importance of the perturbed cell-cycle regulation machinery and demonstrate the sensitivity of DLBCL cell lines to targeted therapy by a pan CDK inhibitor.9,10 Further, the noncanonical TGF-β signaling (ie, the phosphorylation and activation of SMAD1/5 by TGF-β) appears to play a bigger role in controlling TGF-β signals than initially thought. Finally, targeting SMAD5 may at least partially explain the deficient generation of B cells in miR-155 KO mice. However, one should also clearly emphasize the limitations of this study, especially that patient samples were not analyzed. Thus, further experiments to demonstrate the relevance of these findings in humans, particularly whether miR-155 expression impacts RB function and E2F target gene expression, are warranted.
Conflict-of-interest disclosure: The authors declare no competing financial interests.