Key Points
Expression analysis of novel potential regulatory epigenetic factors in hematopoiesis.
Abstract
Hematopoietic differentiation is governed by a complex regulatory program controlling the generation of different lineages of blood cells from multipotent hematopoietic stem cells. The transcriptional program that dictates hematopoietic cell fate and differentiation requires an epigenetic memory function provided by a network of epigenetic factors regulating DNA methylation, posttranslational histone modifications, and chromatin structure. Aberrant interactions between epigenetic factors and transcription factors cause perturbations in the blood cell differentiation program that result in various types of hematopoietic disorders. To elucidate the contributions of different epigenetic factors in human hematopoiesis, high-throughput cap analysis of gene expression was used to build transcription profiles of 199 epigenetic factors in a wide range of blood cells. Our epigenetic transcriptome analysis revealed cell type– (eg, HELLS and ACTL6A), lineage- (eg, MLL), and/or leukemia- (eg, CHD2, CBX8, and EPC1) specific expression of several epigenetic factors. In addition, we show that several epigenetic factors use alternative transcription start sites in different cell types. This analysis could serve as a resource for the scientific community for further characterization of the role of these epigenetic factors in blood development.
Introduction
In hematopoiesis, different types of blood cells are produced from hematopoietic stem cells (HSCs). The complexity of the hematopoietic pathway poses a major challenge in delineating the role of transcription factors and epigenetic regulators in determining cell differentiation, cell fate, and lineage commitment. Understanding the mechanisms that regulate normal hematopoietic differentiation is necessary to determine the causes of hematopoietic pathologies.
In humans, hematopoiesis can be broadly categorized into 2 major lineages: the lymphoid lineage, including B cells, T cells, and natural killer cells, and the myeloid lineage, composed of neutrophils, basophils, eosinophils (collectively called granulocytes), monocytes, macrophages, megakaryocytes, platelets, and erythrocytes. Other blood cells have more ambiguous lineage origins such as dendritic cells (DCs) that can have either myeloid (DC monocytes) or lymphoid (DC plasmacytoid) origin and mast cells, suggested to originate from the myeloid lineage.1
The roles of transcription factors and cytokine signaling in regulating hematopoiesis have been of interest in the recent past.2,3 Lately, epigenetic factors, which can be broadly classified into DNA and chromatin modifiers, have received attention as possible regulators of hematopoiesis. DNA methylation is known to modulate transcription, imprinting, X-chromosome inactivation, and chromatin stability during development and disease.4,5 In humans, there are 3 DNA methyltransferases: 2 de novo methylation enzymes (DNMT3A and B) and 1 maintenance enzyme (DNMT1). Also, several putative DNA demethylases have recently been discovered and may together with DNMTs contribute to control of methylation status and transcriptional regulation.6 Cancers, including hematopoietic malignancies, commonly have perturbed global DNA methylation patterns, contributing to silencing of tumor suppressor genes and activation of oncogenes.5
Chromatin modifiers can be classified into enzymes regulating covalent histone modifications and chromatin remodeling complexes (CRCs) that control chromatin structure dynamics. Chromatin consists of repeating units of nucleosomes, each composed of 147 bp of DNA wrapped around an octameric core of the histone proteins H2A, H2B, H3, and H4. The N-terminal tails of the histones undergo multiple dynamic post-translational modifications that regulate various aspects of chromatin biology, including gene expression. By contrast, CRCs regulate chromatin structure by repositioning, assembling, or disassembling nucleosomes and by exchanging core histones with histone variants.7
Hematologic malignancies can be caused by misregulation of chromatin-modifying enzymes. For example, the genes encoding the histone acetyltransferases, p300 and CBP, are commonly rearranged by chromosomal translocations in leukemia.8 In addition, leukemic cells have been found to have global changes of specific histone modifications.9 Although the role of CRCs in blood development and disease has not been studied extensively, there are strong indications that these enzymes could be major players in human hematopoiesis.10-12 It has been suggested that the SWI/SNF complex is critical for granulopoiesis,13 and overexpression of SMARCA5 may affect normal differentiation of CD34+ progenitors of patients with leukemia.14 Another important family of epigenetic factors involved in normal and leukemic hematopoiesis is the Polycomb group proteins (PcGs). The PcG complexes regulate gene expression through their interactions with DNMTs, histone methyltransferases/demethylases, and histone deacetylases and thereby modulate the proliferation, differentiation, and survival of HSCs.15
In this study, we performed expression profiling of a comprehensive list of epigenetic factors using high-throughput HeliScope cap analysis of gene expression (CAGE) analysis of different hematopoietic cell types.16 HeliScope CAGE is a more sensitive, reliable, and reproducible method compared with microarrays to quantify cDNAs.17 Using this technique and systematic data mining, we found several epigenetic factors are differentially expressed in lineage- and cell-specific patterns, suggesting the existence of characteristic epigenetic regulatory circuits in hematopoiesis. We also identified genes with cell-specific use of alternative transcription start sites (TSSs). Our results highlight potentially novel regulatory functions of epigenetic factors in hematopoiesis.
This work is part of the Functional Annotation Of The Mammalian Genome 5 (FANTOM5) project. Data downloads, genomic tools, and copublished manuscripts are summarized at http://fantom.gsc.riken.jp/5/.
Methods
This study was conducted with approval from the Stockholm Ethical Committee, and informed consent was obtained in accordance with the Declaration of Helsinki.
Hematopoietic cell isolation
Description of types of hematopoietic cells used in this study and their isolation procedures are explained in supplemental Methods on the Blood Web site and http://fantom.gsc.riken.jp/5/. The CAGE library IDs for normal hematopoietic cells and leukemic cell lines are listed in supplemental Table 4.
CAGE
Expression profiling was performed with single molecule sequencing Heliscope CAGE sequencing.15,17 Tag clusters (TCs) were generated for all CAGE libraries16 and normalized to tags per million (TPM). Expression values of robustly expressed TCs within 500 bp of TSSs of RefSeq18 gene models were summed based on the corresponding gene ID. Relative log normalization (RLE) was performed to more correctly compare libraries of different sizes.19
In silico analysis
Data analysis was performed using R (http://www.rproject.org), version 2.15.1. TPM values were analyzed by principal component analysis (PCA) with the “prcomp” function with scaling of variance. For cluster analysis and heatmaps, TPM values were increased by 1 unit and log10 transformed. The “hclust” function, default settings, was used for unsupervised hierarchical clustering. Heatmaps were constructed with the “heatmap.2” function of the “gplots” package, and colors were applied with the “colorRamps” package. To identify differentially expressed genes, CAGE count data were analyzed with the Bioconductor (http://www.bioconductor.org) package “edgeR” using the “exactTest” function. Expression data for the obtained candidate genes were inspected manually. P values for comparisons of gene expression between cells/lineages were calculated using a 2-sided Mann-Whitney U test, using the average score approach for ties.
For analysis of alternative TSSs, TCs within 500 bases from annotated start sites (RefSeq) of epigenetic genes were investigated. Genes with a minimum of 2 TSSs with an average TPM ≥ 10 in any cell type is available in supplemental Table 5. The normalization procedure is described in supplemental Methods.
Quantitative reverse transcription-polymerase chain reaction
cDNA was synthesized using maxima first-strand cDNA synthesis kit (Fermentas) and was analyzed by quantitative polymerase chain reaction (qPCR) with normalization against actin expression. For details, see supplemental Methods.
Results
Hematopoietic cell lineages cluster together based on the expression profiles of epigenetic factors
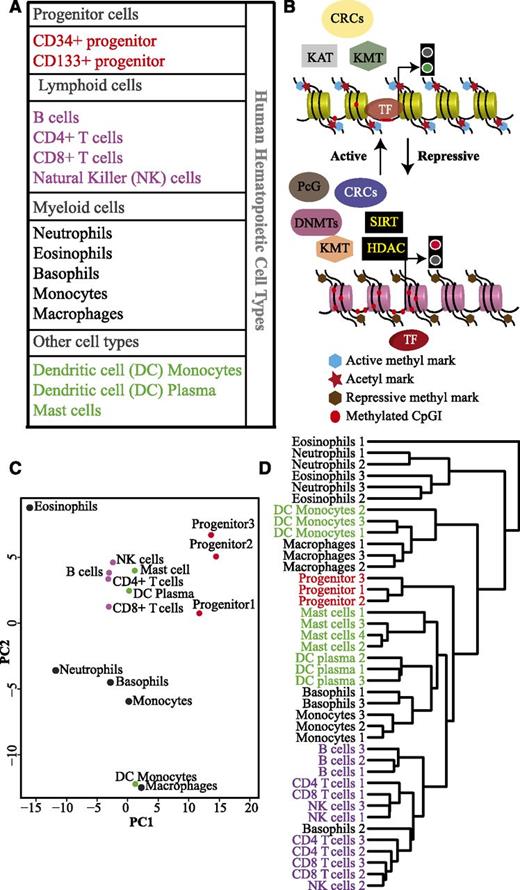
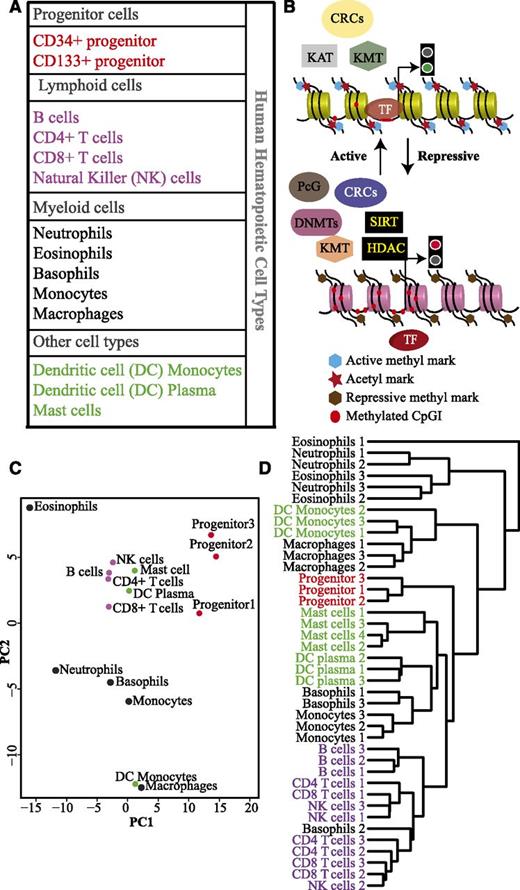
In this study, we analyzed expression profiles of 199 epigenetic factors in 14 different blood cell types (Figure 1A). The progenitor populations were treated as independent samples, because they were isolated based on different surface markers. Transcription profiles of each cell type were created using CAGE analysis, a high-throughput method for sequencing 5′ capped RNAs.17,20,21 The epigenetic factors investigated in this study were classified into functional categories: DNA methyltransferases (DNMTs) and putative DNA demethylases, histone/lysine methyltransferases (HMTs/KMTs), lysine demethylases (KDMs), histone/lysine acetyltransferases (HATs/KATs), histone deacetylases (HDACs), chromatin remodeling complexes (CRCs), histone chaperones, and polycomb group proteins (PcGs) (Figure 1B; supplemental Table 1). To identify similarities and differences in the expression profile of the epigenetic regulators in different hematopoietic cell types, PCA was performed. Interestingly, we observed distinct clusters for the progenitor cells and the lymphoid cells (Figure 1C). Myeloid cell types, however, do not form a single cluster. Basophils and monocytes cluster together, whereas eosinophils cluster with neutrophils in the first component but are separated in the second component. The monocyte-derived cell types, DC monocytes, and macrophages form a separate cluster. Notably, DC plasma cells cluster together with lymphoid cells and DC monocytes with myeloid cells indicating that these cells retain an epigenetic memory of their origin. The 2 different progenitor cell populations cluster together, and therefore, their expression values were averaged and termed as progenitor in some of the subsequent graphs.
Hematopoietic cell types cluster together based on the expression of different epigenetic factors. (A) Table listing the different hematopoietic cell types used in this study. Progenitor cells, lymphoid, and myeloid lineages are shown in red, purple, and black, respectively. Mast cells and dendritic cells (DCs) of myeloid and lymphoid origin are indicated in green. (B) A general scheme of interdependence of different classes of epigenetic factors involved in regulating gene expression. Some of these factors modulate DNA/histone modifications, whereas chromatin remodeling complexes (CRCs) alter chromatin structure to either expose or conceal transcription factor (TF) binding sites. The polycomb complexes interact with DNA/histone modifying factors to regulate the gene expression at the chromatin level. (C) Principal component analysis (PCA) based on expression levels of 199 epigenetic factors in different hematopoietic cell types. Progenitor cells (red dots), lymphoid cells (purple dots), myeloid cells (black dots), mast cells, DC plasma, and DC monocytes (green dots). (D) Unsupervised hierarchical cluster analysis of the different hematopoietic cell types from individual donors (biological replicates) based on the expression of 199 epigenetic factors.
Hematopoietic cell types cluster together based on the expression of different epigenetic factors. (A) Table listing the different hematopoietic cell types used in this study. Progenitor cells, lymphoid, and myeloid lineages are shown in red, purple, and black, respectively. Mast cells and dendritic cells (DCs) of myeloid and lymphoid origin are indicated in green. (B) A general scheme of interdependence of different classes of epigenetic factors involved in regulating gene expression. Some of these factors modulate DNA/histone modifications, whereas chromatin remodeling complexes (CRCs) alter chromatin structure to either expose or conceal transcription factor (TF) binding sites. The polycomb complexes interact with DNA/histone modifying factors to regulate the gene expression at the chromatin level. (C) Principal component analysis (PCA) based on expression levels of 199 epigenetic factors in different hematopoietic cell types. Progenitor cells (red dots), lymphoid cells (purple dots), myeloid cells (black dots), mast cells, DC plasma, and DC monocytes (green dots). (D) Unsupervised hierarchical cluster analysis of the different hematopoietic cell types from individual donors (biological replicates) based on the expression of 199 epigenetic factors.
To test the reproducibility of our data, we repeated the PCA analysis using the expression profiles of 157 of our 199 epigenetic factors present in the Novershtern et al dataset22 (supplemental Figure 4). Both CAGE and microarray data consisting of 199 and 157 epigenetic factors, respectively, showed similar PCA clustering. We also used the CAGE data to perform unsupervised hierarchical clustering based on the expression of epigenetic factors (Figure 1D; supplemental Figure 2). This analysis resulted in a separation of different blood lineages similar to that previously shown when performing clustering based on global transcriptome data.22 Thus, both PCA and cluster analysis provide strong indications that progenitors, lymphocytes, and myeloid cells have distinct expression profiles of epigenetic regulators.
Differential expression of DNA modifying factors in hematopoietic cells
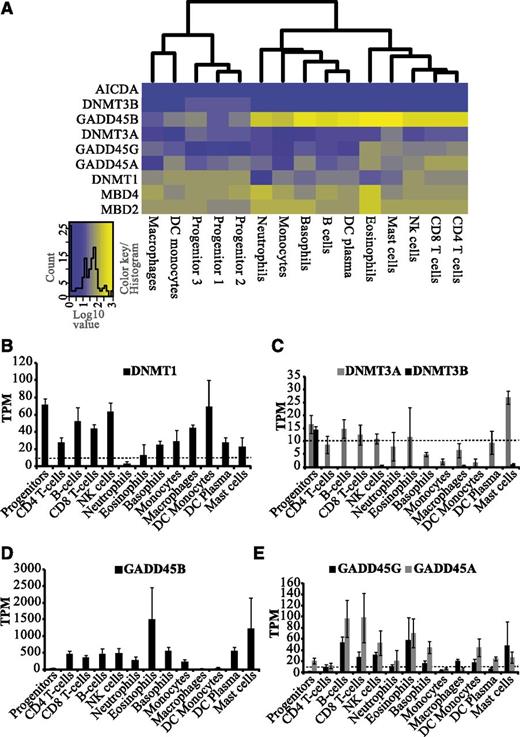
DNA methylation has been demonstrated to play a vital role in hematopoiesis and HSC self-renewal.23 Our analysis reveals distinct expression patterns of DNMT1, DNMT3A, and DNMT3B in hematopoietic cells. DNMT1 expression was relatively high in progenitors, T lymphocytes, natural killer cells, macrophages, and DC monocytes and low in granulocytes (Figure 2A-B). Expression of DNMT3A was low in monocytes, macrophages, and DC monocytes in comparison with the other cell types (Figure 2C). In contrast, DNMT3B expression was only observed in progenitors (Figure 2C; supplemental Figure 3). Our data are in agreement with a previous report showing that DNMT3B is predominantly expressed in progenitor cells and down-regulated during differentiation.4
Distinct expression patterns of DNMTs and potential DNA demethylases in hematopoietic cells. (A) Heatmap and clustering based on transcript levels of DNA methyltransferases and putative DNA demethylases showing gene expression levels from low (blue) to high (yellow) (see color key). The heatmaps were constructed using log10-transformed TPM values from CAGE data. For hematopoietic progenitors, individual samples are included because they were isolated based on different surface markers. For other cell types, replicate averages were used. (B-E) Average expression levels (TPM) of DNA methyltransferases (DNMT1, DNMT3A, and DNMT3B) and potential DNA demethylases (GADD45A, B, and G) are shown in hematopoietic cell types. Dotted line represents the threshold of 10 TPM, above which we considered expression to be significant.
Distinct expression patterns of DNMTs and potential DNA demethylases in hematopoietic cells. (A) Heatmap and clustering based on transcript levels of DNA methyltransferases and putative DNA demethylases showing gene expression levels from low (blue) to high (yellow) (see color key). The heatmaps were constructed using log10-transformed TPM values from CAGE data. For hematopoietic progenitors, individual samples are included because they were isolated based on different surface markers. For other cell types, replicate averages were used. (B-E) Average expression levels (TPM) of DNA methyltransferases (DNMT1, DNMT3A, and DNMT3B) and potential DNA demethylases (GADD45A, B, and G) are shown in hematopoietic cell types. Dotted line represents the threshold of 10 TPM, above which we considered expression to be significant.
Apart from DNMTs, several potential demethylases were also differentially expressed (Figure 2A). The GADD45 proteins have been implicated in DNA demethylation.6 In general, GADD45B transcripts were found to be several-fold higher in blood cells compared with GADD45A and GADD45G (Figure 2D-E). Progenitors, DC monocytes, and macrophages express negligible levels of GADD45B but higher levels of other putative DNA demethylases, such as MBD2 and MBD424 (Figure 2A,D-E; supplemental Figure 4). Transcription profiling of granulopoietic cells has previously shown that GADD45B is highly expressed in neutrophils, whereas GADD45A peaks in metamyleocytes,25 suggesting distinct roles of GADD45A and GADD45B during hematopoiesis.
Cell type–specific expression of histone modifying enzymes in hematopoietic cells
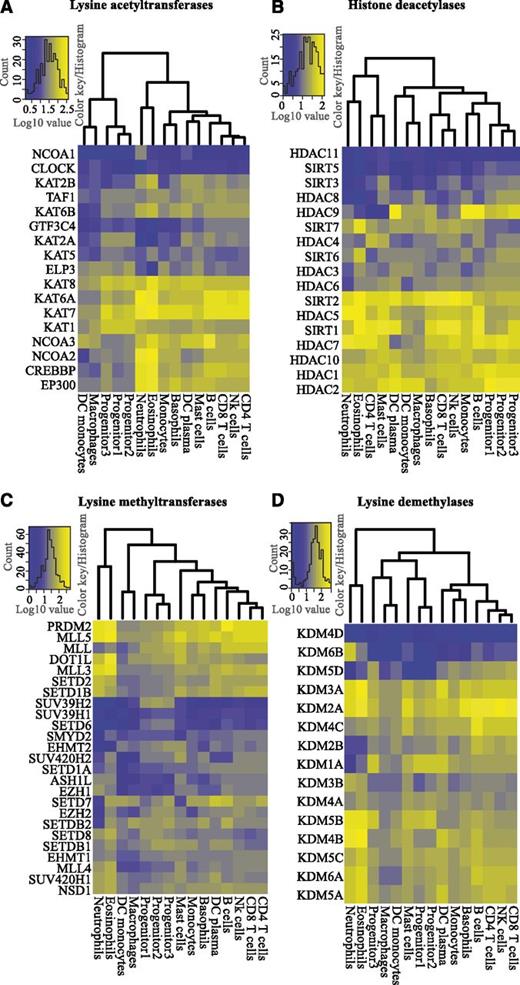
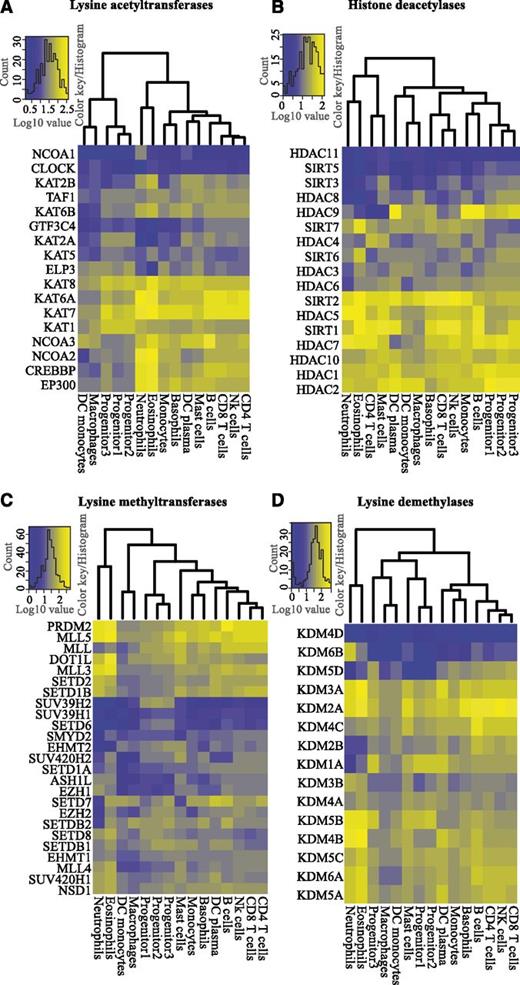
To understand the potential roles of histone-modifying enzymes in hematopoiesis, we performed cluster analysis and constructed heatmaps to determine expression patterns of KATs, HDACs, KMTs, and KDMs in blood cells (Figure 3A-D). We analyzed the expression levels of 17 KATs and found that KAT1, KAT7, and KAT8 were the most prominent transcripts in progenitors, whereas CREBP and EP300 were mainly expressed in mature cell types (Figure 3A; supplemental Figure 5B). Interestingly, NCOA3 and NCOA2 were highly expressed in B cells and neutrophils/eosinophils, respectively, indicating a cell type–specific switch of NCOA expression (supplemental Figure 5C). We performed a similar cluster analysis for 17 HDACs (Figure 3B). HDAC1, HDAC2, HDAC10, SIRT1, HDAC5, and SIRT2 were ubiquitously expressed, suggesting a general role for these enzymes in deacetylation of histones and other proteins in hematopoiesis (Figure 3B), whereas HDAC11 and SIRT5 were not expressed in blood cells. We also observed cell type–specific expression of HDACs. HDAC4, an enzyme overexpressed in childhood T-cell acute lymphoid leukemia (T-ALL)26 and acute promyelocytic leukemia (APL),27 was expressed in T cells and mast cells, whereas HDAC9, associated with childhood acute lymphoid leukemia (ALL),26 was primarily transcribed in B cells, monocytes, and DC plasma cells (supplemental Figure 5D-E).
Differential expression of histone modifying enzymes. Unsupervised hierarchical clustering and heatmaps of the expression levels (log10-transformed TPM values) of histone-modifying enzymes in hematopoietic cells, divided into (A) lysine acetyl transferases (KATs), (B) histone deacetylases (HDACs), (C) lysine methyl transferases (KMTs), and (D) lysine demethylases (KDMs).
Differential expression of histone modifying enzymes. Unsupervised hierarchical clustering and heatmaps of the expression levels (log10-transformed TPM values) of histone-modifying enzymes in hematopoietic cells, divided into (A) lysine acetyl transferases (KATs), (B) histone deacetylases (HDACs), (C) lysine methyl transferases (KMTs), and (D) lysine demethylases (KDMs).
Clustering of CAGE data for KMTs revealed several noteworthy patterns (Figure 3C; supplemental Figure 6). PRDM2 and MLL5, 2 potential tumor suppressors,28,29 were transcribed in all blood cells. However, their expression was higher in most mature blood cells relative to progenitors (supplemental Figure 6A). MLL5 has been implicated to play a role in myeloid malignancies, and MLL5 deletion in mice causes multiple problems in blood cell development.29-31 PRDM2 expression was significantly higher in B cells relative to other lymphoid cells (Figure 3C; supplemental Figure 6A). Interestingly, PRDM2 knockout mice show high incidences of diffuse large B-cell lymphomas and leukemia.28 Dot1L, a H3K79 methyltransferase, is expressed in most human blood cell types (Figure 3C; supplemental Figure 6B). Consistent with this, studies in conditional Dot1L knockout mice have demonstrated its requirement for normal development of all blood lineages.32 In contrast to Dot1L, the expression of SUV39H2 (a H3K9 histone methyltransferase) is specific for progenitor cells (Figure 3C; supplemental Figure 6C).
KDMs are known to be involved in normal and malignant hematopoiesis. KDM1A, a H3K4 demethylase, interacts with transcription factors essential for normal hematopoiesis.33 We find that KDM1A is expressed in all cell types, except in neutrophils and eosinophils (Figure 3D; supplemental Figure 6D). KDM1A overexpression leads to a differentiation block in AML,34 and KDM1A knockdown causes impaired development of multiple hematopoietic lineages.35 The H3K27 demethylases, KDM6A (UTX) and KDM6B (Jmjd3), regulate Hox gene expression.36 Despite a functional overlap, their expression profiles suggest individual roles in hematopoieisis. KDM6A is expressed in most blood cell types, whereas KDM6B is expressed at similar levels only in neutrophils (Figure 3D; supplemental Figure 6E). Although KDM6A has been suggested to have a regulatory function in blood development,37 the precise function of KDM6B remains to be investigated. Thus, our findings demonstrate a blood cell type–specific expression pattern of histone-modifying enzymes.
Expression of SNF2 chromatin remodeling ATPases in hematopoietic cell types
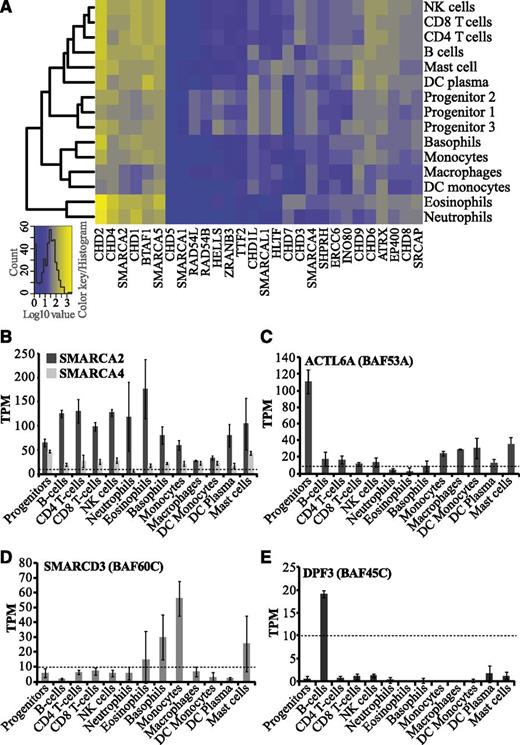
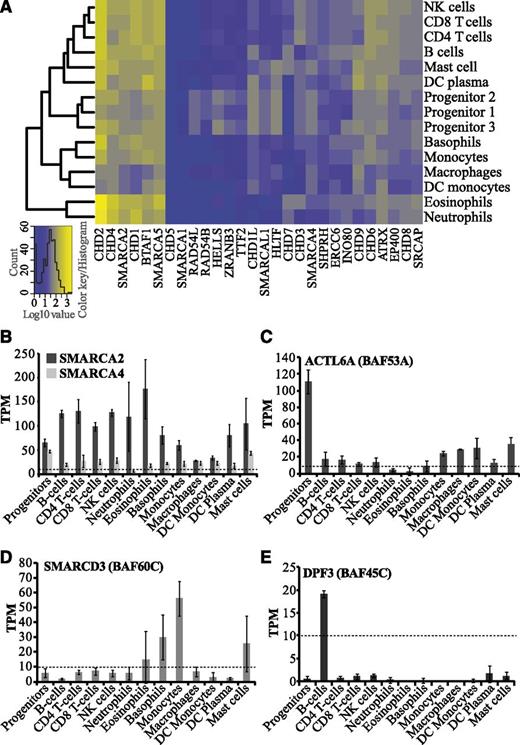
Chromatin remodeling enzymes belong to the SNF2 family, which modulates chromatin structure in an ATP-dependent manner. Aberrant expression and mutations of SNF2 genes have been associated with several developmental disorders and malignancies.7,10 We found that the levels of SNF2 transcripts range from very low/negligible to very high in blood cells, suggesting distinct functions of specific SNF2 enzymes in hematopoiesis (Figure 4A). SMARCA1, the ATPase subunit of the NURF CRC, and CHD5, shows negligible expression in all blood cell types (Figure 4A). In agreement with this, high levels of SMARCA1 and CHD5 have previously been reported only in neuronal tissues.38,39 Several SNF2 ATPases such as BTAF1, CHD2, CHD4, CHD1, SMARCA2, and SMARCA5 are abundantly expressed in most hematopoietic cells. Although CHD4,40 SMARCA2,10 and SMARCA514 have already been reported to be involved in hematopoiesis or leukemia, the functional roles of BTAF1, CHD2, and CHD1 remain elusive.
Expression profiles of SNF2 ATPases and BAF complex subunits in hematopoietic cells. (A) Cluster analysis and heatmap of transcript levels (log10-transformed TPM values) for SNF2 ATPases. (B-E) Bar graphs showing average TPM values for human BAF (SWI/SNF) subunits, which include (B) catalytic subunits (SMARCA2/BRM and SMARCA4/BRG1), (C) ACTL6A (BAF53A), (D) SMARCD3 (BAF60C), and (E) DPF3 (BAF45C). Error bars display standard deviations.
Expression profiles of SNF2 ATPases and BAF complex subunits in hematopoietic cells. (A) Cluster analysis and heatmap of transcript levels (log10-transformed TPM values) for SNF2 ATPases. (B-E) Bar graphs showing average TPM values for human BAF (SWI/SNF) subunits, which include (B) catalytic subunits (SMARCA2/BRM and SMARCA4/BRG1), (C) ACTL6A (BAF53A), (D) SMARCD3 (BAF60C), and (E) DPF3 (BAF45C). Error bars display standard deviations.
The SNF2 ATPases HELLS, ZRANB3, RAD54B, and RAD54L were specifically expressed in progenitor cells (Figure 4A; supplemental Figure 3A-B). HELLS regulates de novo DNA methylation through its interaction with DNMT3B, which also shows progenitor-specific expression in our data (Figure 2C; supplemental Figure 3). Although its precise role is yet to be determined, our results suggest that HELLS may be involved in regulation of human hematopoiesis, possibly together with DNMT3B, because both of these enzymes have a progenitor-specific expression pattern (Figure 2B; supplemental Figure 3).
Combinatorial expression of BAF complex subunits in hematopoietic lineages
The mammalian SWI/SNF complex, also called BAF (BRG1 [SMARCA4] or BRM [SMARCA2] associated factors), can have diverse subunit compositions, which determines its function in establishment and maintenance of cell fate.41 The ATPase subunits, SMARCA4 and SMARCA2, are 2 mutually exclusive catalytic cores of the BAF complex.10 They can associate with different auxiliary subunits to form distinct complexes specific either to different cell/tissue types or to developmental stage.42 Our analysis shows high levels of SMARCA2 in lymphocytes and granulocytes compared with SMARCA4, suggestive of the existence of a predominant form of BAF complex with a SMARCA2 catalytic core in hematopoietic cells (Figure 4B; supplemental Table 1). A SMARCA4-containing BAF complex is involved in development of both T cells and granulocytes.13,43 However, we cannot rule out the existence of 2 distinct forms of BAF complexes, one with SMARCA2 and the other with SMARCA4, in the hematopoietic system. These complexes could have different regulatory functions by interacting with different sets of transcription factors as previously suggested.44
The embryonic stem cell BAF complex (esBAF) consists of the exchangeable accessory subunits ACTL6A (BAF53A), SMARCD2 (BAF60B), and PHF10 (BAF45A), along with other subunits. During neuronal development, these subunits are exchanged for ACTL6B (BAF53B), SMARCD3 (BAF60C), and DPF3 (BAF45C).41 Our data indicate different subunit compositions of the BAF complex in different hematopoietic cell types. We observed higher levels of ACTL6A transcripts in progenitor cells compared with differentiated cells (Figure 4C; supplemental Figure 3A), possibly mimicking the subunit composition of the esBAF complex. It has recently been demonstrated that ACTL6A has a critical role in the survival of hematopoietic progenitors.12 In contrast to ACTL6A, SMARCD3 and DPF3 were found to be expressed exclusively in differentiated cells such as basophils/monocytes and B cells, respectively (Figure 4D-E). We also found several other differentially expressed exchangeable BAF subunits in hematopoietic cell types (supplemental Table 1). This implies that diverse BAF complexes could be involved in regulating cell type–specific gene expression and thereby governing lineage choices.
Hematopoietic lineage-specific expression of epigenetic factors
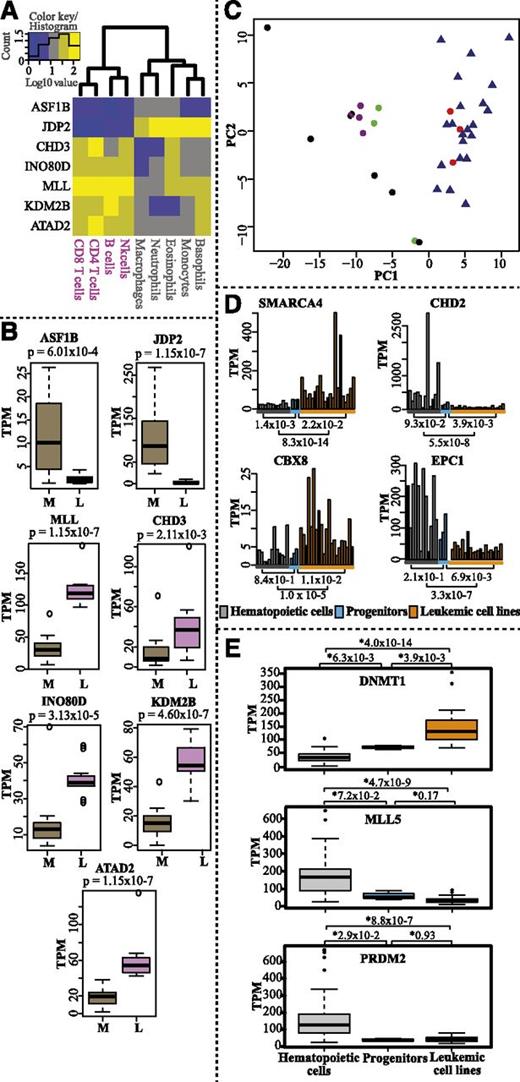
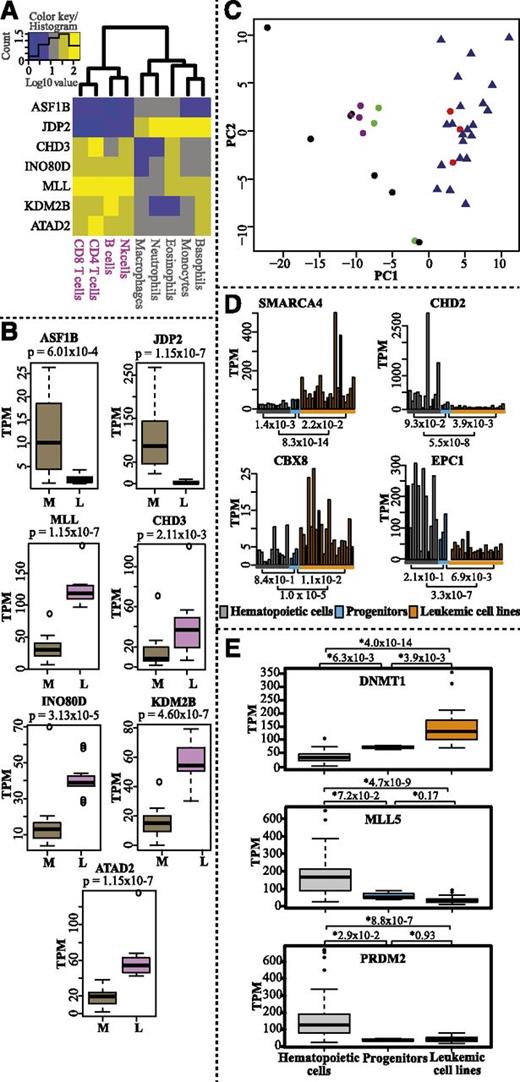
We probed CAGE expression profiles for epigenetic factors that were differentially expressed between the myeloid and the lymphoid lineages. We observed, for example, that ASF1B and JDP2 were highly expressed in the myeloid lineage, whereas CHD3, INO80D, MLL, KDM2B, and ATAD2 were highly expressed in the lymphoid lineage (Figure 5A-B and supplemental Table 2 for complete gene list). ASF1B regulates histone assembly and disassembly in a DNA replication-dependent manner together with CAF-1, whereas JDP2 is a histone chaperone with multiple roles in regulating transcription repression and nucleosome assembly.45 The possible function of ASF1B and JDP2 in myelopoiesis remains to be determined. However, in agreement with our results, 1 study reported that the promoter of JDP2 is hypomethylated and selectively expressed in myeloid cells.46 Mi2/NuRD is a multisubunit CRC with CHD3 (Mi2α) and CHD4 (Mi2β) as catalytic subunits. In the context of the NuRD complex, CHD4 is implicated in B-cell development and normal lineage progression.47 Our analysis indicates that CHD3 may be a determinant for lymphoid lineage choice based on its higher expression levels in the lymphoid lineage compared with the myeloid lineage (Figure 5A-B). Chromosomal aberrations and mutations have identified MLL, an H3K4-specific KMT, as a potent epigenetic regulator of lineage determination.48 Our analysis supports a role for MLL in lymphoid lineage determination. However, the precise mechanism remains to be elucidated.
Hematopoietic lineage-specific expression and comparative expression profiles of epigenetic factors in normal hematopoietic cells, progenitor cells, and leukemic cell lines. (A) Heatmap and hierarchical clustering of genes differentially expressed between myeloid and lymphoid lineages (log10-transformed TPM values). (B) Box plots displaying expression levels (TPM values) of the genes in myeloid (M) and lymphoid (L) lineages. P values were calculated with the Mann-Whitney U test (2 sided). (C) PCA analysis of all progenitor samples (red dots), myeloid cells (black dots), lymphoid cells (purple dots), mast and dendritic cells (green dots), and leukemic cell lines (blue triangles) (supplemental Table 1) based on expression of epigenetic factors (TPM values). (D) Bar charts displaying the average transcript levels (TPM values) of SMARCA4, CBX8, CHD2, and EPC1 in normal mature blood cells (gray bars, average of replicates for each cell type), progenitor replicates (blue bars), and leukemic cell lines (orange bars). For cell lines, single datasets were used. (E) Box plots showing the CAGE expression profiles (TPM values) for DNMT1, MLL5, and PRDM2 in leukemic cell lines compared with normal mature hematopoietic cell types and progenitor cells. P values were calculated using the Mann-Whitney U test (2 sided).
Hematopoietic lineage-specific expression and comparative expression profiles of epigenetic factors in normal hematopoietic cells, progenitor cells, and leukemic cell lines. (A) Heatmap and hierarchical clustering of genes differentially expressed between myeloid and lymphoid lineages (log10-transformed TPM values). (B) Box plots displaying expression levels (TPM values) of the genes in myeloid (M) and lymphoid (L) lineages. P values were calculated with the Mann-Whitney U test (2 sided). (C) PCA analysis of all progenitor samples (red dots), myeloid cells (black dots), lymphoid cells (purple dots), mast and dendritic cells (green dots), and leukemic cell lines (blue triangles) (supplemental Table 1) based on expression of epigenetic factors (TPM values). (D) Bar charts displaying the average transcript levels (TPM values) of SMARCA4, CBX8, CHD2, and EPC1 in normal mature blood cells (gray bars, average of replicates for each cell type), progenitor replicates (blue bars), and leukemic cell lines (orange bars). For cell lines, single datasets were used. (E) Box plots showing the CAGE expression profiles (TPM values) for DNMT1, MLL5, and PRDM2 in leukemic cell lines compared with normal mature hematopoietic cell types and progenitor cells. P values were calculated using the Mann-Whitney U test (2 sided).
Comparative analysis of normal hematopoietic cells with leukemic cell lines
We also analyzed the expression profile of epigenetic factors in 21 leukemic cell lines (supplemental Table 1) and compared them with normal hematopoietic cells. PCA analysis showed that all leukemic cell lines cluster together with progenitor cells (Figure 5C; supplemental Figure 8). This indicates that the progenitors and leukemic cells may share similar epigenetic mechanisms for self-renewal. However, we also identified some genes encoding epigenetic factors with different expression profiles in normal hematopoietic cells, including progenitor cells, and leukemic cell lines (Figure 5D; supplemental Figure 7; supplemental Table 2). The CRCs, SMARCA4 and CHD2, and the PcG proteins CBX8 and EPC1 display different expression levels in leukemic cell lines compared with progenitors and other hematopoietic cells (Figure 5D). Interestingly, CHD2, which is somatically mutated in 8.3% of CLL patients,49 showed higher expression in normal hematopoietic cells than in leukemic cell lines. Although the specific functions of CBX8 and EPC1 in hematopoiesis or in leukemia have not been fully characterized, CBX8 has been shown to be essential for MLL-AF9–induced AML,50 and EPC1 has been shown to be involved in chromosomal translocation in ALL.51 MLL5, PDRM2, and DNMT1 have been reported to be deregulated in cancer. MLL5 and PDRM2 are potential tumor suppressor genes, whereas DNMT1 is known to methylate CpG islands in tumor suppressor gene promoters.4,26,27 Indeed, we observed significantly lower expression of MLL5/PDRM2 and higher expression of DNMT1 in leukemic cell lines compared with normal hematopoietic cells (Figure 5E). In conclusion, we demonstrate expression differences of several epigenetic factors in hematological leukemic cell lines compared with normal hematopoietic cells.
Validation of gene expression patterns of epigenetic factors
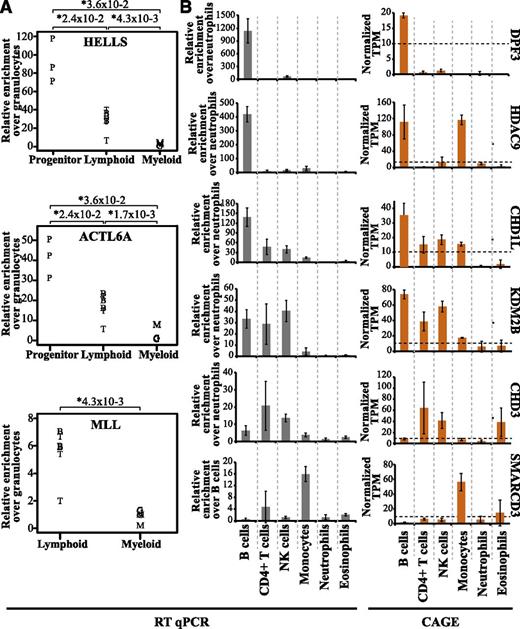
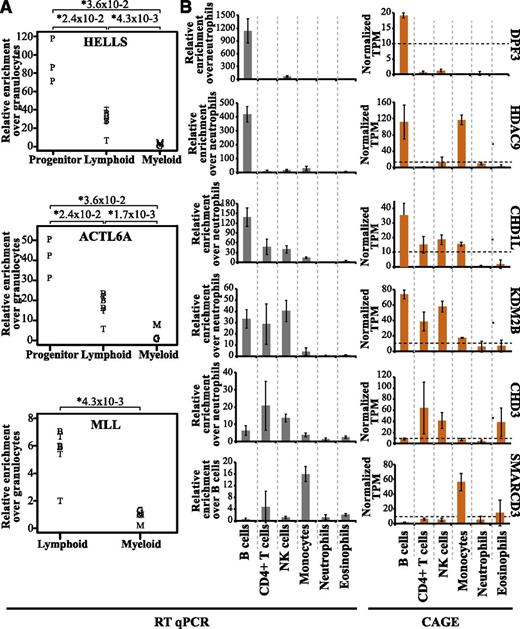
To validate our data, we analyzed the expression of 9 genes (HELLS, ACTL6A, MLL, DPF3, HDAC9, CHD1L, KDM2B, CHD3, and SMARCD3) in selected cell types using qRT-PCR. In agreement with our CAGE results, both HELLS and ACTL6A showed predominant expression in progenitor cells, whereas MLL expression was found to be lymphoid specific (compare Figure 6A with Figure 4C and supplemental Figures 3 and 5B). For DPF3, HDAC9, CHD1L, KDM2B, CHD3, and SMARCD3, the expression pattern between cells types were very similar when comparing CAGE and qRT-PCR analysis (Figure 6B). In addition, we also compared our datasets with the Hematology Expression Atlas (HaemAtlas) generated using mRNA expression array data from several blood cell types.52 Although the HaemAtlas contains fewer cell types, we could confirm analogous gene expression of several epigenetic factors (supplemental Table 3). Collectively, this strengthens the validity of our methodology and data analysis.
Expression patterns of epigenetic factors validated by qRT-PCR. The qRT-PCR transcript levels were normalized to actin gene expression, and the relative fold enrichment was calculated over the cell type with the lowest expression for each gene (y-axis). (A) The relative fold enrichment of transcript levels of HELLS, ACTL6A, and MLL was determined by qRT-PCR. B cells are depicted as B, T cells as T, CD34+ progenitors as P, granulocytes as G, and monocytes as M. (B) Differential expression patterns of 6 epigenetic factors in different cell types validated by qRT-PCR (gray bars) and compared with the CAGE average TPM values (orange bars). The expression of these genes wa validated using total RNA obtained from 3 individual healthy donors separate from those used for CAGE analysis.
Expression patterns of epigenetic factors validated by qRT-PCR. The qRT-PCR transcript levels were normalized to actin gene expression, and the relative fold enrichment was calculated over the cell type with the lowest expression for each gene (y-axis). (A) The relative fold enrichment of transcript levels of HELLS, ACTL6A, and MLL was determined by qRT-PCR. B cells are depicted as B, T cells as T, CD34+ progenitors as P, granulocytes as G, and monocytes as M. (B) Differential expression patterns of 6 epigenetic factors in different cell types validated by qRT-PCR (gray bars) and compared with the CAGE average TPM values (orange bars). The expression of these genes wa validated using total RNA obtained from 3 individual healthy donors separate from those used for CAGE analysis.
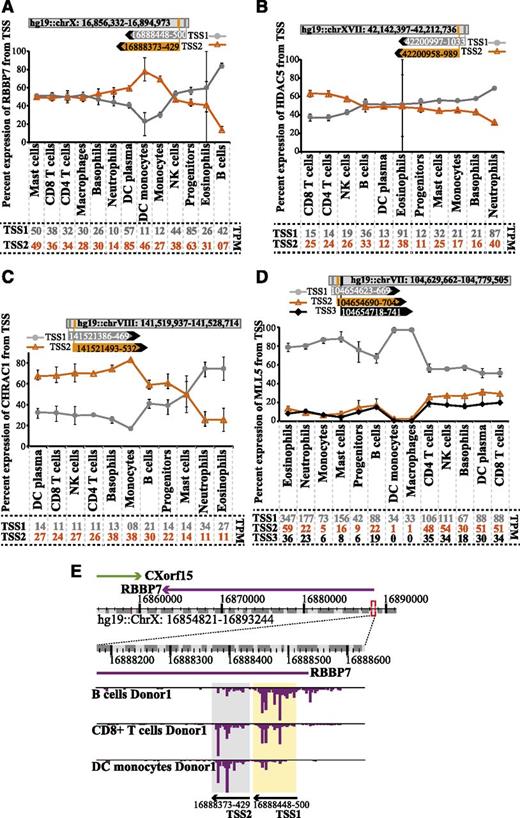
Blood cell type–specific usage of alternative TSSs
CAGE sequencing from the 5′ cap gives base pair resolution of TSSs, allowing for detection of alternative TSSs.17 We identified several epigenetic factors that use alternative TSSs (supplemental Table 5). For some genes such as RBBP7, HDAC5, and CHRAC1 (Figure 7A-C), the dominant TSS was different in different hematopoietic cell types. For other genes, such as MLL5 (Figure 7D), the dominant TSS was the same in all cells but sometimes showed variations in the degree of preference. The alternative TSS for RBBP7 is depicted in the snapshot view of the ZENBU genome browser (Figure 7E).53 TSS1 and TSS2 sites are preferred by B cells and DC monocytes, respectively, whereas CD8+ T cells show uniform use of both TSSs in RBBP7 mRNA expression. Interestingly, most of the other lymphocytes use both TSSs and contribute to the RBBP7 expression (Figure 7A). The different use of alternative TSSs may indicate cell type–specific transcriptional regulation. However, biological outcome of the alternative TSS remains to be elucidated.
Hematopoietic cells show differential preference for alternative TSS use for selected epigenetic factors. (A-D) Graphs displaying the average percent of expression (TPM) from alternative TSS for RBBP7, HDAC5, CHRAC1, and MLL5 in selected cell types. Percentage is based on sum of TSSs >10 TPM in ≥1 cell type. CAGE tag clusters for alternative TSSs for RBBP7 and HDAC5 are mapped on the sense strand, whereas CHRAC1 and MLL5 are mapped to antisense strand TSS1, 2, and 3 (when applicable) and are shown in gray, orange, and black lines, respectively. Error bars shows the minimum and maximum percent expression values for respective TSS and cell type. The coordinates of the gene location and the TSSs are shown on top of each graph (not to scale). Average TPM values for TSSs in each hematopoietic cell type are shown at the bottom of each graph. (E) Example snap shot from ZENBU genome browser showing expression (TPM) of RBBP7 in B cells, CD8+ T cells, and DC monocytes. Expansion of the region surrounding the TSSs in A displays the 2 separate CAGE tag clusters marked in yellow (TSS1) and gray (TSS2).
Hematopoietic cells show differential preference for alternative TSS use for selected epigenetic factors. (A-D) Graphs displaying the average percent of expression (TPM) from alternative TSS for RBBP7, HDAC5, CHRAC1, and MLL5 in selected cell types. Percentage is based on sum of TSSs >10 TPM in ≥1 cell type. CAGE tag clusters for alternative TSSs for RBBP7 and HDAC5 are mapped on the sense strand, whereas CHRAC1 and MLL5 are mapped to antisense strand TSS1, 2, and 3 (when applicable) and are shown in gray, orange, and black lines, respectively. Error bars shows the minimum and maximum percent expression values for respective TSS and cell type. The coordinates of the gene location and the TSSs are shown on top of each graph (not to scale). Average TPM values for TSSs in each hematopoietic cell type are shown at the bottom of each graph. (E) Example snap shot from ZENBU genome browser showing expression (TPM) of RBBP7 in B cells, CD8+ T cells, and DC monocytes. Expansion of the region surrounding the TSSs in A displays the 2 separate CAGE tag clusters marked in yellow (TSS1) and gray (TSS2).
Discussion
In this report, we used CAGE data and systematic analysis to generate a comprehensive map of the expression levels of 199 epigenetic factors in the hematopoietic system. We identified epigenetic factors that were expressed in a cell- or lineage-specific manner and could be potential candidates for cell fate determination. Furthermore, validation of several epigenetic factors in various blood cell types by qPCR strengthens CAGE data analysis. Moreover, we analyzed the expression in leukemic cell lines and found differential expression of several epigenetic factors compared with normal blood cells. Finally, we identify cell-specific alternative TSS use.
DNA methylation is one of the most well-characterized epigenetic mechanisms. It was recently shown to regulate expression of hematopoietic transcription factors such as PU.1 and GATA215 and is known to be involved in regulation of hematopoietic lineage choice.22,45 We found that DNMT1 was weakly expressed in granulocytes compared with the other cell types and that DNMT3B expression was restricted to hematopoietic progenitors consistent with a previous report.4 Interestingly DNMT1 expression was significantly higher in leukemic cell lines, consistent with the findings that the promoters of many tumor suppressor genes are methylated.4 In addition, we identified some putative DNA demethylases that are differentially expressed in different hematopoietic cell types, implying a potential role in hematopoietic differentiation.
Epigenetic regulation by CRCs has been studied for some time, but the complex composition of the CRCs has been brought to light only recently.41 Specific CRCs are known to be assembled with distinct ancillary subunits depending on the developmental program. For example, the human BAF complex changes its subunit composition during the development of neurons from embryonic stem cells.54 Recently, Kkrasteva et al showed that ACTL6A is a crucial BAF subunit for the maintenance of HSCs and hematopoietic progenitor cells.12 Our result suggests different roles of accessory BAF complex subunits in the hematopoietic system as indicated for example by the specific expression of ACTL6A and DPF3 genes in progenitor and B cells, respectively. Thus, our data suggest the existence of a similar mechanism in the hematopoietic system as in neurons.
We also identified epigenetic factors that are specific for either the myeloid or lymphoid lineage, eg, lymphoid-specific MLL and INO80D. Similarly, Allantaz et al demonstrated myeloid-specific expression of the miRNAs mir-27 and mir-223, which target and down-regulate MLL and INO80D mRNAs in myeloid cells.55 MLL has been shown to interact with Pax5, a key transcription factor for B-cell development.56 In agreement with this and MLL’s role in B-cell and lymphoid development, we observe a lymphoid-specific expression of both MLL and INO80D.
We used the single base pair resolution of HeliScope CAGE data to demonstrate the existence and cell-specific preference of alternative TSS for several epigenetic factors in the hematopoietic system. Although the functional characterization of the transcripts generated from alternative TSS remains elusive, our data identify additional complexities in the transcriptional regulation of epigenetic factors in the human hematopoietic system.
The cross-talk and functional interactions between epigenetic regulators that collectively give rise to the different epigenomes of differentiating cells is only beginning to be understood. This study describes the cell- and lineage-specific expression of epigenetic factors in a wide range of blood cells and thereby provides a useful framework toward understanding epigenetic control of hematopoiesis. We identified several known and putative epigenetic regulators of hematopoietic development and disease. Our data also provide an entry point for clinical hematology to predict potential targets for translational medicine.
This article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Iyadh Douagi (Department of Hematology and Regenerative Medicine) for helping with the cell sorting, and Mohsen Karimi and Alf Grandien for the generous gift of totalA isolated from CD34+ progenitor cells for qRT-PCR experiments. The authors thank all members of the Functional Annotation Of The Mammalian Genome 5 (FANTOM5) consortium for contributing to generation of samples and analysis of the dataset and Genome Network Analysis Service (GeNAS) for data production.
The Lennartsson group was supported by the Åke Olsson Foundation for Hematology. The Ekwall group was supported by the Swedish Cancer Foundation, Swedish Research Council, Göran Gustafsson Foundation, and Knut and Alice Wallenberg Foundation. FANTOM5 was made possible by a Research Grant for RIKEN Omics Science Center from the Japanese Ministry of Education, Culture, Sports, Science and Technology MEXT to Yoshihide Hayashizaki, a Research Grant from MEXT to RIKEN Preventive Medicine and Diagnosis Innovation Program, a grant of the Innovative Cell Biology by Innovative Technology (Cell Innovation Program) from the MEXT (to Yoshihide Hayashizaki), and a Grant from MEXT to RIKEN Center for Life Science Technologies.
Authorship
Contribution: P.P. and M.R. performed data analysis and wrote the manuscript; E.A. performed the initial CAGE analysis for the epigenetic factors and assisted in writing the manuscript; M.I. was responsible for CAGE data production; T.L. was responsible for tag mapping; H.K. managed the data handling; C.O.D. and A.R.R.F. were responsible for FANTOM5 management and the concept; and A.L. and K.E. assisted in writing the manuscript and planned and coordinated the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andreas Lennartsson, Department of Biosciences and Nutrition, NOVUM, Karolinska Institutet, Halsov 7-9, 14183 Huddinge, Sweden; e-mail: andreas.lennartsson@ki.se; and Karl Ekwall, Department of Biosciences and Nutrition, NOVUM, Karolinska Institutet, Halsov 7-9, 14183 Huddinge, Sweden; e-mail: karl.ekwall@ki.se.
References
Author notes
P.P. and M.R. contributed equally to this work.
A.L. and K.E. contributed equally to this work.
*RIKEN Omics Science Center ceased to exist as of April 1, 2013 due to RIKEN reorganization.