Key Points
Hypoxia mediates TKI resistance.
Hypoxia enhances CML stem cell maintenance.
Abstract
C-abl oncogene 1, nonreceptor tyrosine kinase (ABL1) kinase inhibitors such as imatinib mesylate (imatinib) are effective in managing chronic myeloid leukemia (CML) but incapable of eliminating leukemia stem cells (LSCs), suggesting that kinase-independent pathways support LSC survival. Given that the bone marrow (BM) hypoxic microenvironment supports hematopoietic stem cells, we investigated whether hypoxia similarly contributes to LSC persistence. Importantly, we found that although breakpoint cluster region (BCR)-ABL1 kinase remained effectively inhibited by imatinib under hypoxia, apoptosis became partially suppressed. Furthermore, hypoxia enhanced the clonogenicity of CML cells, as well as their efficiency in repopulating immunodeficient mice, both in the presence and absence of imatinib. Hypoxia-inducible factor 1 α (HIF1-α), which is the master regulator of the hypoxia transcriptional response, is expressed in the BM specimens of CML individuals. In vitro, HIF1-α is stabilized during hypoxia, and its expression and transcriptional activity can be partially attenuated by concurrent imatinib treatment. Expression analysis demonstrates at the whole-transcriptome level that hypoxia and imatinib regulate distinct subsets of genes. Functionally, knockdown of HIF1-α abolished the enhanced clonogenicity during hypoxia. Taken together, our results suggest that in the hypoxic microenvironment, HIF1-α signaling supports LSC persistence independent of BCR-ABL1 kinase activity. Thus, targeting HIF1-α and its pathway components may be therapeutically important for the complete eradication of LSCs.
Introduction
Chronic myeloid leukemia (CML) is a disease of hematologic stem cells caused by the reciprocal translocation t(9:22).1,2 This produces the chimeric breakpoint cluster region (BCR)–c-abl oncogene 1, nonreceptor tyrosine kinase (ABL1) oncoprotein that drives the pathogenesis of CML, and as a consequence, BCR-ABL1 represents the primary target of therapy.1 The development of the tyrosine kinase inhibitor (TKI), imatinib mesylate (imatinib, Gleevec), has drastically improved the treatment of CML with the majority of chronic phase (CP) patients achieving complete cytogenetic responses.3 However, although imatinib is effective in controlling CML and preventing progression to blast crisis (BC), it is not curative. Most patients who achieve complete cytogenetic responses continue to have detectable BCR-ABL1 transcripts,4 and in almost all cases, CML stem cells can still be detected in patient bone marrow (BM).5 Even for patients with complete molecular responses and no detectable BCR-ABL1 transcripts, cessation of TKI therapy frequently results in disease relapse.6,7 This suggests that despite TKI treatment, leukemic stem cells (LSCs) are able to survive and persist in patients, necessitating long-term TKI therapy. Identifying the mechanisms that support the persistence of LSCs is therefore of utmost importance for their eradication.
The mechanism of LSC persistence is not well-characterized. Although BCR-ABL1-dependent factors, including kinase domain mutations8 and BCR-ABL1 overexpression,9 have been proposed to play a part, this is not conclusively supported by both in vitro work and clinical observations. For example, several studies have shown that primitive quiescent CML cells are intrinsically resistant to TKIs.10,11 Further, in patients with stable complete cytogenetic responses, the presence of BCR-ABL1-kinase domain mutations is infrequent and does not predict relapse.12 Together, these studies10-12 indicate that LSCs may not require BCR-ABL1 for persistence and may thus resemble normal hematopoietic stem cells (HSCs) in their requirements for survival.13 Of note, studies in normal hematopoiesis have indicated that the BM microenvironment, which includes the stem cell niche, is crucial for normal HSC function.14 Accordingly, and because LSCs likely arise from HSCs or primitive progenitors, it is plausible that the BM microenvironment also contributes to LSC persistence. Indeed, recent studies indicate that physiologic growth factor signaling, by stimulating prosurvival pathways in CML progenitors, may render them independent of BCR-ABL1-mediated survival signals.15-17
Physiological hypoxia is said to occur when the oxygen (O2) tension falls below 2%.18 In humans, including patients with leukemia, the average BM O2 has been measured at 6% to 7%,19,20 whereas in mouse, the oxygen tension is estimated to be below 1% in the stem cell niche.21,22 Of note, hypoxia has long been regarded as a pathological phenomenon in ischemic solid tumors that is associated with radio- and chemoresistance.23,24 Further, recent studies in embryonic18 and hematopoietic25 systems have revealed that physiologic hypoxia also plays important roles in regulating stem cell functions.26 Although hypoxia is emerging as a key factor in the biology of leukemia,27 its role in CML LSCs and therapy resistance has not been thoroughly investigated. Although earlier studies have demonstrated that primary CML cells and CML cell lines can survive hypoxic conditions in vitro,28-30 and that hypoxia confers both stem-like characteristics and imatinib resistance in cell lines,29,30 it is unclear whether such features extend to primary CML LSCs.
To address these issues, we characterized the interplay between physiologic hypoxia and imatinib in primary human CML stem and progenitor populations, including their ability to function as LSCs. We found that although imatinib inhibits BCR-ABL1 effectively, hypoxia promotes CML cell maintenance, primarily through the upregulation of hypoxia-inducible factor 1 α (HIF1-α). We also uncover a set of prosurvival genes that are preferentially upregulated by hypoxia in CML versus normal cord blood CD34+ cells. Our study indicates that cosuppression of BCR-ABL1 and HIF1-α or HIF1-α-regulated targets represents a rational strategy for elimination of LSCs.
Materials and methods
Primary samples
Primary CP CML samples were obtained from patients at the Singapore General Hospital. Written informed consent was obtained from all patients under protocols approved by the institutional review board. Cord blood samples were obtained from the Singapore Cord Blood Bank. This study was conducted in accordance with the Declaration of Helsinki.
Cell culture, selection of CD34+ subpopulations, and hypoxia treatment
Mononuclear cells were purified by Ficoll-Paque (Sigma-Aldrich) density gradient centrifugation. CD34+ cells were purified using either a CD34 MicroBead kit or CD34 multiSort/CD38 MicroBead kits (Miltenyi Biotec) and subsequently grown in serum-free media supplemented with cytokines.31 Hypoxic treatment (0.5% oxygen, 5% carbon dioxide) was performed in a humidified incubator (Innova CO-170; New Brunswick Scientific). Cell numbers were determined by trypan blue exclusion and flow cytometry.
Colony-forming assay
Cells were plated in drug-free methylcellulose (H4434; StemCell Technologies). The number of colonies (colony-forming unit-granulocyte macrophages and colony-forming unit-erythroid) was scored 14 days later.
Annexin V staining and fluorescence in situ hybridization
These were performed as previously described.32
Intracellular phospho-CrkL staining
Staining was performed according to protocols from Cell Signaling Technology. Cells were stained with Viability Dye eFluor 780 (eBioscience), followed by CD34-fluorescein isothiocyanate and CD38-allophycocyanin (APC) (BD Biosciences). Isotype and phosphorylated–v-crk avian sarcoma virus CT10 oncogene homolog-like (pCrkL) signals were established using normal rabbit immunoglobulin G (IgG; sc-3888; Santa Cruz) and anti-pCrkL (1:50 dilution; Tyr207; 3181; Cell Signaling Technology), respectively. Cells were then stained with a secondary antibody (Alexa Fluor 594 goat anti-rabbit IgG heavy + light chain ([H+L]); Invitrogen) and analyzed by flow cytometry.
Long-term culture-initiating cell-limiting dilution assay
Limiting dilution was performed using reagents and protocols from Stemcell Technologies. Cells were plated on irradiated M2-10B4 cells in 96-well plates at 5 different densities with 8 replicates each. At the end of 7 weeks of incubation, wells were scored as positive or negative for the presence of colonies, and the frequency of long-term culture-initiating cells (LTC-ICs) was calculated using L-Calc. Colonies from each condition were pooled and analyzed for the presence of BCR-ABL by fluorescence in situ hybridization (FISH).
Engraftment of immunodeficient mice
All animal experiments were approved and performed in accordance with the guidelines required by the SingHealth Institutional Animal Care and Use Committee. CD34+ CML cells (2 × 106 cells/mouse) were treated in vitro before they were injected via tail vein into irradiated (240 cGy) immunodeficient nonobese diabetic/severe combined immunodeficiency interleukin 2 receptor γ chain-deficient mice [NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ or NSG mice] The Jackson Laboratory). Mice were euthanized after 6 weeks, and the contents of BM (both femurs and tibias), peripheral blood, and spleen were checked for human engraftment by staining with anti-human CD45-APC and anti-mouse CD45-fluorescein isothiocyanate (both from BD Biosciences). Engrafted cells from each mouse were subjected to FISH analysis for the presence of BCR-ABL1.
Immunohistochemistry and immunofluorescence
Immunohistochemistry was performed using HIF1-α (ADI-OSA-602-E; Enzo) and HIF2-α (Ab8365; Abcam) antibodies. Immunofluorescence was performed on fixed cells on micro cover glasses (12 mm; VWR). Isotype and HIF1-α signals were established using normal rabbit IgG (sc-3888; Santa Cruz Biotechnology) and anti-HIF1-α (Ab51608; Abcam), respectively. Cells were than stained with a secondary antibody (Alexa Fluor 594 goat anti-rabbit IgG [H+L]), mounted (P36931 ProLong Gold antifade reagent; Invitrogen), and visualized by confocal microscopy (LSM710; Carl Zeiss).
Microarray analysis
Using the Illumina TotalPrep Amplification kit (Ambion), 120 ng total RNA was subjected to cDNA synthesis. The cDNA was purified using the MinElute Reaction Cleanup kit (Qiagen). In vitro transcription was performed on the cDNA to generate biotin-labeled cRNA, according to manufacturer recommendations, followed by purification using the RNeasy Mini kit (Qiagen). The integrity and size distribution of the cRNA libraries were verified by RNA Nano 6000 chip profiling on the 2100 Bioanalyzer (Aligent). High-quality cRNA libraries were then hybridized on Human HT-12 beadchips (Illumina) and scanned on the BeadArray Reader (Illumina) at scan factor 2. The raw microarray data were normalized using the cross-correlation method.33 Differential gene expression was identified on the basis of a fold change cutoff of more than 1.5 compared with the dimethylsulfoxide (DMSO)/normoxia control for each patient and each time. The microarray data were deposited in the Gene Expression Omnibus database (accession number GSE48294).
Statistical analysis
Data were reported as mean ± standard error of the mean. Statistical significance was determined by Student t test.
Additional methods can be found in the supplemental information, available on the Blood Web site.
Results
Hypoxia enhances CML progenitor function in the presence and absence of imatinib
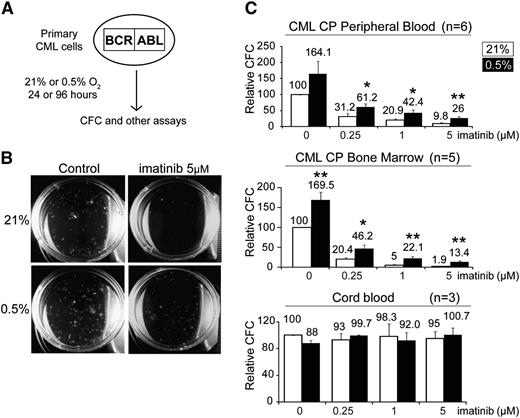
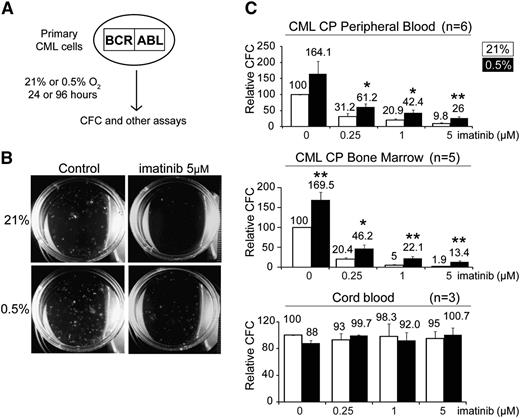
To investigate the effect of hypoxia on primary patient-derived CML progenitors, we first performed colony-forming assays to enumerate the number of committed CML progenitors under hypoxic conditions. CML cells were first treated under normoxic or hypoxic conditions, after which they were plated in drug-free methylcellulose, and colony-forming cells (CFCs) were counted at 14 days (Figure 1A-B). This 2-step procedure allowed us to assess the specific effect of the various conditions on CFCs exclusively during the initial culture period.31 We found that hypoxia by itself resulted in a 2-fold increase in the number of CFCs (Figure 1C; supplemental Table 1). In addition, hypoxia also enhanced the ability of CML progenitors to persist in the presence of imatinib by 2- to fivefold (Figure 1C). Importantly, we observed the same trend in CP samples obtained from peripheral blood (n = 6) or BM (n = 5), indicating that the effects of hypoxia are intrinsic to CML progenitors (Figure 1C). Furthermore, the increase in the number of CML progenitors was apparent even when cells were not subjected to the initial 96-hour in vitro culture period (supplemental Figure 1A). Interestingly, we also observed that the effect on CFCs was dependent on the degree of hypoxia, as lower oxygen concentrations (0.5% vs 5%) resulted in greater increases in CFC numbers (supplemental Figure 1B). In contrast, hypoxia and imatinib had no effect on cord blood CFCs (Figure 1C). Our findings demonstrate that hypoxia specifically enhances the in vitro maintenance of CML CFC function by twofold and that this is associated with a 2- to fivefold relative resistance to imatinib compared with normoxia.
Hypoxia enhances the colony forming capacity of committed CML progenitors. (A) Schematic illustrating the experimental procedure used to assay the effect of hypoxia and imatinib on progenitor cells. CML progenitors were exposed to imatinib with or without hypoxia in serum-free media supplemented with growth factors for 96 hours. Cells were then harvested, washed, and assayed for CFCs, LTC-ICs, engraftment potential, and other assays, as described in the text. (B) Representative images taken at 14 days of a colony-forming assay, after a 96-hour treatment period. (C) CML (n = 6 and 5) cells obtained from peripheral blood or BM and normal cord blood cells (n = 3) (unselected or CD34+ selected) were treated under 21% or 0.5% O2 for 96 hours. The cells were washed and plated in drug-free methylcellulose for 2 weeks. The numbers of CFCs are expressed as percentages of the untreated group (0 µM imatinib) incubated at 21% O2 and are indicated on the top of each column. For experiments performed with CP CD34+ cells, the CFC frequency per 1000 CD34+ cells is 185.5 ± 29.9 (supplemental Table 1). Error bars show standard of the mean. *P < .05 and **P < .01 denote Student t test statistical significance over 21% O2 without imatinib controls.
Hypoxia enhances the colony forming capacity of committed CML progenitors. (A) Schematic illustrating the experimental procedure used to assay the effect of hypoxia and imatinib on progenitor cells. CML progenitors were exposed to imatinib with or without hypoxia in serum-free media supplemented with growth factors for 96 hours. Cells were then harvested, washed, and assayed for CFCs, LTC-ICs, engraftment potential, and other assays, as described in the text. (B) Representative images taken at 14 days of a colony-forming assay, after a 96-hour treatment period. (C) CML (n = 6 and 5) cells obtained from peripheral blood or BM and normal cord blood cells (n = 3) (unselected or CD34+ selected) were treated under 21% or 0.5% O2 for 96 hours. The cells were washed and plated in drug-free methylcellulose for 2 weeks. The numbers of CFCs are expressed as percentages of the untreated group (0 µM imatinib) incubated at 21% O2 and are indicated on the top of each column. For experiments performed with CP CD34+ cells, the CFC frequency per 1000 CD34+ cells is 185.5 ± 29.9 (supplemental Table 1). Error bars show standard of the mean. *P < .05 and **P < .01 denote Student t test statistical significance over 21% O2 without imatinib controls.
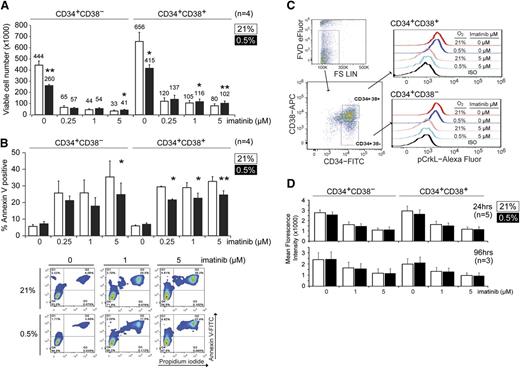
Hypoxia reduces proliferation of CML progenitors and impairs TKI-induced apoptosis
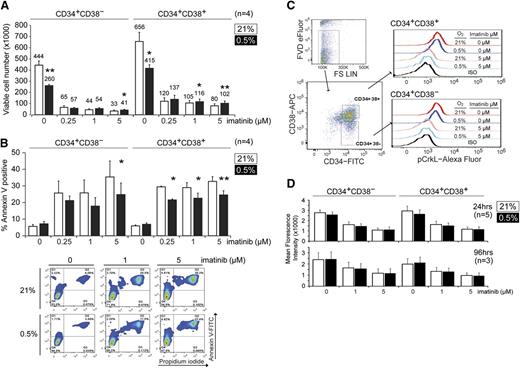
To understand how hypoxia might be enhancing CFC maintenance and imatinib resistance, we next examined the growth of the CML CD34+CD38+ (enriched for committed progenitors) and CD34+CD38− (enriched for stem cell progenitors) subpopulations under normoxic (21% O2) or hypoxic (0.5% O2) conditions. Here, we determined the change in cell numbers by trypan blue exclusion and flow cytometry after 4 days of culture. We found that both CD34+CD38+ and CD34+CD38− CML progenitors exhibited 30% to 50% slower growth under hypoxia (Figure 2A). We also observed that imatinib greatly reduced the growth of CML cells, regardless of the oxygen concentration, although there was a small but statistically significant increase in viable CD34+CD38+ and CD34+CD38− cells after imatinib treatment in hypoxic vs normoxic conditions (Figure 2A). Because changes in cell number may be caused by changes in either proliferation and/or cell death, we next monitored the degree of apoptosis by Annexin V and PI staining. We found that hypoxia did not significantly alter the proportion of apoptotic cells in the absence of drug but that it consistently suppressed imatinib-induced apoptosis in both CD34+CD38− and CD34+CD38+ populations at clinically achievable imatinib concentrations (Figure 2B).34 To exclude the possibility that hypoxia was impairing the ability of imatinib to inhibit BCR-ABL1 kinase, we also examined the level of pCrkL, a surrogate marker for BCR-ABL1 kinase activity,11 in CML progenitors under hypoxia. In both CD34+CD38− and CD34+CD38+ subpopulations, imatinib reduced the level of pCrkL to a similar extent, regardless of oxygen concentration (Figure 2C-D). Next, we used carboxyfluorescein diacetate succinimidyl ester tracing to measure proliferation.31 Consistent with the decreased growth observed under hypoxia, we found that both CD34+CD38− and CD34+CD38+ CML progenitors proliferated less at 0.5% vs 21% O2 and that the addition of imatinib further inhibited cell division (supplemental Figure 2). Together, our findings indicate that hypoxia inhibits the proliferation of committed and primitive CML progenitors while also impairing the ability of imatinib to induce apoptosis in these cells, despite effective BCR-ABL1 inhibition.
Hypoxia suppresses growth and imatinib-induced apoptosis in CML progenitors. (A) CP CML CD34+CD38− or CD34+CD38+ cells (1 × 105 cells each, n = 4) were treated with imatinib at 21% or 0.5% O2 for 96 hours, and the total number of viable cells was determined by trypan blue exclusion and flow cytometry. Cell numbers are indicated on the top of each column. (B) Cells were treated as in panel A, and the percentage of apoptotic cells was measured by Annexin V staining using flow cytometry (n = 4). Representative flow cytometry dot plots are shown. (C) Flow cytometric evaluation of pCrkL in CML cells. CD34+ cells were cultured for 24 or 96 hours and subjected to flow cytometric analysis. Viable cells were identified with the fixable viability dye (FVD; eFluor), and pCrkL levels were measured by intracellular staining in both CD34+CD38− and CD34+CD38− subpopulations. Flow cytometry dot plots and histograms show the results after 24 hours treatment. The background signals were determined in the same populations stained with the appropriate isotype controls. (D) Mean fluorescence intensity of pCrkL in multiple samples is shown (n = 5 and 3). *P < .05, **P < .01.
Hypoxia suppresses growth and imatinib-induced apoptosis in CML progenitors. (A) CP CML CD34+CD38− or CD34+CD38+ cells (1 × 105 cells each, n = 4) were treated with imatinib at 21% or 0.5% O2 for 96 hours, and the total number of viable cells was determined by trypan blue exclusion and flow cytometry. Cell numbers are indicated on the top of each column. (B) Cells were treated as in panel A, and the percentage of apoptotic cells was measured by Annexin V staining using flow cytometry (n = 4). Representative flow cytometry dot plots are shown. (C) Flow cytometric evaluation of pCrkL in CML cells. CD34+ cells were cultured for 24 or 96 hours and subjected to flow cytometric analysis. Viable cells were identified with the fixable viability dye (FVD; eFluor), and pCrkL levels were measured by intracellular staining in both CD34+CD38− and CD34+CD38− subpopulations. Flow cytometry dot plots and histograms show the results after 24 hours treatment. The background signals were determined in the same populations stained with the appropriate isotype controls. (D) Mean fluorescence intensity of pCrkL in multiple samples is shown (n = 5 and 3). *P < .05, **P < .01.
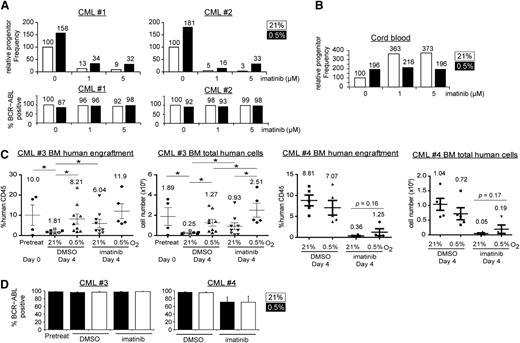
Hypoxia promotes CML stem cell maintenance
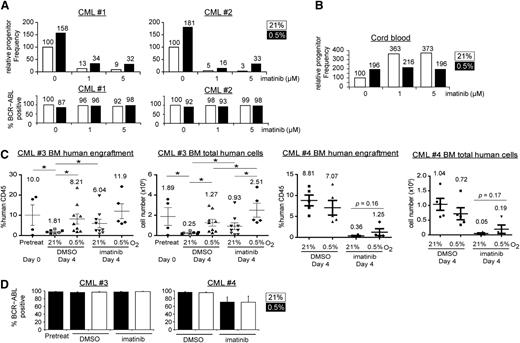
Colony-forming assays reflect primarily the clonogenic capacity of more mature committed progenitors, but not that of the more primitive progenitors, which are enriched for LSCs. To test whether hypoxia promotes the maintenance of primitive progenitors, we performed LTC-IC-limiting dilution assays.31 Here, we treated CML cells and incubated them on stromal layers for an additional 5 weeks before plating for colony formation. In line with the colony-forming assay results, hypoxia increased the progenitor frequency, both in the presence or absence of imatinib (Figure 3A, n = 2; supplemental Table 1). We confirmed that the colonies recovered from the LTC-IC were of CML origin, as determined by BCR-ABL1 FISH (Figure 3A, 87%-100%; average, 96%). Interestingly, hypoxia alone also increased the progenitor frequency of cord blood cells (Figure 3B, n = 2), suggesting that hypoxia promotes the maintenance of both normal cells and LSCs.
Hypoxia increases clonogenicity and engraftment of CML stem cells. The relative frequencies of primitive CML (A) or cord blood progenitors (B) were determined by LTC-IC limiting dilution assays for 2 independent samples. Cells were treated under 21% or 0.5% O2 for 96 hours and were harvested for long-term, drug-free culture. The progenitor frequency was calculated on the basis of colony formation and was scaled against 100% for the 0 µM imatinib group incubated at 21% O2. The cord blood results (B) show the average of 2 samples with similar trends (n = 2). For the CML samples, colonies from each treatment condition were pooled, and the percentage of BCR-ABL1-positive cells was determined by FISH, with percentages indicated on the top of each column. (C) BM engraftment of CD34+ CML cells in NSG mice. CD34+ CML cells (2 × 106 cells/mouse) from 2 CP CML patients (CML#3 and CML#4) were treated with 21% or 0.5% O2 and DMSO or imatinib for 96 hours and were injected into sublethally irradiated mice. The percentage and the total number of engrafted human CD45+ cells in the BM of recipient mice are shown in the left and right panels for each patient sample, respectively. Open symbols (CML#3) and filled symbols (both CML#3 and CML#4) represent experiments using 5 μM or 1 μM imatinib, respectively. Data were available for only 3 groups with open symbols (untreated at 21% O2, untreated at 0.5% O2, and imatinib-treated at 21% O2). Each point represents data from a single mouse. Horizontal bars indicate mean with numbers shown for each condition, and error bars indicate standard error of the mean. (D) The average percentage of BCR-ABL1 positive cells in each group of mice was determined by FISH. *P < .05.
Hypoxia increases clonogenicity and engraftment of CML stem cells. The relative frequencies of primitive CML (A) or cord blood progenitors (B) were determined by LTC-IC limiting dilution assays for 2 independent samples. Cells were treated under 21% or 0.5% O2 for 96 hours and were harvested for long-term, drug-free culture. The progenitor frequency was calculated on the basis of colony formation and was scaled against 100% for the 0 µM imatinib group incubated at 21% O2. The cord blood results (B) show the average of 2 samples with similar trends (n = 2). For the CML samples, colonies from each treatment condition were pooled, and the percentage of BCR-ABL1-positive cells was determined by FISH, with percentages indicated on the top of each column. (C) BM engraftment of CD34+ CML cells in NSG mice. CD34+ CML cells (2 × 106 cells/mouse) from 2 CP CML patients (CML#3 and CML#4) were treated with 21% or 0.5% O2 and DMSO or imatinib for 96 hours and were injected into sublethally irradiated mice. The percentage and the total number of engrafted human CD45+ cells in the BM of recipient mice are shown in the left and right panels for each patient sample, respectively. Open symbols (CML#3) and filled symbols (both CML#3 and CML#4) represent experiments using 5 μM or 1 μM imatinib, respectively. Data were available for only 3 groups with open symbols (untreated at 21% O2, untreated at 0.5% O2, and imatinib-treated at 21% O2). Each point represents data from a single mouse. Horizontal bars indicate mean with numbers shown for each condition, and error bars indicate standard error of the mean. (D) The average percentage of BCR-ABL1 positive cells in each group of mice was determined by FISH. *P < .05.
To further confirm the in vivo relevance of hypoxia on LSCs, we determined the effect of physiological hypoxia on the ability of CML LSCs to engraft NSG mice.35,36 CD34+ CML cells from 5 different patients with CP CML (2 × 106 cells/mouse) were treated in vitro for 96 hours before being injected into sublethally irradiated NSG mice. Six weeks after injection, the mice were euthanized and engraftment of human cells was assessed in BM, peripheral blood, and spleen. Among the 5 samples used, we were only able to successfully engraft CP CML cells from 2 samples (CML#3 and CML#4). For CML#3, we found that hypoxia consistently enhanced CML cell engraftment in all 3 hematopoietic compartments and that CML cells engrafted most efficiently in BM (Figure 3C; supplemental Figure 3). Specifically, we found that samples maintained in vitro at 21% oxygen exhibited impaired engraftment compared with samples that were injected immediately before any in vitro culture, and that hypoxia treatment maintained the ability of CML progenitors to engraft mice (Figure 3C). Interestingly, the engraftment ability of the imatinib-treated groups was consistently higher than that of the corresponding DMSO control groups. The decreased engraftment of the DMSO 21% O2 control group suggested gradual loss of engraftment potential during the in vitro treatment, whereas engraftment potential could be maintained by either imatinib and/or hypoxia, at least for the CML cells of this particular patient. Again, the engrafted cells in each mouse were found to be of CML origin, as confirmed by BCR-ABL1 FISH (Figure 3D, 90%-100%; average, 97%). For CML#4, we again observed a trend that hypoxic incubation promotes BM engraftment in the imatinib-treated group (0.36% in imatinib-treated cells grown in 21% O2 vs 1.25% in imatinib-treated cells grown in 0.5% O2; P = .16) (Figure 3C). Taken together, our in vitro and in vivo studies of CML stem cell function, despite patient-to-patient variability with respect to engraftment potential, are consistent with a role for physiologic hypoxia in CML stem cell maintenance. Next, we set out to understand the molecular underpinnings of this process.
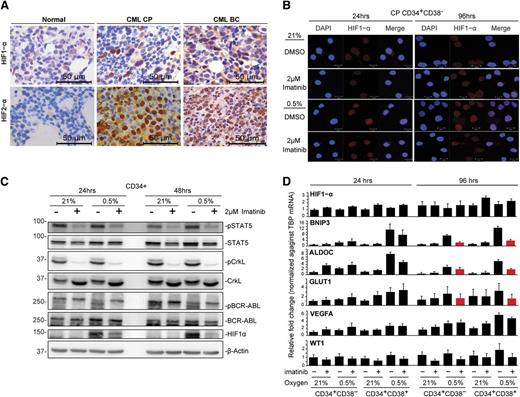
HIF1 activation by hypoxia and BCR-ABL1 is only partially attenuated by imatinib
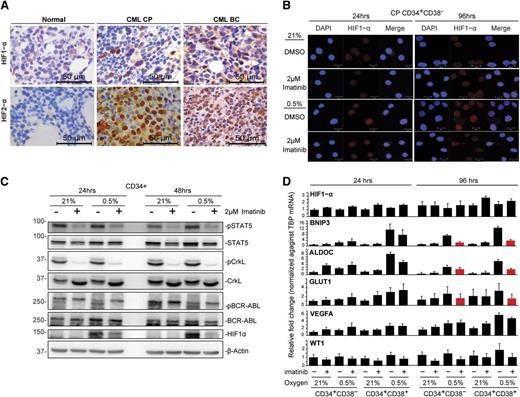
The cellular hypoxic response is primarily driven by HIFs, and both HIF1-α and HIF-2α play key roles in cancer stem cells.37,38 We therefore sought to analyze HIF expression in CML BM from patients in CP and BC, using immunohistochemistry. In CP samples, we observed scattered HIF1-α expression in cells corresponding to immature myeloid precursors, whereas in BC samples, HIF1-α expression was more widespread and consistent with the increased blast population in BC (Figure 4A; supplemental Figure 4A; antibody controls in supplemental Figure 4B). Interestingly, we observed ubiquitous HIF2-α expression in both CP and BC BM (Figure 4A). We conclude that both HIF1-α and HIF2-α are expressed in CML progenitors in vivo.
Hypoxia induces HIF1-α expression in CML progenitors that can be partially attenuated by imatinib. (A) Representative immunohistochemical staining for HIF1-α and HIF2-α proteins in normal, CP, and BC CML BM specimens. (B) Immunofluorescence detection of HIF1-α protein expression in CD34+CD38− and CD34+CD38+ populations. Results show representative images of 4 independent experiments. Scale bars represent 10 µm. (C) The levels of HIF1-α, pBCR-ABL1, pSTAT5, and pCrkL in CD34+ CML samples were detected by western blotting. Representative images of 2 independent experiments are shown. (D) The expression of HIF1-α, hypoxia-regulated genes (BNIP3, VEGFA, ALDOC, and GLUT1), and the BCR-ABL1-regulated gene WT1 in CD34+CD38− and CD34+CD38− subpopulations is shown. Expression was quantified by real-time quantitative polymerase chain reaction after normalization against the housekeeping gene, TBP (n = 3). Red bars highlight genes induced by hypoxia and downregulated by concurrent imatinib treatment.
Hypoxia induces HIF1-α expression in CML progenitors that can be partially attenuated by imatinib. (A) Representative immunohistochemical staining for HIF1-α and HIF2-α proteins in normal, CP, and BC CML BM specimens. (B) Immunofluorescence detection of HIF1-α protein expression in CD34+CD38− and CD34+CD38+ populations. Results show representative images of 4 independent experiments. Scale bars represent 10 µm. (C) The levels of HIF1-α, pBCR-ABL1, pSTAT5, and pCrkL in CD34+ CML samples were detected by western blotting. Representative images of 2 independent experiments are shown. (D) The expression of HIF1-α, hypoxia-regulated genes (BNIP3, VEGFA, ALDOC, and GLUT1), and the BCR-ABL1-regulated gene WT1 in CD34+CD38− and CD34+CD38− subpopulations is shown. Expression was quantified by real-time quantitative polymerase chain reaction after normalization against the housekeeping gene, TBP (n = 3). Red bars highlight genes induced by hypoxia and downregulated by concurrent imatinib treatment.
Because HIF1-α has been suggested to play important roles in CML,39,40 we next analyzed HIF1-α expression in CML progenitors in vitro. Using immunofluorescence and western blot, we found that activated nuclear HIF1-α could only be observed under hypoxia (Figure 4B-C; control in supplemental Figure 4C) and disappeared on reexposure to normoxia (supplemental Figure 4D), and further, that hypoxia-induced activation of HIF1-α was partially attenuated by imatinib (Figure 4B-C). We also found that HIF1-α downregulation by imatinib was largely posttranscriptional, as the level of HIF1-α transcripts remained relatively constant across treatments at different times (Figure 4D, n = 3). Several HIF1-α regulated genes, including BNIP3, VEGFA, ALDOC, and GLUT1,41 were consistently upregulated by short- and long-term hypoxia, an effect that was variably attenuated by imatinib (Figure 4D). Thus, at 96 hours, imatinib was mostly able to prevent the hypoxia-mediated increase in BNIP3, ALDOC, and GLUT1, but not that of VEGF (Figure 4D). Importantly, and consistent with the ability of imatinib to inhibit BCR-ABL1 in hypoxia (Figure 2C), imatinib consistently downregulated phosphorylated signal transducer and activator of transcription 5A (pSTAT5) and pCrkL (Figure 4C; supplemental Figure 4E) and WT1 expression, a gene known to be upregulated in CML cells in a BCR-ABL1-dependent manner (Figure 4D; supplemental Figure 4F).42 Together, our data demonstrate that BCR-ABL1 augments hypoxia-induced HIF1-α activation in CML progenitors and that imatinib only partially attenuates HIF1-α activation and HIF target gene transcription.
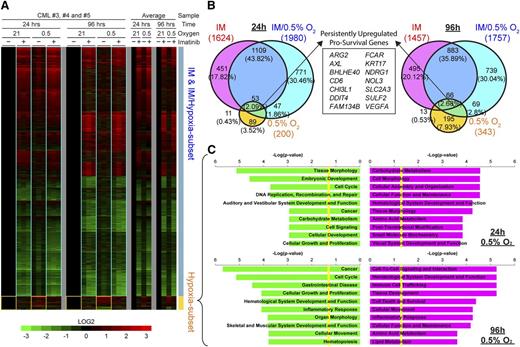
Gene expression profiling shows that hypoxia transactivates cell survival and cell adhesion factors in CML progenitors
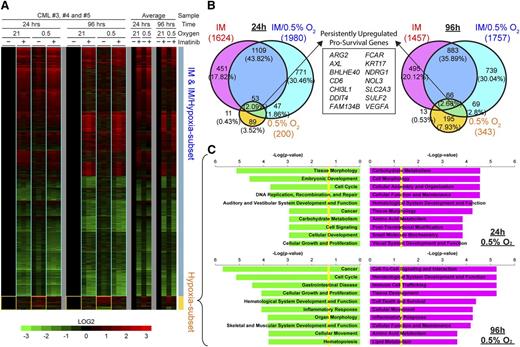
To better understand the relationship between hypoxia and imatinib exposure at the gene regulation level, we performed microarray analysis on CP CD34+ CML progenitors. Cells were treated for 24 or 96 hours, and global transcriptional changes were identified after comparison with control treatments. Both imatinib and hypoxia alone induced disparate transcriptional changes, with imatinib regulating a much larger subset of targets than hypoxia (Figure 5A-B). During combined imatinib and hypoxia treatment, there was transactivation of both hypoxia and imatinib target genes, with the overall transcriptional profile being predominantly similar to that of imatinib alone (Figure 5A-B). There were few synergistic or antagonistic effects between imatinib and hypoxia, as evidenced by the small number of coregulated targets (4.38% and 6.01% of the total differentially expressed genes at 24 and 96 hours, respectively; Figure 5B). Importantly, many of the hypoxia-induced target genes were attenuated by imatinib, which concurs with our observation that imatinib inhibits BCR-ABL1 kinase activity effectively (Figure 2C; supplementary Figure 5) but also inhibits HIF1-α activity partially (Figure 4B-C).
The hypoxia-driven transcriptional response in CML progenitor cells is distinct from that elicited by imatinib. (A) Microarray heat maps show changes in gene expression for CP-CD34+ progenitors from 3 individuals (triplicate columns) after imatinib (+) or DMSO (−) control treatments in 0.5% or 21% O2 for 24 or 96 hours. Color scale shows fold-change upregulation (red) or downregulation (green) of gene expression for each sample compared with their individual untreated controls (DMSO, 21% O2) on a Log2 scale. The overall changes in gene expression for each treatment are shown as averages for all 3 patients (single columns). Yellow boxes demarcate target genes specifically induced by hypoxia, whereas blue boxes show similar transcriptional response for imatinib treatments with or without hypoxia. (B) Proportional Venn diagrams show unique and coregulated target genes induced by imatinib and/or hypoxia treatments after 24 or 96 hours. The number of differentially expressed genes is stated for each condition, and the corresponding percentage of the total number of genes for each time is denoted in parentheses. The box shows 14 survival genes consistently upregulated by hypoxia with and without imatinib at 24 and 96 hours. (C) Ingenuity Pathway Analysis of the upregulated (red) and downregulated (green) hypoxia subset target genes at 24 and 96 hours, respectively, showing the top 10 most significantly affected biological functions for each condition. Yellow lines indicate threshold cutoff of P < .05 for Fisher's exact test.
The hypoxia-driven transcriptional response in CML progenitor cells is distinct from that elicited by imatinib. (A) Microarray heat maps show changes in gene expression for CP-CD34+ progenitors from 3 individuals (triplicate columns) after imatinib (+) or DMSO (−) control treatments in 0.5% or 21% O2 for 24 or 96 hours. Color scale shows fold-change upregulation (red) or downregulation (green) of gene expression for each sample compared with their individual untreated controls (DMSO, 21% O2) on a Log2 scale. The overall changes in gene expression for each treatment are shown as averages for all 3 patients (single columns). Yellow boxes demarcate target genes specifically induced by hypoxia, whereas blue boxes show similar transcriptional response for imatinib treatments with or without hypoxia. (B) Proportional Venn diagrams show unique and coregulated target genes induced by imatinib and/or hypoxia treatments after 24 or 96 hours. The number of differentially expressed genes is stated for each condition, and the corresponding percentage of the total number of genes for each time is denoted in parentheses. The box shows 14 survival genes consistently upregulated by hypoxia with and without imatinib at 24 and 96 hours. (C) Ingenuity Pathway Analysis of the upregulated (red) and downregulated (green) hypoxia subset target genes at 24 and 96 hours, respectively, showing the top 10 most significantly affected biological functions for each condition. Yellow lines indicate threshold cutoff of P < .05 for Fisher's exact test.
Because hypoxia impairs imatinib-induced apoptosis and enhances clonogenicity, we determined the hypoxia-induced targets that might be responsible. Detailed examination of persistently upregulated targets by hypoxia at both 24 and 96 hours with and without imatinib highlighted the enhancement of 14 antiapoptotic/survival genes (Figure 5B, overlap between 0.5% O2 and imatinib/0.5% O2; supplemental Table 2). Next, we conducted gene ontology analysis on targets (Figure 5A, yellow boxes; Figure 5C; supplemental Table 3) that were predominantly dysregulated in hypoxia treatments and partially reduced by imatinib but could still be independently responsible for sustaining CML progenitors. Carbohydrate metabolism was the most significantly upregulated, a typical feature of glycolytic adaption under hypoxia in the acute 24-hour phase. Cell cycle and cellular growth and proliferation signatures were generally downregulated, which is consistent with the decreased cell numbers and proliferative index measurements we observed (Figure 2A; supplemental Figure 2). Interestingly, the biological functions of cell-to-cell signaling and interaction, as well as hematological system and development, were upregulated with an overrepresentation of genes involved in increased adhesion, migration, and infiltration of hematopoietic lineages in long-term cultures. Overall, this analysis indicates a possible role for hypoxia in leukemogenesis through the concerted transactivation of multiple survival and adhesion factors that may contribute to decreased apoptosis and improved colony-forming potential and engraftment.
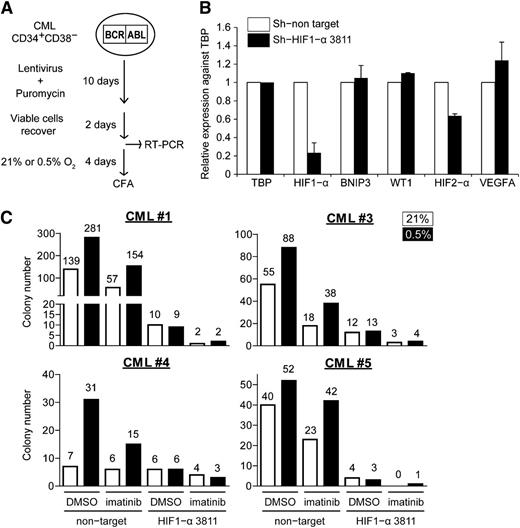
Knockdown of HIF1-α expression abolishes the enhanced clonogenicity of CML progenitors under hypoxia
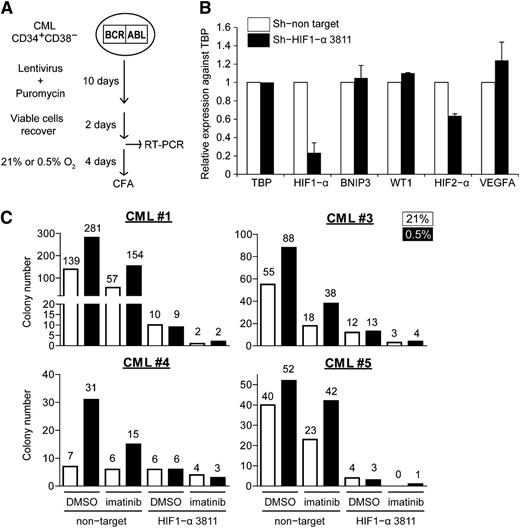
Imatinib partially attenuates HIF1-α signaling, but residual HIF1-α transcriptional activity may still be sufficient to support the survival and maintenance of CML cells under hypoxia. To determine whether HIF1-α plays a role in supporting CML progenitors under imatinib and hypoxia, we knocked down HIF1-α expression under these conditions. Here, CML CD34+38− cells were transduced with lentivirus-expressing control or HIF1-α-specific short hairpin RNAs (shRNAs; 2 validated clones, sh-HIF1-α 3811 and sh-HIF1-α 10819), followed by puromycin selection (Figure 6A). We were able to recover enough cells from those transduced with control and sh-HIF1-α 3811 (supplemental Table 4) and confirmed the specific knockdown of HIF1-α in these cells by quantitative real-time polymerase chain reaction (Figure 6B; supplemental Figure 6). As expected, hypoxia increased the number of CFCs in the control cells, but this was abolished in the HIF1-α knockdown cells, both in the presence and absence of imatinib (Figure 6C). These results suggest that HIF1-α is primarily responsible for the maintenance of CML progenitors under hypoxic conditions, both in the presence and absence of imatinib.
Knockdown of HIF1-α reduces clonogenicity of CML progenitors. (A) Schematic showing the procedure for lentiviral transduction of HIF1-α shRNA vectors. (B) Relative expression of HIF1-α, BNIP3, WT1, HIF2-α, and VEGFA normalized against the housekeeping gene TBP in lentiviral-transduced cells (n = 2). (C) Lentiviral-transduced cells were treated with 21% or 0.5% oxygen, with or without 1 µM imatinib for 96 hours and plated for colony formation. The number of colonies was normalized against the number of cells plated on methylcellulose (1400, 7333, 2100, and 1400 puromycin-selected cells for CML#1, CML#3, CML#4, and CML#5, respectively). The data show the results of 4 independent samples. Colony numbers are indicated on the top of each column.
Knockdown of HIF1-α reduces clonogenicity of CML progenitors. (A) Schematic showing the procedure for lentiviral transduction of HIF1-α shRNA vectors. (B) Relative expression of HIF1-α, BNIP3, WT1, HIF2-α, and VEGFA normalized against the housekeeping gene TBP in lentiviral-transduced cells (n = 2). (C) Lentiviral-transduced cells were treated with 21% or 0.5% oxygen, with or without 1 µM imatinib for 96 hours and plated for colony formation. The number of colonies was normalized against the number of cells plated on methylcellulose (1400, 7333, 2100, and 1400 puromycin-selected cells for CML#1, CML#3, CML#4, and CML#5, respectively). The data show the results of 4 independent samples. Colony numbers are indicated on the top of each column.
Identification of prosurvival genes differentially upregulated in CML vs normal cord blood CD34+ cells
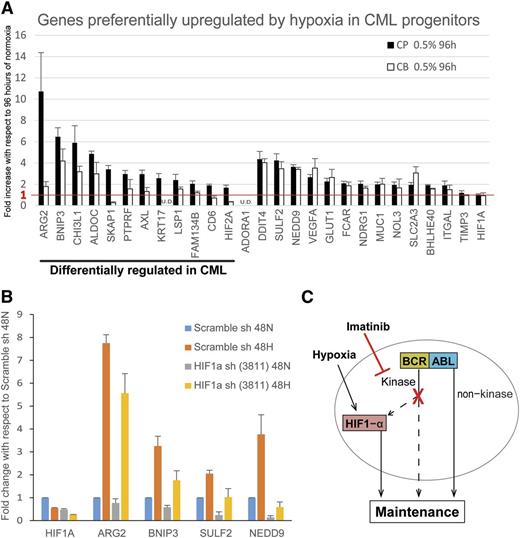
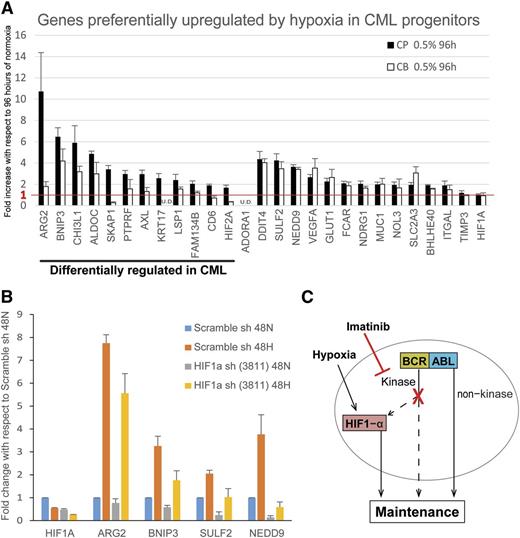
Because hypoxia enhances colony formation of CML progenitors but not normal progenitors (Figure 1C), as well as improves CML progenitor survival in the presence of imatinib (Figure 2B), our data suggested hypoxia may differentially upregulate prosurvival genes in CML vs cord blood progenitors. Specifically, we compared the expression of 27 hypoxia-induced genes (identified in Figure 5 and supplemental Table 3) between cord blood and CP CML samples. As depicted in Figure 7A, we found that 14 genes were equivalently upregulated in cord blood and CML cells, whereas 12 genes were preferentially upregulated in CML vs cord blood cells. Notably, among these 12 genes, 5 (ARG2, CHI3L1, FAM134B, KRT17, and CD6) have prosurvival functions and/or are associated with transformation and poorer clinical outcome in human cancers.43-49 Next, we confirmed that a subset of these genes is dependent on HIF1-α for hypoxia-induced upregulation. As shown in Figure 7B, knockdown of HIF1-α resulted in abrogation of the hypoxia-induced upregulation of ARG2, BNIP3, SULF2, and NEDD9.
Prosurvival genes are differentially upregulated in CML cells vs normal cord blood cells. (A) CP CML and cord blood (CB) CD34+ progenitors (n = 3 each) were treated under normoxia or hypoxia for 96 hours and the RNA harvested for real-time polymerase chain reaction analysis by Fluidigm array. Results show the fold increase by hypoxia with respect to that of normoxia. The red line indicates the cutoff for genes with no hypoxic upregulation. Expression was normalized to that of TBP. Error bars represent standard of the mean. U.D., undetectable. (B) K562 cells were transduced with scrambled shRNA control or HIF1-α shRNA (n = 3). Cells were treated under normoxia or hypoxia for 48 hours, and the RNA were harvested for RT-PCR analysis by Fluidigm array. Results show the hypoxia-induced fold change in expression with respect to that of scrambled control treated with normoxia. Expression was normalized to TBP. Error bars represent standard of the mean. (C) Model showing the maintenance of CML progenitors under hypoxia and imatinib in the hypoxic BM environment.
Prosurvival genes are differentially upregulated in CML cells vs normal cord blood cells. (A) CP CML and cord blood (CB) CD34+ progenitors (n = 3 each) were treated under normoxia or hypoxia for 96 hours and the RNA harvested for real-time polymerase chain reaction analysis by Fluidigm array. Results show the fold increase by hypoxia with respect to that of normoxia. The red line indicates the cutoff for genes with no hypoxic upregulation. Expression was normalized to that of TBP. Error bars represent standard of the mean. U.D., undetectable. (B) K562 cells were transduced with scrambled shRNA control or HIF1-α shRNA (n = 3). Cells were treated under normoxia or hypoxia for 48 hours, and the RNA were harvested for RT-PCR analysis by Fluidigm array. Results show the hypoxia-induced fold change in expression with respect to that of scrambled control treated with normoxia. Expression was normalized to TBP. Error bars represent standard of the mean. (C) Model showing the maintenance of CML progenitors under hypoxia and imatinib in the hypoxic BM environment.
Discussion
CML LSCs are thought to serve as a reservoir for resistance and relapse, and the inability of BCR-ABL1 TKIs to eliminate LSCs may explain why most patients require long-term therapy.50 These notions suggest the existence of BCR-ABL1-independent factors mediating LSC maintenance and survival.10-13 Indeed, recent work has identified extracellular signals from stromal cells and growth factors as important contributors to LSC survival that confer BCR-ABL1-independence.11,13 Recent studies also underscore the importance of physiologic hypoxia in maintaining HSC function.26 However, the relationship between hypoxia and CML LSCs remains poorly understood. In the current work, we explored the effect of physiologic hypoxia on CML progenitor and LSC function, as well as the relationship between hypoxia, LSC function, and imatinib exposure. By doing so, we identify physiologic hypoxia as an important contributor to CML LSC persistence, despite BCR-ABL1 inhibition, and suggest therapeutic approaches to extinguish LSC function.
As a first step in establishing therapeutic strategy for LSCs, we evaluated the effect of hypoxia on committed and primitive progenitor function, as well as LSCs in the presence and absence of imatinib. We found that hypoxia enhanced the ability of primary CP CML cells to function in all 3 assays and conclude that physiologic hypoxia contributes to CML progenitor maintenance and LSC function. By performing the experiments in the presence of clinically relevant imatinib concentrations, we find that hypoxia protects against imatinib-induced apoptosis in both CD34+CD38+ and CD34+CD38− CML progenitors, despite BCR-ABL1 kinase inhibition, and that hypoxia maintains both LTC-IC formation and engraftment capacity. By assessing cell division of CD34+CD38+ and CD34+CD38− CML progenitors, we find that hypoxia slows cell division. These observations are consistent with previous reports describing an association between increased proliferation or loss of quiescence and so-called stem cell exhaustion.51-53 The decreased proliferation is also congruent with the idea that imatinib-resistant, quiescent CML progenitors are the populations that contribute to disease persistence.54 Consistent with the effects of hypoxia on cell division, we found that hypoxia downregulated genes involved in cell cycle and cellular growth and proliferation.
Decreased intracellular levels of imatinib, possibly through the actions of drug transporter pumps, have been suggested as a mechanism for imatinib resistance.9 However, the above might not be relevant here, as BCR-ABL1 kinase activity remains suppressed by imatinib under hypoxia. To measure the effect of hypoxia on BCR-ABL1 kinase activity, we monitored the level of pCrkL in the CML stem and committed populations. We found that even under prolonged treatments (96 hours), hypoxia alone does not reduce the levels of pCrkL in both stem and committed progenitors (Figure 2). This is in contrast to previous reports that in CML cell lines, hypoxia reduces the level of BCR-ABL1 phosphorylation29 or BCR-ABL1 protein.30 The reason behind this discrepancy is unclear, but it may be a result of differences in model systems between CML cell lines and primary human CML cells. In support of this possibility, we found that CML cell lines were less able to form colonies under hypoxia (unpublished data), further emphasizing the difference in hypoxic responses between primary and CML cell lines.
Given that cells within the stem cell niche communicate through cytokines, and cytokines can induce BCR-ABL1-independent imatinib resistance,16,17,55 it would be of interest to determine the combined effect of hypoxia and cytokines on CML progenitors. Our unpublished data also indicate that hypoxia and stromal cell coculture enhance colony formation, with or without imatinib. Thus, it is likely that several components of the BM microenvironment, including hypoxia, cytokines, and cell–cell interactions,56 play a part in LSC maintenance.
We also explored the molecular mechanisms underlying the effects of hypoxia on CML progenitors. Because in most cell types the canonical hypoxic response is elicited by HIFs, we specifically explored the role of HIFs.41 First, we show in primary CML LSCs that HIF1-α is stabilized and localized to the nucleus only under hypoxia, in contrast to a previous report using mouse cell lines.40 We further demonstrate that imatinib can partially attenuate the expression of HIF1-α, and by silencing HIF1-α, we show that the maintenance of CML progenitors under hypoxia is dependent on HIF1-α expression.
Interestingly, despite the transient nature of HIF1-α expression produced by our protocol (supplemental Figure 4D), the functional effects were long-lasting. Several mechanisms may account for the enduring effects of transient HIF1-α expression and include induction of epigenetic changes57 and, possibly, HIF1-mediated expression of long-lived proteins. The situation in CML cells is also reminiscent of normal HSCs, as a previous report using human BM cells also showed that severe combined immunodeficiency-repopulating cells are preferentially preserved under hypoxic incubation.58 Our findings are also in line with recent studies in mouse CML models demonstrating that HIF1-α is required for the long-term maintenance of LSCs.39 Interestingly, these investigators concluded that LSCs seem to be more dependent on the HIF1-α pathway for survival than HSCs.39 Although our work did not specifically address the role of HIF in BC CML, we observed massive expression of HIF1-α in BC CML BM specimens. Notably, others have reported that in BC cell lines selected for imatinib resistance, BCR-ABL1 upregulation is associated with enhanced HIF1-α signaling.59 Interestingly, these biological effects of HIF1-α were evident under normoxic conditions, and therefore support our observation that HIF1-α silencing can impair CML colony formation under normoxia as well as hypoxia (Figure 6C). It also has been reported that engraftment of acute myeloid leukemia cells results in the appearance of widespread areas of hypoxia in the BM, whereas engraftment of normal HSCs only produces discrete areas of hypoxia.60 As BCR-ABL1 kinase activity augments HIF1-α signaling under hypoxia, we predicted, and indeed observed, a much higher level of HIF1-α activity in CML BC cells compared with CP, as BC cells overexpress BCR-ABL1.61
Further mechanistic insights were revealed through our transcriptome analysis. Decreased cell death during hypoxia treatment of CML cells may be a result of the upregulation of antiapoptotic factors that counteract imatinib eradication. Indeed, antiapoptotic function is a prominent feature of hypoxia that mediates survival during ischemic stress.62 Separately, the hypoxic regulation of diverse cell adhesion factors is known to enhance binding of leukocytes to endothelial cells,63 which is important for extravasation and may explain the enhancement of clonogenicity and engraftment of hypoxia-treated CML progenitors. These findings connect multiple mechanisms involving survival and adhesion factors driven by hypoxia with the establishment of long-term CML resistance against imatinib.
In summary, our study suggests that under physiologic hypoxia, HIF-dependent signaling is incompletely abolished by imatinib and is able to support LSC maintenance and potency (Figure 7C). Thus, a combination of anti-BCR-ABL1 and anti-HIF1-α therapy might represent a promising strategy to eliminate LSCs and cure CML. Some of the hypoxia-regulated genes in CML cells (Figures 5B and 7A; supplemental Tables 2 and 3), particularly those extensively involved in survival and cell adhesion, could also represent therapeutic targets in CML progenitors, especially if direct targeting of HIF1-α should prove too toxic for normal hematopoiesis.64
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE48294).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful for insightful discussions with Dr Chai Chou. We thank Zheng Lin and Norazean Zaiden for technical support and Dr Sharon Lim for advice on immunofluorescence. This study was supported by grants from the Singapore Ministry of Health’s National Medical Research Council, Biomedical Research Council of the Agency for Science, Technology and Research, Singapore National Research Foundation, Singapore Ministry of Education under the Research Center of Excellence Programme, Swedish Research Council, the Swedish Cancer Society, and the European Union.
Authorship
Contribution: K.P.N. and K.L.L. designed and performed research, interpreted data, and wrote the paper; W.H. and A.M. performed experiments; S.Y.T. interpreted data; C.T.H.C. contributed vital material and clinical information, and participated in discussions; L.P. designed research and analyzed data; and S.T.O. designed research, interpreted data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: S. Tiong Ong, Duke-National University of Singapore Graduate Medical School, 8 College Rd, Singapore, 169857; e-mail: sintiong.ong@duke-nus.edu.sg.