Key Points
CD68-GFP reporter mice show GFP transgene expression in both monocytes and tissue resident macrophage populations.
Adoptively transferred CD68-GFP monocytes maintain GFP expression after recruitment in an ongoing inflammatory response.
Abstract
The recruitment of monocytes and their differentiation into macrophages at sites of inflammation are key events in determining the outcome of the inflammatory response and initiating the return to tissue homeostasis. To study monocyte trafficking and macrophage differentiation in vivo, we have generated a novel transgenic reporter mouse expressing a green fluorescent protein (GFP) under the control of the human CD68 promoter. CD68-GFP mice express high levels of GFP in both monocyte and embryo-derived tissue resident macrophages in adult animals. The human CD68 promoter drives GFP expression in all CD115+ monocytes of adult blood, spleen, and bone marrow; we took advantage of this to directly compare the trafficking of bone marrow–derived CD68-GFP monocytes to that of CX3CR1GFP monocytes in vivo using a sterile zymosan peritonitis model. Unlike CX3CR1GFP monocytes, which downregulate GFP expression on differentiation into macrophages in this model, CD68-GFP monocytes retain high-level GFP expression for 72 hours after differentiation into macrophages, allowing continued cell tracking during resolution of inflammation. In summary, this novel CD68-GFP transgenic reporter mouse line represents a powerful resource for analyzing monocyte mobilization and monocyte trafficking as well as studying the fate of recruited monocytes in models of acute and chronic inflammation.
Introduction
Immune cells of the mononuclear phagocyte system (MPS) play a key role in host immune responses. Tissue-resident macrophages play a sentinel role in initiating acute inflammatory responses and, together with monocyte-derived macrophages that differentiate from recruited monocytes, they regulate local inflammatory responses. Macrophages play an important role in initiating wound repair and restoring tissue homeostasis after infection or tissue injury.1,2 In some cases, tissue homeostasis is not restored, and macrophages can promote continuing tissue damage and chronic inflammation, such as in rheumatoid arthritis and atherosclerosis.3 Monocytes are circulating CD115+ myeloid cells that develop from the common monocyte precursor in the bone marrow downstream of the macrophage dendritic cell (DC) precursor in the bone marrow before being released into the bloodstream.4-7 Factors that enhance monocyte release from bone marrow into blood include chemokines derived from sites of inflammation,8-11 and recent studies have identified hyperglycemia and hyperlipidemia as important factors that regulate monocytosis by acting on bone marrow progenitor cells.12-16 Blood monocytes fall into 2 populations—Ly6Chi and Ly6Clo subsets that have been predicted to have different roles in vivo because of differential expression of chemokine receptors and different behaviors in intravital microscopy studies in which Ly6Clo monocytes have a vascular patrolling behavior.17-19
Human CD68 and its murine homolog, macrosialin (Cd68), are both heavily glycosylated type I transmembrane proteins that belong to the lysosomal/endosomal-associated membrane proteins.20 The exact function of CD68 has yet to be confirmed, but a role in antigen processing and as a scavenger receptor have been previously proposed.20,21 In terms of expression, CD68 has been reported to be restricted to cells of myeloid lineage, specifically monocyte/macrophages.22 CD68 tissue staining is commonly used as a marker for infiltrating monocyte-derived macrophages in atherosclerotic lesions and other sites of chronic inflammation.23
The majority of studies that involve monitoring leukocyte trafficking and inflammatory cell recruitment use transgenic animals in which specific cell types have been labeled by genetic “knock-in” strategies that add a fluorescent protein gene into a gene locus active in a specific cell type. This strategy is exemplified by the CX3CR1 GFP knock-in mouse24 and more recently the CCR2 RFP knock-in mouse.25 Both of these reporter mice have been widely used to study trafficking of different monocyte subsets during resting and inflammatory conditions. Both of these transgenic reporter strains rely on chemokine receptor gene regulatory elements to label monocyte subsets and both rely on continued chemokine receptor expression to label macrophages that differentiate from monocytes in vivo. By using the powerful gene regulatory elements that we have identified in the promoter and intron 1 of the human CD68 gene, we sought to efficiently label all tissue resident macrophage populations as well as all monocyte-derived macrophages found at sites of inflammation.
In this study we demonstrate that CD68-GFP mice have high-level GFP reporter gene expression in monocytes in blood, bone marrow, and spleen as well as tissue-resident macrophages. We have used CD68-GFP mice to investigate monocyte trafficking and macrophage differentiation in vivo. Importantly, we demonstrate that adoptively transferred CD68-GFP monocytes that traffic to sites of inflammation retain high-level GFP expression after differentiation into macrophages in situ.
Methods
Materials
All cell culture media and buffers were obtained from PAA systems (Yeovil, United Kingdom) unless otherwise specified. All laboratory chemicals were obtained from Sigma-Aldrich (Gilligham, Dorset, United Kingdom). Chemokines and other chemoattractants were purchased from Peprotech (London, United Kingdom) and R&D Systems (Abingdon, Oxford, United Kingdom).
Molecular cloning and generation of CD68-GFP mice
A complementary DNA fragment encoding enhanced green fluorescent protein (EGFP) (from pEGFP-N1 vector, Invitrogen) was subcloned through pSL301 to change the restriction sites and then cloned into the 1265 vector containing human CD68 promoter (−2.9 kb).26 DNA was excised from the cloning vector and injected into C57BL/6J mouse oocytes. Transgenic offspring were genotyped by polymerase chain reaction (PCR) using the following primers: TY1: 5′ TTC TCG GCT CTG TGA ATG ACA 3′ and 5′ CAG CCC TCT CTT GGA AAG GAG G 3′. Founder mice were backcrossed with C57BL/6J mice (F1-F4 progeny) providing equal numbers of transgene positive offspring and littermate controls (subsequently referred to as WT).
Animals
All animal studies were conducted with approval from the Dunn School of Pathology Local Ethical Review Committee and in accordance with the UK Home Office regulations (Guidance on the Operation of Animals, Scientific Procedures Act, 1986). Male (10 to 14 weeks) C57BL/6J mice were from Harlan Laboratories, Oxfordshire, United Kingdom; CX3CR1GFP mice24 were kindly provided by David Jackson and Louise Johnson (Weatherall Institute of Molecular Medicine, University of Oxford).
Adoptive transfer GFP monocytes into zymosan-induced peritonitis
Monocytes were isolated from bone marrow using EasySep Mouse Monocyte Enrichment kit (Stem Cell Technologies). Briefly, femurs and tibias were harvested and flushed through with ice cold phosphate-buffered saline (PBS). Bone marrow cell suspensions were passed through a 70-µm cell strainer to obtain a single cell suspension. Red blood cells were removed by hypotonic lysis (Pharmlyse, BD) according to the manufacturer's instructions. The bone marrow cell suspension was treated with the EasySep reagents and monocytes isolated by depletion using an EasyPlate magnet (Stem Cell Technologies). Cells were applied to the magnet in a 96-well plate for 5 minutes, followed by a second selection for a further 2 minutes because this was found to obtain a higher yield of cells than the single 10-minute isolation recommended by the manufacturer. The purity of the resulting populations was confirmed by flow cytometry using CD45 (BD, Bioscience), CD11b (BD, Bioscience), 7/4 (Serotec), and Ly-6G (Biolegend). Bone marrow isolations were typically found to yield 1 to 2 × 106 cells at a purity of 75% to 85% monocytes.
C57BL/6J mice were administered 500 µL of 100 µg zymosan A in PBS (Sigma-Aldrich) intraperitoneally 30 minutes before IV administration of 1.5 × 106 isolated monocytes (70% to 80% GFP-positive, in 200 µL). After 16 or 72 hours, mice were euthanized and peritoneal exudates were collected by peritoneal lavage with 5 mL of ice cold sterile PBS-2 mM EDTA. Total cell counts and cellular composition of peritoneal exudate were determined as previously described.27 Details of macrophage isolations, culture, and flow cytometric analysis can be found in the supplemental Methods and materials on the Blood Web site.
Statistical analysis
All quantitative data are reported as mean ± standard error of the mean (SEM) of n observations. Statistical evaluation was performed using 1- or 2-way analysis of variance (Prism 6 GraphPad Software, San Diego, CA) followed by Dunnett’s or Bonferroni’s multiple comparison post hoc test, with P < .05 as statistically significant.
Results
Generation of CD68-GFP mice and phenotypic analysis
CD68-GFP transgenic mice expressing EGFP under control of the human CD68 promoter intron 1 cassette26,28 were generated by pro-nuclear injection and genotyped to confirm GFP transgene transmission (Figure 1A). Mice were fertile and produced an expected Mendelian ratio of transgenic and nontransgenic litter mates with no obvious developmental defects. Confocal microscopy of blood smears confirmed genotyping, with positive GFP expression observed in peripheral blood leukocytes highlighting a simple method for phenotyping these mice (Figure 1B). Fluorescent in situ hybridization (FISH) analysis was carried out to identify chromosomal site(s) of transgene integration into the mouse genome. Figure 1C shows a single site of integration on chromosome 3 for both CD68 vector and GFP transgene.
GFP expression in monocyte and macrophage populations. (A) The 2940-bp hCD68 promoter and intron 1 cassette were cloned upstream of EGFP and transgene transmission confirmed by PCR analysis of DNA from ear snips. (B) Confocal microscopy of a blood smear from a CD68-GFP mouse showed positive GFP expression in blood monocytes. (C) FISH analysis of metaphase chromosome preparations using a CD68 probe (red signal) and chromosome paint (green) made from mouse fibroblasts cultures counterstained with DAPI (blue). The green arrow indicates the transgene site integration on chromosome 3. (D) GFP mRNA expression was confirmed by RT-PCR analysis of tissue lysates from brain, lung, spleen, thymus, and liver of CD68-GFP and WT. (Dashed vertical line has been inserted to indicate a repositioned gel lane.) (E) Western blot analysis for GFP protein expression in tissue lysates (30 µg/tissue) from brain, lung, spleen, and thymus of CD68-GFP and WT mice. (F) To determine transgene copy number, 100 ng of DNA was isolated from the thymus of male CD68-GFP and male WT mice, amplified using standard PCR, and run on agarose gel side-by-side with the pEGFP-N1 vector. A male GFP knock-in (KI) mouse that carries a single copy of the GFP transgene on the X chromosome and a female homozygote GFP KI mouse that carries 2 copies of the GFP transgene on the X chromosome were used as copy number controls. (G) Transgene copy number was quantified with quantitative PCR using a 10-fold serial dilution standard curve of the pEGFP-N1 vector. (H) Table shows mean relative copy number ± SEM.
GFP expression in monocyte and macrophage populations. (A) The 2940-bp hCD68 promoter and intron 1 cassette were cloned upstream of EGFP and transgene transmission confirmed by PCR analysis of DNA from ear snips. (B) Confocal microscopy of a blood smear from a CD68-GFP mouse showed positive GFP expression in blood monocytes. (C) FISH analysis of metaphase chromosome preparations using a CD68 probe (red signal) and chromosome paint (green) made from mouse fibroblasts cultures counterstained with DAPI (blue). The green arrow indicates the transgene site integration on chromosome 3. (D) GFP mRNA expression was confirmed by RT-PCR analysis of tissue lysates from brain, lung, spleen, thymus, and liver of CD68-GFP and WT. (Dashed vertical line has been inserted to indicate a repositioned gel lane.) (E) Western blot analysis for GFP protein expression in tissue lysates (30 µg/tissue) from brain, lung, spleen, and thymus of CD68-GFP and WT mice. (F) To determine transgene copy number, 100 ng of DNA was isolated from the thymus of male CD68-GFP and male WT mice, amplified using standard PCR, and run on agarose gel side-by-side with the pEGFP-N1 vector. A male GFP knock-in (KI) mouse that carries a single copy of the GFP transgene on the X chromosome and a female homozygote GFP KI mouse that carries 2 copies of the GFP transgene on the X chromosome were used as copy number controls. (G) Transgene copy number was quantified with quantitative PCR using a 10-fold serial dilution standard curve of the pEGFP-N1 vector. (H) Table shows mean relative copy number ± SEM.
Reverse transcription (RT)-PCR analysis verified GFP messenger RNA (mRNA) expression across several adult tissues including liver, brain, spleen, and lung (Figure 1D). Furthermore, western blot analysis corroborated the mRNA profile, with GFP protein expression (27 kDa) observed in brain, lung, spleen, and thymus (Figure 1E). Analysis of gene copy number by quantitative genomic DNA PCR with 100 ng of DNA from 5 CD68-GFP animals showed the copy number of the CD68-GFP heterozygote animals to be 4 copies (Figure 1F-G). DNA from a line of transgenic animals generated by insertion of a single copy of the GFP transgene into the Hprt gene on the mouse X chromosome was used as a positive control (E.M., A.J.I., Jyoti Patel, Gemma E. White, D.R.-K., D.R.G., and K.M.C., manuscript in preparation [produced using genoway.com Quick-knock-in technology]). Male mice by definition have a single copy of the eGFP gene and homozygous female animals from this line have 2 eGFP genes per genome.
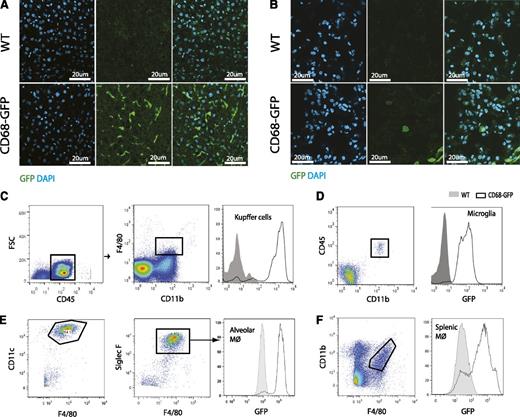
Immunofluorescence and flow cytometric analysis was carried out to define specific cellular localization of transgene GFP expression in tissue-resident macrophages. GFP expression was clearly evident in Kupffer cells of CD68-GFP mice (CD45+/CD11b+/F4/80+) liver sections (Figure 2A); this was confirmed in tissue flow cytometry analysis (Figure 2C). Similar GFP transgene expression was observed in lung (Figure 2B) and primary alveolar macrophages (CD11c+/Siglec-F+/F4/80Int) recovered by bronchial alveolar lavage (Figure 2E). Microglial cells (CD45Int+/CD11b+) (Figure 2D) and splenic macrophages (CD11b+/F4/80+) (Figure 2F) were also shown to express the GFP transgene in tissue flow cytometry analysis of adult brain and spleen. Costaining for macrophage restricted antigens Cd68 (macrosialin) and Galectin-3 showed strong association with GFP expression (supplemental Figure 1A-B). Similar GFP costaining patterns were observed in alveolar macrophages (supplemental Figure 1C-D). Partial colocalization with Cd68 and GFP was also observed in spleen and thymus (supplemental Figure 1E-F).
Immunofluorescence and flow cytometric analysis of GFP expression in tissue-resident macrophage populations. Immunofluorescence analysis for endogenous GFP expression in frozen tissue sections of (A) liver and (B) lung from CD68-GFP and WT mice. Flow cytometric analysis of GFP expression in (C) Kupffer cells (CD45+/CD11b+/F4/80+). (D) Microglia (CD45+/CD11bInt+), (E) alveolar macrophages (CD45+/CD11c+/Siglec-F+/F4/80Int+) and (F) splenic macrophages (CD11b+/ F4/80+) from CD68-GFP and WT mice. Results are representative of 3 mice per group.
Immunofluorescence and flow cytometric analysis of GFP expression in tissue-resident macrophage populations. Immunofluorescence analysis for endogenous GFP expression in frozen tissue sections of (A) liver and (B) lung from CD68-GFP and WT mice. Flow cytometric analysis of GFP expression in (C) Kupffer cells (CD45+/CD11b+/F4/80+). (D) Microglia (CD45+/CD11bInt+), (E) alveolar macrophages (CD45+/CD11c+/Siglec-F+/F4/80Int+) and (F) splenic macrophages (CD11b+/ F4/80+) from CD68-GFP and WT mice. Results are representative of 3 mice per group.
GFP expression during mouse development
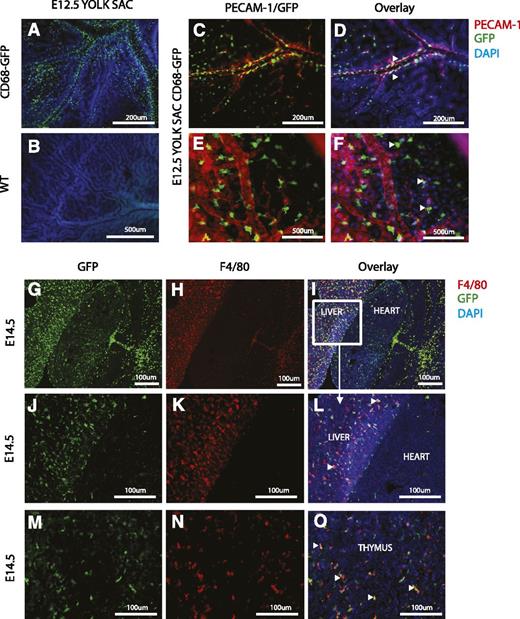
In mice, the earliest macrophages develop in the yolk sac from embryonic day 8 (E8), distinct from the hematopoietic progeny that emerge within the hematogenic endothelium between E10.5 and E12.5.29 Accordingly, GFP cells were detected as early as E8.5 in yolk sacs from the CD68-GFP reporter mice (data not shown). Whole mount staining of yolk sacs at E12.5 revealed many GFP-expressing cells, which were absent in WT control embryos (Figure 3A-B). Strikingly, analysis of the developing yolk sac vasculature revealed GFP-positive monocyte/macrophages along the abluminal surface of mature vessels, which were costained with the endothelial marker PECAM-1 (Figure 3C-D). GFP-positive macrophages were also observed at apparent sites of newly developing/sprouting blood vessels (Figure 3E-F).
Immunofluorescence analysis of GFP expression during mouse development. (A-F) Whole-mount staining of yolk sacs from CD68-GFP (A,C-F) and WT (B) embryos at E12.5 to detect the presence of GFP+ cells (monocytes and macrophages) by immunofluorescence with an anti-GFP monoclonal antibody (green). Costaining with PECAM-1 (red), an endothelial cell marker, shows association of GFP+ cells with vessels (arrowheads in D,F). Nuclei are stained with DAPI (blue). (G-O) Frontal cryosections from CD68-GFP mouse embryos at E14.5. Immunofluorescence staining shows GFP+ cells (green) in the liver, heart, and thymus (G,J,M). F4/80+ cells (red) are abundant in the liver and thymus, but fewer F4/80 macrophages are seen in the heart at this stage in development (H,K,N). Arrowheads in panels L and O mark cells that are both GFP and F4/80 positive. Nuclei are stained with DAPI (blue).
Immunofluorescence analysis of GFP expression during mouse development. (A-F) Whole-mount staining of yolk sacs from CD68-GFP (A,C-F) and WT (B) embryos at E12.5 to detect the presence of GFP+ cells (monocytes and macrophages) by immunofluorescence with an anti-GFP monoclonal antibody (green). Costaining with PECAM-1 (red), an endothelial cell marker, shows association of GFP+ cells with vessels (arrowheads in D,F). Nuclei are stained with DAPI (blue). (G-O) Frontal cryosections from CD68-GFP mouse embryos at E14.5. Immunofluorescence staining shows GFP+ cells (green) in the liver, heart, and thymus (G,J,M). F4/80+ cells (red) are abundant in the liver and thymus, but fewer F4/80 macrophages are seen in the heart at this stage in development (H,K,N). Arrowheads in panels L and O mark cells that are both GFP and F4/80 positive. Nuclei are stained with DAPI (blue).
GFP-positive cells were widely detected in the embryo proper from E10.5, with expression in the developing heart and brain and dorsal root ganglia (supplemental Figure 2). GFP-positive macrophages increased in number over the course of development and were abundant in the heart and liver from E14.5 (Figure 3G,J). F4/80 was used as an additional macrophage marker and good association with GFP expression was found in the developing liver and thymus; however, some cells remained only positive for F4/80 or GFP (Figure 3J-O). Interestingly, F4/80 staining was completely absent in the developing heart when compared with GFP expression (Figure 3G-L). Collectively, these data suggest that GFP transgene expression in CD68-GFP mice faithfully and effectively tracks the earliest emergence of macrophages and that this transgenic mouse could be a useful tool for studying the recruitment of monocytes/macrophages to sites of vasculogenesis.
Comparative analysis of GFP expression in leukocyte populations from CD68-GFP and CX3CR1GFP reporter mice
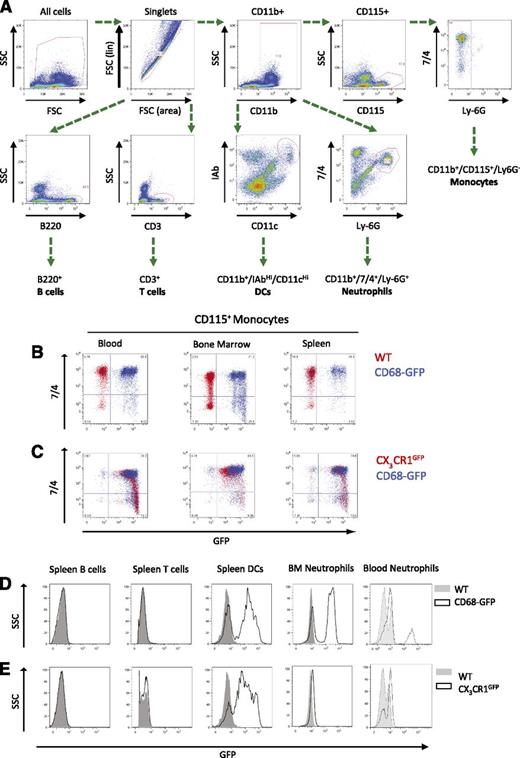
Tissue-resident macrophages in the adult mouse may arise from embryonic progenitor cells, fetal monocytes, or actively recruited monocytes.11,18 The recruitment of monocytes and subsequent retention and differentiation into macrophages is a key feature of many inflammatory conditions. To investigate the expression of the CD68-GFP transgene in other leukocyte populations, we performed detailed flow cytometry of major hematopoietic tissues using the gating scheme outlined in Figure 4A. Murine monocytes present in blood and the major monocyte reservoirs of the bone marrow and spleen express GFP. A 7/4 expression was used to discriminate between monocyte subsets,30 and we found that both the CD11b+/CD115+/Ly-6G−/7/4LO and CD11b+/CD115+/Ly-6G−/7/4HI monocyte populations displayed comparable levels of GFP transgene expression (Figure 4B). Analysis of Ly-6C is also commonly used to differentiate between mouse monocyte subsets; accordingly, we confirmed that Ly-6C and 7/4 identify the same populations of blood monocytes and that our observations of GFP expression can be applied to the Ly-6CHI and Ly-6CLO populations (supplemental Figure 3). The CX3CR1GFP mouse has been widely used as the transgenic of choice to investigate monocyte recruitment and trafficking.11,31,32 In this study, we carried out a side-by-side comparative analysis of CX3CR1GFP and CD68-GFP leukocyte populations to assess the level of GFP transgene expression in monocytes and macrophages. GFP expression was observed in monocytes isolated from CX3CR1GFP and CD68 GFP mice, respectively (Figure 4C). Similar to CX3CR1GFP mice, lymphocyte subsets (B and T cells) in the CD68-GFP mice were negative for GFP; however, GFP expression was detected in a fraction of splenic DCs (Figure 4D) which was also observed in CX3CR1GFP mice (Figure 4E). CD68-GFP mice were also shown to express GFP in a minority population of blood neutrophils and a larger proportion of bone marrow neutrophils (Figure 4D). Further analysis of the expression of GFP in the neutrophil population revealed altered expression of CXCR2 and CXCR4 between the GFP+ and GFP– neutrophil populations in CD68GFP mice, with greater expression of the bone marrow–associated receptors in the GFP+ neutrophils (supplemental Figure 4).
Comparative flow cytometric analysis of GFP expression in blood, bone marrow, and spleen of CD68-GFP and CX3CR1GFP mice. (A) The gating and staining strategy used to characterize various leukocyte populations (spleen was used to set antibody panel; isotype controls used are listed in “Methods”). (B-C) Monocytes from blood, bone marrow, and spleen displayed GFP expression in both CD68-GFP and CX3CR1GFP mice. WT mice were used as negative controls for GFP expression and shown overlaid as red dots (B). (D-E) CD68-GFP mice express GFP in a subset of bone marrow/blood neutrophils and splenic DCs, but expression was absent in lymphocytes. (Representative flow cytometry plots are shown; analysis was carried out in 3 to 4 mice.)
Comparative flow cytometric analysis of GFP expression in blood, bone marrow, and spleen of CD68-GFP and CX3CR1GFP mice. (A) The gating and staining strategy used to characterize various leukocyte populations (spleen was used to set antibody panel; isotype controls used are listed in “Methods”). (B-C) Monocytes from blood, bone marrow, and spleen displayed GFP expression in both CD68-GFP and CX3CR1GFP mice. WT mice were used as negative controls for GFP expression and shown overlaid as red dots (B). (D-E) CD68-GFP mice express GFP in a subset of bone marrow/blood neutrophils and splenic DCs, but expression was absent in lymphocytes. (Representative flow cytometry plots are shown; analysis was carried out in 3 to 4 mice.)
To further evaluate any expression of GFP in minor cell populations, a reverse-gating experiment was performed. Peritoneal exudate cells, blood, bone marrow, and spleen cell suspensions were first gated for GFP expression and then GFP+ cells were interrogated for expression of other lineage markers (supplemental Figure 5A-D). The majority of cells expressing GFP in peritoneal cavity, blood, bone marrow, and spleen are the myeloid cell populations highlighted in Figure 4. However, expression of GFP was also seen in a small population of peritoneal B1 cells (supplemental Figure 5A), although this was a minority population of the total peritoneal B-cell population. Expression in a small population of B cells was also seen in blood and spleen (supplemental Figure 5B,D), although a minority population as seen by comparison with Figure 4D. GFP expression was seen in eosinophils, but not in mast cells, basophils, or NK cells (supplemental Figure 5E-F).
Human CD68 promoter drives high-level GFP transgene expression in resident peritoneal macrophages
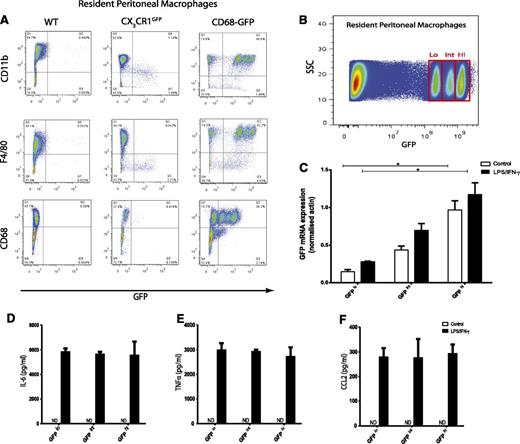
It has previously been reported that the CX3CR1GFP mouse does not express GFP in certain resting tissue resident macrophage populations, including Kupffer cells, alveolar macrophages, and peritoneal macrophages.11,24 Although CD68-GFP mice showed GFP transgene expression in Kupffer cells, alveolar macrophages, and microglial cells (Figure 2; supplemental Figure 1), we decided to examine GFP expression in resident peritoneal macrophages. Peritoneal macrophages, defined as CD11b+/F4/80+/CD68+, isolated from the peritoneal cavity of nonstimulated animals were highly positive for GFP when compared with WT and CX3CR1GFP mice by flow cytometry analysis (Figure 5A). Closer examination revealed 3 distinct GFP populations (referred to as GFPhi, int, lo) in the peritoneal macrophage pool of the CD68-GFP mouse. The levels of GFP expression were not associated with cell clumping because all cells analyzed were confirmed as singlets by analysis of pulse width (data not shown). Cell-cycle analysis of all 3 GFP populations showed that all cells were in a G0/G1 “resting” phase, which indicates no active cell division (supplemental Figure 6A-C). Identical transgene copy number was also found across all 3 populations (supplemental Figure 6D-E). GFP mRNA levels were measured in these 3 sorted populations by quantitative RT-PCR. The mRNA levels correlated with GFP protein expression with GFP transgene mRNA being expressed at 4-fold higher levels in GFPhi cells compared with GFPlo cells (Figure 5B-C). Stimulation for 12 hours with lipopolysaccharide (LPS) and interferon--γ showed no differences in pro-inflammatory cytokine or chemokine production among the 3 GFP peritoneal populations (Figure 5D-F). These data provide evidence that normal macrophage function is not impaired by GFP transgene expression.
Resident peritoneal macrophages from CD68-GFP mice express high levels of GFP. (A) Flow cytometry analysis of resident peritoneal macrophages isolated from CD68-GFP mice revealed 3 distinct populations with different levels of GFP expression. These 3 populations were positive for CD11b, F4/80, and CD68 (myeloid/macrophage markers). In contrast, resident peritoneal macrophages isolated from CX3CR1GFP mice displayed no GFP expression, although they were positive for CD11b, F4/80, and CD68. (Representative flow cytometry plots are shown; analysis was carried out in 5 to 8 mice.) (B) Three GFP resident macrophages populations (GFPhi, int, lo) from a pool of 10 CD68-GFP mice were sorted on the basis of GFP expression. (C) Total RNA was isolated, and quantitative RT-PCR was carried out to measure GFP mRNA expression in control cells and cells featuring LPS and interferon-γ. (D-F) Cytokine and chemokine levels were measured in LPS/interferon-γ–stimulated supernatants. (D) IL-6 (E) tumor necrosis factor-α (TNF-α), and (F) CCL2 levels showed no difference across all 3 populations. Data are mean ± SEM of 3 independent experiments, with 3 technical replicates per experiment. Statistical significance was assessed by 1-way analysis of variance and Dunnett’s multiple comparison posttest. ND, not detectable.
Resident peritoneal macrophages from CD68-GFP mice express high levels of GFP. (A) Flow cytometry analysis of resident peritoneal macrophages isolated from CD68-GFP mice revealed 3 distinct populations with different levels of GFP expression. These 3 populations were positive for CD11b, F4/80, and CD68 (myeloid/macrophage markers). In contrast, resident peritoneal macrophages isolated from CX3CR1GFP mice displayed no GFP expression, although they were positive for CD11b, F4/80, and CD68. (Representative flow cytometry plots are shown; analysis was carried out in 5 to 8 mice.) (B) Three GFP resident macrophages populations (GFPhi, int, lo) from a pool of 10 CD68-GFP mice were sorted on the basis of GFP expression. (C) Total RNA was isolated, and quantitative RT-PCR was carried out to measure GFP mRNA expression in control cells and cells featuring LPS and interferon-γ. (D-F) Cytokine and chemokine levels were measured in LPS/interferon-γ–stimulated supernatants. (D) IL-6 (E) tumor necrosis factor-α (TNF-α), and (F) CCL2 levels showed no difference across all 3 populations. Data are mean ± SEM of 3 independent experiments, with 3 technical replicates per experiment. Statistical significance was assessed by 1-way analysis of variance and Dunnett’s multiple comparison posttest. ND, not detectable.
Adoptively transferred CD68-GFP monocytes are recruited to sites of inflammation and maintain GFP expression after differentiation to macrophages
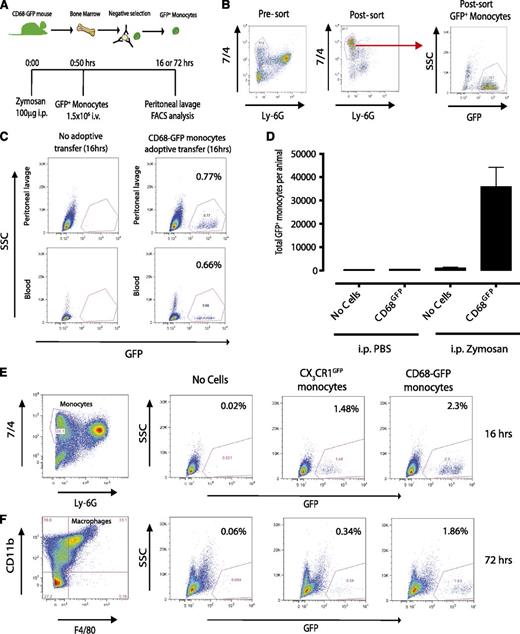
To determine whether the CD68-GFP mouse would be a useful resource to study in vivo monocyte trafficking and macrophage differentiation, we carried out adoptive transfer studies in a model of sterile inflammation. Figure 6A summarizes the experimental design used. Isolation of bone marrow monocytes with negative immunomagnetic selection typically provided a yield of 70% monocytes (defined as 7/4+/Ly6G−), of which 70% to 80% were positive for GFP (Figure 6B). Adoptive transfer of 1.5 × 106 CD68-GFP+ monocytes IV to C57BL/6J recipient mice 30 minutes after challenge with 100 µg zymosan resulted in 3 to 4 × 104 GFP+ monocytes (0.7% to 2.3%) being recruited to the peritoneal cavity at 16 hours (Figure 6C-E). These recruited monocytes were shown to be retained at the site of inflammation and differentiate to macrophages (characterized as CD11b+/F4/80+) at 72 hours (Figure 6F). In a parallel series of experiments, CX3CR1GFP monocytes adoptively transferred into a sterile peritonitis model were also detectable at 16 hours, but downregulated GFP expression (∼4.5-fold reduction) by 72 hours posttransfer (Figure 6E-F). Because the increase in cellular autofluorescence during macrophage differentiation, gates for GFP-positive cells were set according to the appropriate GFP-negative control for each time point.33
Adoptive transfer of CD68-GFP and CX3CR1GFP monocytes. (A-B) Monocytes were isolated from CD68-GFP bone marrow using negative immunomagnetic selection (see “Methods”). Isolated monocytes were characterized as 7/4high/Ly6Glow, with a typical yield of 70% monocytes, of which 70% to 80% were positive for GFP. (C) A total of 1.5 × 106 isolated CD68-GFP monocytes were adoptively transferred into C57BL/6J mice by IV injection 30 minutes after intraperitoneal injection with 100 μg zymosan. Mice were euthanized at 16 hours, and peritoneal lavage and blood samples were taken. Adoptively transferred GFP+ monocytes were present in peritoneal lavage (range 2 to 3 × 104) and blood of animals that received zymosan. (D) Recruited cell numbers are expressed as mean ± SEM of 6 mice. (E-F) Representative flow cytometry plots of peritoneal lavage from C57BL/6J mice that received adoptively transferred GFP positive monocytes from CD68-GFP or CX3CR1GFP mice (IV) during ongoing zymosan-induced peritonitis. (E) Mice were euthanized at 16 hours, and recruited monocytes were analyzed. (F) Mice were euthanized at 72 hours, and GFP+ monocytes that had differentiated into macrophages were identified using F4/80 and CD11b.
Adoptive transfer of CD68-GFP and CX3CR1GFP monocytes. (A-B) Monocytes were isolated from CD68-GFP bone marrow using negative immunomagnetic selection (see “Methods”). Isolated monocytes were characterized as 7/4high/Ly6Glow, with a typical yield of 70% monocytes, of which 70% to 80% were positive for GFP. (C) A total of 1.5 × 106 isolated CD68-GFP monocytes were adoptively transferred into C57BL/6J mice by IV injection 30 minutes after intraperitoneal injection with 100 μg zymosan. Mice were euthanized at 16 hours, and peritoneal lavage and blood samples were taken. Adoptively transferred GFP+ monocytes were present in peritoneal lavage (range 2 to 3 × 104) and blood of animals that received zymosan. (D) Recruited cell numbers are expressed as mean ± SEM of 6 mice. (E-F) Representative flow cytometry plots of peritoneal lavage from C57BL/6J mice that received adoptively transferred GFP positive monocytes from CD68-GFP or CX3CR1GFP mice (IV) during ongoing zymosan-induced peritonitis. (E) Mice were euthanized at 16 hours, and recruited monocytes were analyzed. (F) Mice were euthanized at 72 hours, and GFP+ monocytes that had differentiated into macrophages were identified using F4/80 and CD11b.
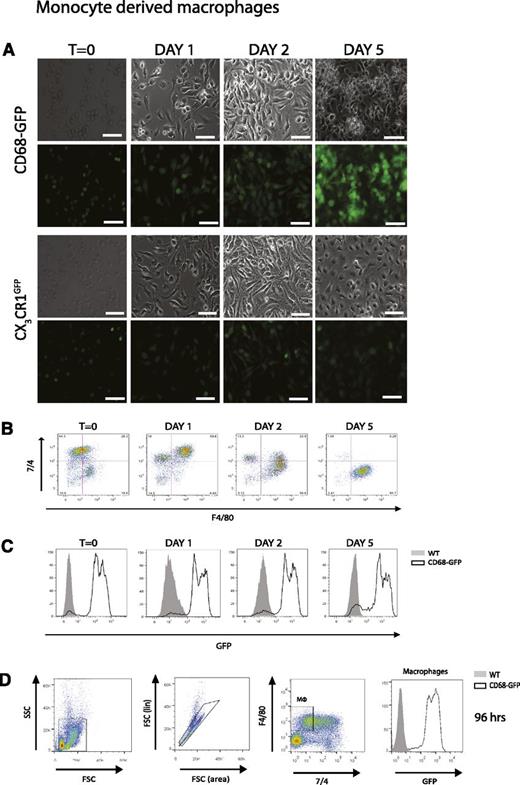
To confirm our in vivo observations, we cultured isolated bone marrow monocytes from CD68-GFP and CX3CR1GFP mice in vitro in macrophage colony-stimulating factor–containing media. Using immunofluorescence and flow cytometric analysis, we clearly demonstrate that CD68-GFP monocytes retain and indeed increase GFP expression during differentiation from monocytes to macrophages in vitro, whereas CX3CR1GFP monocytes downregulate GFP expression during macrophage colony-stimulating factor–mediated differentiation (Figure 7A-C). In vivo, CD68-GFP mice challenged with 100 µg zymosan for 96 hours showed that the repopulating F4/80+ macrophages during the resolution phase of inflammation were highly positive for GFP transgene expression (Figure 7D).
CD68-GFP mice display increased GFP expression during monocyte-to-macrophage differentiation compared with CX3CR1GFP mice. Bone marrow monocytes isolated from CD68-GFP and CX3CR1GFP mice using negative immunomagnetic selection were cultured in vitro in macrophage colony-stimulating factor containing media. (A) Images taken across 5 days indicate GFP expression is maintained and increased as CD68-GFP monocytes differentiate. In contrast, monocytes isolated from CX3CR1GFP reporter mice showed loss of GFP fluorescence over this time course. Representative images are shown; similar results were obtained with monocytes from 2 independent mice (scale bar = 40 µm). (B-C) Flow cytometry analysis was also carried out to confirm GFP expression levels and macrophage differentiation using F4/80 and 7/4. (D) CD68-GFP and WT mice were injected intraperitoneally with 100 µg zymosan. Mice were euthanized at 96 hours, and a representative flow cytometry plot of peritoneal macrophages using F4/80 and 7/4 and GFP expression is shown (Analysis was carried out in 3 to 4 mice.)
CD68-GFP mice display increased GFP expression during monocyte-to-macrophage differentiation compared with CX3CR1GFP mice. Bone marrow monocytes isolated from CD68-GFP and CX3CR1GFP mice using negative immunomagnetic selection were cultured in vitro in macrophage colony-stimulating factor containing media. (A) Images taken across 5 days indicate GFP expression is maintained and increased as CD68-GFP monocytes differentiate. In contrast, monocytes isolated from CX3CR1GFP reporter mice showed loss of GFP fluorescence over this time course. Representative images are shown; similar results were obtained with monocytes from 2 independent mice (scale bar = 40 µm). (B-C) Flow cytometry analysis was also carried out to confirm GFP expression levels and macrophage differentiation using F4/80 and 7/4. (D) CD68-GFP and WT mice were injected intraperitoneally with 100 µg zymosan. Mice were euthanized at 96 hours, and a representative flow cytometry plot of peritoneal macrophages using F4/80 and 7/4 and GFP expression is shown (Analysis was carried out in 3 to 4 mice.)
We wanted to confirm that there was no effect on macrophage function as a result of carrying the GFP transgene. To address this, we carried out chemotaxis assays using Bio-Gel–elicited macrophages, which showed no significant differences between CD68-GFP and WT macrophages in response to a range of CC chemokines (supplemental Figure 7A-B).
Human CD68 promoter drives high-level GFP transgene expression in chronic inflammation
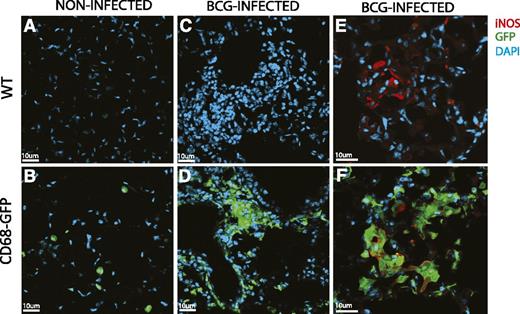
To confirm whether the CD68-GFP is a useful model for probing models of chronic inflammation, we performed intranasal bacille Calmette-Guérin (BCG) infections in CD68-GFP and WT mice. Mice were harvested 4 weeks after infection and had a clear ongoing lung inflammation typified by areas of dense leukocyte accumulation (Figure 8). Uninfected naive CD68-GFP animals showed widely distributed single green cells (Figure 8B), identified as mCd68-expressing alveolar macrophages in supplemental Figure 1C. BCG-infected animals show localized dense accumulations of GFP-expressing leukocytes (Figure 8D). Similar areas identified by 4,6 diamidino-2-phenylindole (DAPI) staining in the WT animals show no GFP signal (Figure 8A,C). The expression of inducible nitric oxide synthase (iNOS) identifies cells polarized to an M1 antimicrobial phenotype.1 No iNOS expression was seen in uninfected lung sections, whereas discrete foci of iNOS expression were seen in the BCG-infected lungs (Figure 8E) and iNOS expression was seen to be localized to a population of GFP-expressing cells (Figure 8F).
CD68-GFP macrophage BCG infection. BCG Pasteur was administered intranasally at 4 × 105 colony-forming unit to WT and CD68-GFP mice. After 4 weeks, mice were euthanized, and immunofluorescence analysis for transgene GFP expression in frozen lung tissue sections of naive (A-B) and BCG-infected (C-D) CD68-GFP and WT mice was performed. (E-F) Costaining with iNOS (red) was performed and showed strong association with endogenous GFP expression.
CD68-GFP macrophage BCG infection. BCG Pasteur was administered intranasally at 4 × 105 colony-forming unit to WT and CD68-GFP mice. After 4 weeks, mice were euthanized, and immunofluorescence analysis for transgene GFP expression in frozen lung tissue sections of naive (A-B) and BCG-infected (C-D) CD68-GFP and WT mice was performed. (E-F) Costaining with iNOS (red) was performed and showed strong association with endogenous GFP expression.
Discussion
We have generated and characterized a novel transgenic reporter mouse expressing a GFP transgene under the control of the human CD68 promoter. GFP transgene expression can readily be detected in embryonic, fetal, and adult tissues in a pattern consistent with expression by tissue resident macrophages. Side-by-side flow cytometry analysis of hematopoietic cells of CD68-GFP and CX3CR1GFP mice showed similar but nonidentical patterns of transgene expression with high-level GFP expression in monocyte-derived resident peritoneal macrophages seen only in CD68-GFP animals (Figure 5). Additionally, we followed CD68-GFP and CX3CR1GFP monocyte differentiation in vitro (Figure 7) and monocyte adoptive transfer experiments in a sterile peritonitis model in vivo (Figure 6). Taken together, our experiments show that CD68-GFP monocyte-derived macrophages retain high-level GFP transgene expression after monocyte differentiation in vitro and in vivo.
The 2.9-kb human CD68 promoter and 83-bp first intron expression cassette has been used to direct expression of a range of different transgenes at a high level in macrophages in vivo, including SR-A,34 interleukin-10,28 PGC1 β,35 matrix metallopeptidase-9,36 FcγIIb,37 Fox-P1,38 acyloxyacyl hydrolase,39 peroxisome proliferator-activated receptor γ,40 and hPOP3.41 In previous work, we used chromatin precipitation experiments to show that the ETS transcription factor family members PU.1, Elf-1, and Fli-1 are bound to the CD68 proximal promoter in myeloid THP-1 cells, but these transcription factors are not bound in primary B-lymphocytes because of interactions with IRF-4.42 The myeloid vs lymphoid specificity of the human CD68 proximal promoter is recapitulated in our transgenic mice (Figure 4D), in which a 3-kb fragment of the human CD68 promoter and intron 1 are integrated into mouse chromosome 3. The strong similarity of expression pattern between the human CD68 promoter–driven transgene and the endogenous Cd68 macrosialin gene (supplemental Figure 1) is likely from evolutionary conservation of binding sites for myeloid lineage–specific transcription factors.43
During embryonic development, GFP-positive cells were observed from E8.5 (data not shown). GFP-positive cells were present throughout the yolk sac at E12.5. This correlates with what has previously been shown with the MacGreen mouse.44 A strong correlation of CD68-GFP–positive cells and developing vessels was observed (Figure 3). Macrophages are known to be involved in the regulation of angiogenesis1,45 and play a role as vascular fusion cells during in the anastomosis of tip and stalk cells.46 Macrophages begin to populate the embryonic liver from E10 and continue to grow in number throughout development.44,47 We found that both F4/80+ and GFP+ macrophages were abundant in the liver at E14.5. There were notably more GFP+ cells in the developing heart than F4/80+ cells. Previous histological studies have shown that from E14 F4/80+ macrophages are present throughout the thymus.47 We found that both F4/80+ and GFP+ macrophages were abundant within the thymus at E14.5. There was clearly strong correlation between GFP and F4/80 expression in the liver heart and thymus; however, a proportion of cells remained only positive for GFP or F4/80. These observations correspond with what has previously been reported in both the MacGreen44 and the MacBlue48 reporter mice, in which transgene expression did not fully map onto F4/80 expression. It has previously been noted that F4/80 is not detectable in several tissue macrophage populations, which may be the case here.49-51
A study carried out by Pillai et al generated a transgenic mouse using human CD68 promoter to drive expression of the reverse tetracycline activator rTA-M2.52 This mouse was then crossed with a second reporter that had GFP under the control of a tetracycline responsive element promoter. The study reported that GFP expression did not completely map onto F4/80 or Csf1r expression in the bone marrow. However, double staining on tissue-resident macrophages was not carried out in this study.52 Our findings clearly indicate high levels of GFP transgene expression in resident macrophage populations, including Kupffer cells, microglia, and peritoneal, splenic, and alveolar macrophages.
Interestingly, peritoneal macrophages isolated from the CD68-GFP mouse fell into 4 distinct populations, expressing different levels of GFP. Macrophage markers including F4/80, CD11b, and CD68 were all expressed at comparable levels across all populations. No differences were observed in cell cycle, transgene copy number, and pro-inflammatory cytokine production after ex vivo stimulation with LPS in the 3 GFP+ populations. A small proportion of resident peritoneal cells (∼15%) that were positive for F4/80, CD11b, and CD68 remained negative for GFP transgene expression. One possible explanation for these differences could be that the CD68-driven transgene expression is detecting a subpopulation of resident peritoneal cells that is not detected by the panel of antibodies widely used to define murine peritoneal macrophages CD11b, F4/80, and CD68. Alternatively, we can speculate that epigenetic changes within the transgene chromatin may account for the observed differences in GFP expression in the 3 peritoneal macrophage populations. Epigenetic silencing has been observed for viral promoters in transgenic mice.53
Flow cytometric analysis revealed GFP transgene expression in 100% of bone marrow monocytes (Figure 4B) and ∼60% of bone marrow–derived neutrophils (Figure 4D). A recent study from Amanzada et al demonstrated CD68 mRNA expression in neutrophils isolated from patients with inflammatory bowel disease.54 The observation that a greater percentage of bone marrow neutrophils expressed GFP than within the mobilized blood neutrophil population may indicate carryover of GFP expression from a shared progenitor population. Flow cytometric phenotyping using antibodies directed against the CXCR2 and CXCR4 chemokine receptors showed that GFP expression by neutrophils in the blood of CD68-GFP mice may be able to inform of neutrophil subsets, with the newly mobilized CXCR4LO neutrophils showing high GFP expression compared with GFP− CXCR4 expressing senescent neutrophil population55 (supplemental Figure 4). Other “macrophage-specific” markers including Emr1 (F4/80) and Csf1r mRNA have also been found in murine neutrophil granulocytes,56 supporting the view that the generation of a truly macrophage specific reporter may be unattainable (reviewed elsewhere22 ). Given our less than complete understanding of myeloid differentiation programs in vitro and in vivo, it is interesting to speculate on the differences seen in human CD68 promoter-driven GFP transgene expression with CX3CR1 knock-in GFP expression. It is possible that pronuclear injection of a cell type–specific promoter driving a GFP transgene has given higher absolute levels of transgene expression than can be obtained with knock-in approaches and that this may allow for better detection on monocyte-derived macrophages in sites of chronic inflammation. Evidence for this assertion comes from our analysis of the chronic inflammation seen in BCG-infected lungs 4 weeks after infection (Figure 8).
Monocyte recruitment, macrophage differentiation, retention, and egress from inflammatory lesions in vivo are an active area of research. Current transgenic reporter lines including CX3CR1-GFP,24 CX3CR1Cre,11 csf1r-EGFP (MacGreen),44 and CCR2-RFP25 have been used to study monocyte recruitment and trafficking during steady-state and different models of chronic inflammation. In this study, we demonstrate that CD68-GFP reporter mice that express high levels of the GFP transgene under steady-state conditions offer significant advantages over other monocyte/macrophage reporter mice. We believe that CD68-GFP mice will find application in studying monocyte recruitment to sites of chronic inflammation and local differentiation into macrophages.
This article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank David Jackson and Louise Johnson for providing the CX3CR1GFP mice and all reviewers for their helpful comments.
This work was supported by the British Heart Foundation (grants RG/10/15/28578, FS/11/82/29332, FS/13/4/30045, and FS/12/69/30008), the National Institutes of Health (grants HL076746 and DK094641) (A.C.) , and a National Institutes of Health Director’s Pioneer Award (DP1AR064158). FISH analysis was performed by the Chromosome Dynamics Core, funded by Wellcome Trust Core Award Grant Number (090532/Z/09/Z).
Authorship
Contribution: A.J.I., E.M., S.N., T.S.K., D.R.-K., S.B., N.S., D.R.G., H.M., E.S., and D.E.W.M. performed experiments; A.J.I. and E.M. analyzed results and made the figures; A.J.I., E.M., K.M.C, A.C., and D.R.G. designed the research; and A.J.I., E.M., and D.R.G. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David R. Greaves, Sir William Dunn School of Pathology, University of Oxford, South Parks Rd, Oxford, OX1 3RE; e-mail: david.greaves@path.ox.ac.uk; and Ajay Chawla, Cardiovascular Research Institute, Departments of Physiology and Medicine, University of California, San Francisco, CA; e-mail: ajay.chawla@ucsf.edu.
References
Author notes
A.J.I. and E.M. contributed equally to this study.