Key Points
A novel RARα fusion gene, TBLR1-RARα, was found in rare cases of APL with t(3;17) chromosomal translocation.
TBLR1-RARα exhibited diminished transcriptional activity by recruiting more corepressors compared with RARα.
Abstract
The majority of acute promyelocytic leukemia (APL) cases are characterized by the PML-RARα fusion gene. Although the PML-RARα fusion gene can be detected in >98% of APL cases, RARα is also found to be fused with other partner genes, which are also related to all-trans retinoic acid (ATRA)-dependent transcriptional activity and cell differentiation. In this study, we identified a novel RARα fusion gene, TBLR1-RARα (GenBank KF589333), in a rare case of APL with a t(3;17)(q26;q21),t(7;17)(q11.2;q21) complex chromosomal rearrangement. To our knowledge, TBLR1-RARα is the 10th RARα chimeric gene that has been reported up to now. TBLR1-RARα contained the B-F domains of RARα and exhibited a distinct subcellular localization. It could form homodimers and also heterodimers with retinoid X receptor α. As a result, TBLR1-RARα exhibited diminished transcriptional activity by recruitment of more transcriptional corepressors compared with RARα. In the presence of pharmacologic doses of ATRA, TBLR1-RARα could be degraded, and its homodimerization was abrogated. Moreover, when treated with ATRA, TBLR1-RARα could mediate the dissociation and degradation of transcriptional corepressors, consequent transactivation of RARα target genes, and cell differentiation induction in a dose- and time-dependent manner.
Introduction
Acute promyelocytic leukemia (APL) is a special disease entity of acute myeloid leukemia. With the clinical use of all-trans retinoic acid (ATRA), APL turns into the most curable form of acute myeloid leukemia. The majority of APL cases are characterized by the fusion between the promyelocytic leukemia (PML) gene and the retinoic acid receptor α (RARα) gene, which is the consequence of t(15;17)(q22;q21) chromosomal translocation.1 Although the PML-RARα fusion gene can be detected in >98% of APL patients,2 RARα is also found to be fused with other partner genes,3-10 such as PML zinc finger (PLZF), nucleophosmin (NPM), and so forth. The RARα portion within fusion proteins is conserved, containing B-F domains of RARα, which cover the DNA-binding and ligand-binding motifs, so that RARα can be combined with retinoid X receptor α (RXRα) to bind the retinoic acid responsive element (RARE).1,11 As a result, the fusion proteins usually form heterodimers with RXRα.9,12 A critical property that all RARα fusion proteins have in common is the ability to self-associate, forming homodimers.13 The chimeric proteins, such as PML-RARα, PLZF-RARα, and NPM-RARα, can robustly recruit transcriptional corepressors, including nuclear receptor corepressor/silencing mediator of retinoid and thyroid receptors (N-CoR/SMRT) and histone deacetylases (HDACs), to RARE and consequently result in ectopic repression of RARα target genes.1,13,14
Transducin β-like 1 X-linked receptor 1 (TBLR1) gene, located at chromosome 3q26.32, encodes an F-box/beta-transducin (WD-40) repeat-containing protein, which contains a lissencephaly type-1-like homology motif (LisH domain) in its N-terminal region and 8 WD-40 repeats in its C-terminal region.15 Overexpression of TBLR1 was validated in lung cancer and breast cancer and was also found enriched in human hematopoietic stem cells.16-18 Moreover, recurrent abnormality of TBLR1 gene locus was highlighted in ets variant 6-runt-related transcription factor 1 (ETV6-RUNX1)-positive acute lymphoblastic leukemia and high-risk childhood acute lymphoblastic leukemia, and a novel rearrangement associated with TBLR1, TBLR1/TP63, was described in B-cell non-Hodgkin lymphoma recently.19-21
Here, we reported a novel RARα fusion gene, TBLR1-RARα, which is generated by a t(3;17)(q26;q21) chromosomal rearrangement in a rare variant APL patient. In this study, we identified the molecular characterization of TBLR1-RARα, illustrated the feature of the chimeric protein generated by TBLR1-RARα, clarified its pathogenesis in APL, and investigated its sensitivity to ATRA treatment. As far as we know, this is the first study demonstrating the involvement of the TBLR1 gene in APL.
Materials and methods
Case report
Patient 1, a 63-year-old man, was admitted because of fatigue, fever, and skin ecchymosis. The blood count showed a hemoglobin level of 63 g/L; white blood cell count of 14.14 × 109/L, including 49% promyelocytes; and platelet count of 72 × 109/L. Bone marrow (BM) smear showed hypercellularity with 83% hypergranular promyelocytes (Figure 1A). BM histochemistry showed the typical pattern of APL (Figure 1B-D). Leukemia cells expressed myeloperoxidase, CD13, and CD9; partially expressed CD117, CD15, CD33, and CD64; but lacked expression of CD34 and HLA-DR. The patient was diagnosed with APL; however, both reverse transcription-polymerase chain reaction (RT-PCR) and fluorescence in situ hybridization (FISH) failed to detect the PML-RARα fusion gene from the BM (Figure 1E). Karyotype analysis revealed 46,XY,t(3;17)(q26;q21),t(7;17)(q11.2;q21)[14]/46,XY[1] (Figure 1F).
Morphologic and cytogenetic analysis of BM sample of the APL case with t(3;17)(q26;q21),t(7;17)(q11.2;q21) translocation. (A) The May-Giemsa staining. BM cells were shown to be blocked at the hypergranular promyelocytic stage lacking Auer rods. (B) Myeloperoxidase staining. (C) Specific esterase staining. (D) Nonspecific esterase staining. (B-D) Cellular histochemical staining examination exhibited high positive rates of blast cells. (E) FISH analysis. PML probe (orange signal) and RARα probe (green signal) were used. No PML-RARα was detected. (F) Karyotype analysis. 46,XY,t(3;17)(q26;q21),t(7;17)(q11.2;q21) was found in 14 metaphases among 15 metaphases detected. (G) Karyotype analysis of relapsed BM cells. A more complex recurring chromosomal transcription, 46,XY,t(3;17)(q26;q21),add(5)(q32),del(6)(q22q24),t(7;17)(q11.2;q21),add(10)(q26),del(11)(p14)[13]/46,XY[1], was observed in 13 metaphases among 14 metaphases detected. The rearrangements and break points in panels F and G were indicated by arrows.
Morphologic and cytogenetic analysis of BM sample of the APL case with t(3;17)(q26;q21),t(7;17)(q11.2;q21) translocation. (A) The May-Giemsa staining. BM cells were shown to be blocked at the hypergranular promyelocytic stage lacking Auer rods. (B) Myeloperoxidase staining. (C) Specific esterase staining. (D) Nonspecific esterase staining. (B-D) Cellular histochemical staining examination exhibited high positive rates of blast cells. (E) FISH analysis. PML probe (orange signal) and RARα probe (green signal) were used. No PML-RARα was detected. (F) Karyotype analysis. 46,XY,t(3;17)(q26;q21),t(7;17)(q11.2;q21) was found in 14 metaphases among 15 metaphases detected. (G) Karyotype analysis of relapsed BM cells. A more complex recurring chromosomal transcription, 46,XY,t(3;17)(q26;q21),add(5)(q32),del(6)(q22q24),t(7;17)(q11.2;q21),add(10)(q26),del(11)(p14)[13]/46,XY[1], was observed in 13 metaphases among 14 metaphases detected. The rearrangements and break points in panels F and G were indicated by arrows.
Remission induction therapy was conducted with ATRA (20-30 mg/m2 per day from days 1 to 24), cytarabine (50 mg/m2 per day from days 7 to 13), and mitoxantrone (MTZ, 5 mg/m2 per day from days 11 to 14). However, the treatment was suspended on day 24 because of severe pulmonary infection. On day 32, BM smear showed induction failure with 58% promyelocytes. Eleven days after suspension, another course of treatment was conducted with arsenic trioxide (As2O3, 10 mg/d for 28 days) and MTZ (5 mg/m2 per day from days 6 to 8). Forty-six days after discontinuation of As2O3, complete remission (CR) was documented. A set of consolidation therapies was administered subsequently. Ten months after diagnosis, the patient relapsed with 81% of promyelocytes in BM. The karyotype analysis revealed a more complex chromosomal translocation: 46,XY,t(3;17)(q26;q21),add(5)(q32),del(6)(q22q24),t(7;17)(q11.2;q21),add(10)(q26),del(11)(p14)[13]/46,XY[1] (Figure 1G). The patient was treated in a local hospital and died 1 month after relapse.
We also investigated another 2 APL patients harboring t(3;17) chromosomal translocation; their karyotypes were reported as follows: 45-46,XY,t(3;17)(q27;q22),del(1)(q21),-7,del(7)(p13),del(7)(q22q34)[cp16]/46,XY[3] (patient 2) and 46,XX,t(3;17)(q21;q25),t(15;17)(q22;q12)[3]/46,XX[5] (patient 3). In patient 3, PML-RARα transcript was also detected by RT-PCR.
All patients gave informed consent, and studies using patient samples were approved by the ethical advisory board of the Institute of Hematology and Blood Diseases Hospital. This study was conducted in accordance with the Declaration of Helsinki.
Cell lines and reagents
The 293T, CV-1, and HEK293 cells were maintained in Dulbecco’s Modified Eagle Medium; K562, U937, HL-60, and NB4 cells were maintained in RPMI 1640 medium; and all were supplemented with 10% fetal bovine serum (HyClone). ATRA and polybrene were purchased from Sigma-Aldrich.
5′-rapid amplification of cDNA ends (5′-RACE)
The 5′-RACE was performed to amplify the unknown chimeric fusion transcript. BM samples were obtained from the above-mentioned APL patient 1, and total RNA was extracted from BM mononuclear cells (BMMNCs) with RNAiso Plus (TaKaRa). The 5′-RACE was performed with a SMARTer RACE complementary DNA (cDNA) Amplification kit (Clontech) according to manufacturer’s instructions. Total RNA was used to synthesize the first-strand cDNA, and the universal primer A mix and gene-specific primer (5′-GCGAGGGAGGGCTGGGCACTATCT-3′) was used to amplify the unknown fusion partner of RARα. PCR products were purified, sequenced, and compared with GenBank sequences.
RNA isolation and RT-PCR
TBLR1-RARα messenger RNA (mRNA) and its reciprocal RARα-TBLR1 mRNA transcripts were assessed by RT-PCR. Total RNA from patients’ BMMNCs, TBLR1-RARα transfected K562 cells, and K562 cells were isolated. cDNA was synthesized using a reverse transcription kit (Invitrogen). A LongAmp Taq PCR Kit (New England Biolabs) was used for PCR amplification following the manufacturer’s instructions. Primers used for PCR were listed as follows: (1) TBLR1-RARα (199 bp): 5′-ATGCCGTAATGCCTGATG-3′ and 5′-GAACTGCTGCTCTGGGTCT-3′; (2) TBLR1-RARα (full-length): 5′-ATTGGCGGTGACCGGATATTCAGTTGCA-3′ and 5′-AGGGCTGTGTCCATGTGGCGTGG-3′; (3) RARα-TBLR1: 5′-GTGCCTCCCTACGCCTTCTTCT-3′ and 5′-ACATCCCCATCCACTTCCATCA-3′; and (4) glyceraldehyde-3-phosphate dehydrogenase (GAPDH): 5′-GAAGGTGAAGGTCGGAGTC-3′ and 5′-GAAGATGGTGATGGGATTTC-3′.
Plasmid constructs
Full-length cDNA of TBLR1-RARα was cloned into both pcDNA3.1 expression vector (Invitrogen) with either Myc or Flag tag fused to carboxy termini of TBLR1-RARα and pCDH1-MCS1-EF1-copGFP lentiviral vector (System Biosciences). Wild-type RARα was cloned from pFLAG-CMV4-RARα plasmid,8 and PML-RARα from pcDNA3.1(+)-PML-RARα plasmid.22 Both fragments were subcloned into pcDNA3.1 expression vector with Flag tag fused to their carboxy termini. RXRα cloned from pSV Sport RXRα23 (Addgene plasmid 8882) was subcloned into pcDNA3.1 expression vector. The empty vector pcDNA3.1 and pCDH1-MCS1-EF1-copGFP were used as control vectors.
Coimmunoprecipitation (Co-IP) and immunoblotting analysis
Total cellular lysates were obtained with radio immunoprecipitation assay lysis buffer; cytoplasmic and nuclear proteins were extracted separately with a Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime Biotechnology) according to the manufacturer’s instructions. Myc-tag monoclonal antibody (MoAb; Cell Signaling Technology) was used for Co-IP assays. After being separated and transferred to a polyvinylidene difluoride membrane, the protein was analyzed by immunoblotting using the following primary antibodies: anti-Flag MoAb from Sigma-Aldrich; anti-NCoR polyclonal antibody (PoAb), anti-SMRT PoAb, anti-mammalian SIN3 transcription regulator homolog A (mSIN3A) PoAb, anti-HDAC3 PoAb, and anti-TBL1 PoAb from Abcam; anti-H2B PoAb from Millipore; and anti-RXRα PoAb and anti-RARα PoAb from Santa Cruz Biotechnology. Anti-β-actin MoAb (Sigma-Aldrich) and anti-H3 PoAb (Abcam) were used as internal controls. The immunoreactive proteins were visualized using SuperSignal chemiluminescent detection system (Pierce).
Immunofluorescence analysis
The 293T cells or HEK293 cells treated with or without 1 μM ATRA were transiently transfected with Myc- or Flag-tagged expression plasmids. Then the cells were permeabilized and sequentially incubated with primary antibodies; secondary antibodies, including DyLight 488 donkey anti-rabbit IgG antibody and DyLight 649 goat anti-mouse IgG antibody (Biolegend); and 4-6-diamidino-2-phenylindole (DAPI; Invitrogen). Images were obtained with a laser scanning confocal microscope (Leica SP2).
Luciferase assay
The transcriptional activity of TBLR1-RARα was assessed by luciferase assay. A combination of RARE Cignal reporter (Cignal Pathway Reporter Kits; Qiagen), including inducible transcription factor responsive construct and constitutively expressing Renilla luciferase construct, and respective vectors (PML-RARα, RARα, and TBLR1-RARα expression vectors) were transiently cotransfected into CV-1 cells or HEK293 cells. The cells were then incubated with different concentrations of ATRA for 48 hours. The luciferase assay was performed using Dual Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions and detected by Lumat LB 9507 (Berthold Technologies). The ratio between firefly and Renilla luciferase was used to normalize the transfection efficiency.
Lentivirus production and transduction
The 293T cells were transfected with lentiviral vector constructs and pPACK packaging plasmids mix (System Biosciences) by calcium phosphate precipitation method. Cells were then infected with prepared lentivirus and sorted by BD FACS Aria II System for green fluorescent protein (GFP)-positive cells.
Cell differentiation analysis
ATRA-treated NB4 cells and GFP-positive U937 cells were incubated with allophycocyanin-conjugated anti-human antibody CD11b (BD Pharmingen) and examined by BD FACS LSR II flow cytometer. An allophycocyanin-conjugated isotype-matched IgG antibody (BD Pharmingen) was used as negative control. The percentage of CD11b-positive cells was analyzed using FlowJo software. For morphologic analysis, cytospin slides of each sample were stained with Wright-Giemsa staining solution.
Statistical analysis
Data were shown as mean ± standard deviation (SD). The significance of differences between 2 groups was determined by Student t test. Data analyses were performed using SPSS 19.0 software package. Statistical significance threshold was set at 0.05; asterisks indicate significant differences (*P < .05; **P < .01; and ***P < .001).
Results
Molecular cloning and identification of TBLR1-RARα fusion transcript in the novel variant APL with t(3;17)(q26;q21) translocation
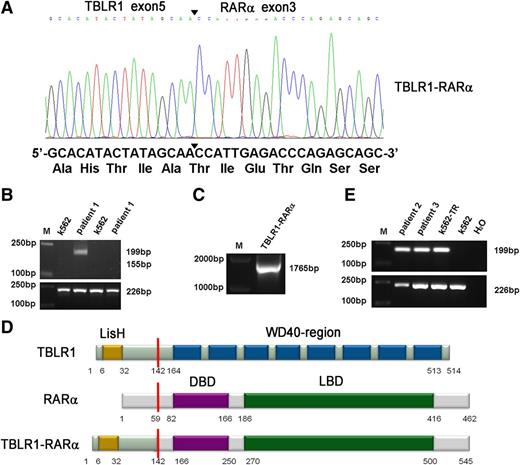
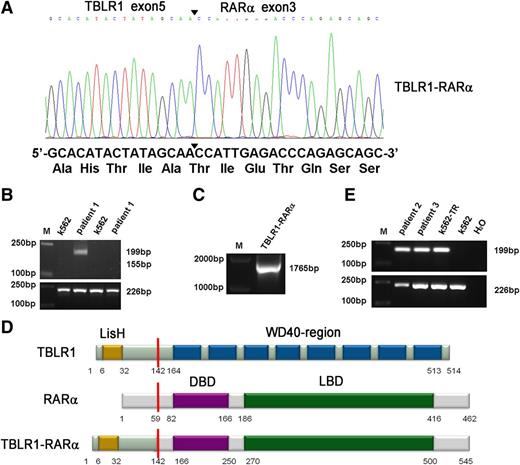
A new chromosomal rearrangement was observed by karyotype analysis without PML-RARα fusion signal detected by RT-PCR or FISH analysis. To identify the unknown chimeric fusion transcript, we performed 5′-RACE according to the manufacturer’s instructions, and the PCR products amplified from patient 1’s BMMNCs were cloned into pMD18-T simple vector and sequenced directly. As a result, TBLR1 was identified as a novel fusion partner of RARα gene. In TBLR1-RARα fusion transcript, the RARα portion, like in other forms of RARα fusion genes,1,7-11 started from its exon 3 and fused to exon 5 of TBLR1 (Figure 2A). To further confirm the presence of TBLR1-RARα fusion transcript, RT-PCR was performed using TBLR1- and RARα-specific primers. The predicted 199-bp PCR product was obtained from patient 1’s BMMNC cDNA (Figure 2B, lane 3), and the full-length PCR product of TBLR1-RARα was also amplified and sequenced from patient 1 (Figure 2C). The sequence analysis revealed that the full-length fusion transcript of TBLR1-RARα spanning from start codon to exon 5 (688 nt) of TBLR1 cDNA (reference NM_024665.4) was fused to RARα cDNA from exon 3 to stop codon forming a 1638-bp in-frame fusion transcript (GenBank KF589333), which is predicted to encode a 545-aa fusion protein, containing a LisH domain in the TBLR1 portion (6-32 aa) and a DNA-binding domain (166-250 aa) and a ligand-binding domain (270-500 aa) in the RARα portion (Figure 2D).1,11,15 The reciprocal RARα-TBLR1 mRNA was not detected by RT-PCR in our study (Figure 2B, lane 5). TBLR1-RARα fusion transcript was also detected in the other 2 APL patients with t(3;17) chromosomal translocation (Figure 2E, lanes 2 and 3) and further confirmed by sequencing.
Molecular analysis and identification of the TBLR1-RARα fusion transcript. (A) Partial sequence analysis of the TBLR1-RARα transcript. TBLR1 exon 5 was fused to exon 3 of RARα. The junction site of TBLR1 and RARα is highlighted by a bold arrowhead. The TBLR1-RARα encodes an in-frame fusion transcript; its partial DNA sequence and the corresponding translated amino acid sequence are shown. (B) RT-PCR analysis of TBLR1-RARα fusion transcript and reciprocal RARα-TBLR1 fusion transcript. TBLR1-RARα fusion transcript (lane 3) was amplified from cDNA derived from the patient 1’s BMMNCs at diagnosis with an internal control (GAPDH) by RT-PCR, whereas the reciprocal RARα-TBLR1 fusion transcript (lane 5) was not detected. cDNA derived from K562 was used as a negative control. (C) Amplification of full-length TBLR1-RARα by RT-PCR. Full-length TBLR1-RARα was also detected from patient 1’s BM samples. (D) Schematic representation of TBLR1, RARα, and TBLR1-RARα proteins. TBLR1-RARα fusion protein contains 545 amino acids (aa), including a LisH domain (6-32 aa) in the TBLR1 portion and a DNA-binding domain (DBD; 166-250 aa) and a ligand-binding domain (LBD; 270-500 aa) in the RARα portion. The break point is indicated by the red line. (E) RT-PCR analysis of TBLR1-RARα fusion transcripts in the other 2 APL patients harboring t(3;17) chromosomal translocation. TBLR1-RARα fusion transcripts (lanes 2 and 3) were amplified from cDNAs derived from the APL patients’ BMMNCs at diagnosis with an internal control (GAPDH) by RT-PCR. cDNA derived from K562 was used as a negative control, and cDNA derived from K562 transfected with TBLR1-RARα (K562-TR) was used as a positive control.
Molecular analysis and identification of the TBLR1-RARα fusion transcript. (A) Partial sequence analysis of the TBLR1-RARα transcript. TBLR1 exon 5 was fused to exon 3 of RARα. The junction site of TBLR1 and RARα is highlighted by a bold arrowhead. The TBLR1-RARα encodes an in-frame fusion transcript; its partial DNA sequence and the corresponding translated amino acid sequence are shown. (B) RT-PCR analysis of TBLR1-RARα fusion transcript and reciprocal RARα-TBLR1 fusion transcript. TBLR1-RARα fusion transcript (lane 3) was amplified from cDNA derived from the patient 1’s BMMNCs at diagnosis with an internal control (GAPDH) by RT-PCR, whereas the reciprocal RARα-TBLR1 fusion transcript (lane 5) was not detected. cDNA derived from K562 was used as a negative control. (C) Amplification of full-length TBLR1-RARα by RT-PCR. Full-length TBLR1-RARα was also detected from patient 1’s BM samples. (D) Schematic representation of TBLR1, RARα, and TBLR1-RARα proteins. TBLR1-RARα fusion protein contains 545 amino acids (aa), including a LisH domain (6-32 aa) in the TBLR1 portion and a DNA-binding domain (DBD; 166-250 aa) and a ligand-binding domain (LBD; 270-500 aa) in the RARα portion. The break point is indicated by the red line. (E) RT-PCR analysis of TBLR1-RARα fusion transcripts in the other 2 APL patients harboring t(3;17) chromosomal translocation. TBLR1-RARα fusion transcripts (lanes 2 and 3) were amplified from cDNAs derived from the APL patients’ BMMNCs at diagnosis with an internal control (GAPDH) by RT-PCR. cDNA derived from K562 was used as a negative control, and cDNA derived from K562 transfected with TBLR1-RARα (K562-TR) was used as a positive control.
TBLR1-RARα fusion proteins self-associate into homodimers and associate with RXRα into heterodimers
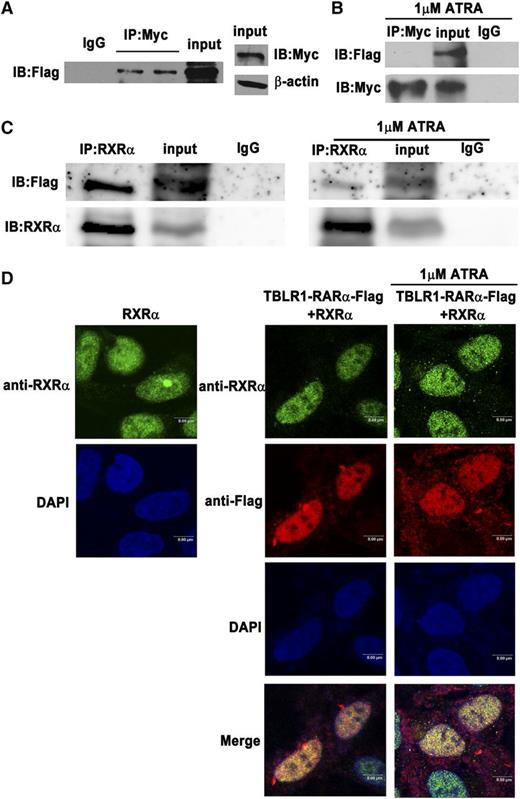
Until now, all RARα fusion proteins reported have shared at least one feature in common, that is, the ability to self-associate and to form homodimers.1,13 TBLR1 was reported to form homodimers through its LisH domain.24 In order to examine whether TBLR1-RARα can form homodimers, Co-IP assays were performed. Like other RARα fusion proteins, Flag-tagged TBLR1-RARα could coimmunoprecipitate with Myc-tagged TBLR1-RARα (Figure 3A), which indicated that TBLR1-RARα could self-associate and form homodimers. Whether ATRA has an effect on homodimer formation was further examined by Co-IP assays. As shown in Figure 3B, TBLR1-RARα was unable to form homodimers after 1 μM ATRA treatment of 48 hours, which revealed that ATRA interfered with homodimerization of TBLR1-RARα. Heterodimerization with RXRα is important for RARα fusion proteins to bind RARE.6,9,12 To investigate whether TBLR1-RARα could associate with RXRα, Co-IP assay was performed, and the result showed that Flag-tagged TBLR1-RARα could coimmunoprecipitate with and colocalize with RXRα (Figure 3C, left; Figure 3D). In addition, treatment of 1 μM ATRA for 48 hours could not completely disassociate the TBLR1-RARα/RXRα complexes (Figure 3C, right; Figure 3D).
Identification of homodimerization and heterodimerization of TBLR1-RARα fusion proteins by Co-IP. The 293T cells were transfected with both Myc- and Flag-tagged TBLR1-RARα expression plasmids. (A) Homodimerization of TBLR1-RARα was detected in 293T cells by Co-IP. β-actin was used as an internal control. (B) Homodimerization could not be detected in 293T cells after treatment with 1 μM ATRA for 48 hours by Co-IP. (C) RXRα and Flag-tagged TBLR1-RARα expression plasmids were transfected into 293T cells with or without 1 μM ATRA treatment of 48 hours. Flag-tagged TBLR1-RARα could coimmunoprecipitate with RXRα (left). Heterodimers could still be detected in the presence of ATRA treatment (right). (D) Immunofluorescence analysis of 293T cells transfected with RXRα expression plasmid alone (left) or together with Flag-tagged TBLR1-RARα plasmid (right). The latter was incubated with or without 1 μM ATRA for 48 hours. Flag and RXRα antibodies were used as primary antibodies, and DAPI was used for nuclear staining. At least 100 cells were counted for colocalization analysis. Bars represent 8 μm. IB, immunoblotting; IgG, control IP with isotype antibody; Input, nonimmunoprecipitated cell lysates; IP, immunoprecipitation.
Identification of homodimerization and heterodimerization of TBLR1-RARα fusion proteins by Co-IP. The 293T cells were transfected with both Myc- and Flag-tagged TBLR1-RARα expression plasmids. (A) Homodimerization of TBLR1-RARα was detected in 293T cells by Co-IP. β-actin was used as an internal control. (B) Homodimerization could not be detected in 293T cells after treatment with 1 μM ATRA for 48 hours by Co-IP. (C) RXRα and Flag-tagged TBLR1-RARα expression plasmids were transfected into 293T cells with or without 1 μM ATRA treatment of 48 hours. Flag-tagged TBLR1-RARα could coimmunoprecipitate with RXRα (left). Heterodimers could still be detected in the presence of ATRA treatment (right). (D) Immunofluorescence analysis of 293T cells transfected with RXRα expression plasmid alone (left) or together with Flag-tagged TBLR1-RARα plasmid (right). The latter was incubated with or without 1 μM ATRA for 48 hours. Flag and RXRα antibodies were used as primary antibodies, and DAPI was used for nuclear staining. At least 100 cells were counted for colocalization analysis. Bars represent 8 μm. IB, immunoblotting; IgG, control IP with isotype antibody; Input, nonimmunoprecipitated cell lysates; IP, immunoprecipitation.
TBLR1-RARα exhibits a distinct subcellular localization
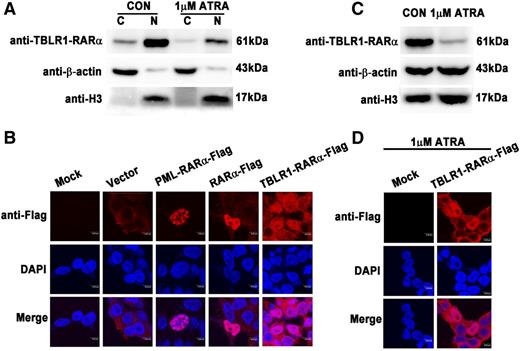
The characteristics of subcellular distribution of RARα fusion proteins are different from each other and may contribute to the pathogenesis of APLs.1,14,25 In our study, TBLR1-RARα was predominantly found in nucleus and a small amount of it in cytoplasm by immunoblotting (Figure 4A). Immunofluorescence analysis showed PML-RARα primarily localized in a microspeckled nuclear pattern, whereas RARα exhibited a diffuse nuclear pattern (Figure 4B), which was consistent with previous reports.1,14,25 However, TBLR1-RARα demonstrated a distinct subcellular localization pattern and was diffusely distributed both in nucleus and cytoplasm.
Distinct subcellular localization of TBLR1-RARα fusion protein. (A) Immunoblotting analysis of cytoplasmic and nuclear components of 293T cells transfected with Flag-tagged TBLR1-RARα. TBLR1-RARα was predominantly expressed in nucleus and a small amount of it in cytoplasm and was downregulated both in nucleus and cytoplasm after treatment with 1 μM ATRA. (B) Immunofluorescence analysis of 293T cells transfected with expression plasmids of vehicle (vector), PML-RARα-Flag, RARα-Flag, and TBLR1-RARα-Flag, respectively. Flag antibody was used as primary antibody, and DAPI for nuclear staining. At least 100 cells were counted for localization analysis. (C) The expression of TBLR1-RARα was decreased in 1 μM ATRA–treated 293T cells by immunoblotting. β-actin and H3 were used as internal controls for cytoplasmic and nuclear proteins. (D) Immunofluorescence analysis of 293T cells incubated with 1 μM ATRA for 48 hours. Bars (B,D) represent 8 μm. C, cytoplasm; N, nucleus.
Distinct subcellular localization of TBLR1-RARα fusion protein. (A) Immunoblotting analysis of cytoplasmic and nuclear components of 293T cells transfected with Flag-tagged TBLR1-RARα. TBLR1-RARα was predominantly expressed in nucleus and a small amount of it in cytoplasm and was downregulated both in nucleus and cytoplasm after treatment with 1 μM ATRA. (B) Immunofluorescence analysis of 293T cells transfected with expression plasmids of vehicle (vector), PML-RARα-Flag, RARα-Flag, and TBLR1-RARα-Flag, respectively. Flag antibody was used as primary antibody, and DAPI for nuclear staining. At least 100 cells were counted for localization analysis. (C) The expression of TBLR1-RARα was decreased in 1 μM ATRA–treated 293T cells by immunoblotting. β-actin and H3 were used as internal controls for cytoplasmic and nuclear proteins. (D) Immunofluorescence analysis of 293T cells incubated with 1 μM ATRA for 48 hours. Bars (B,D) represent 8 μm. C, cytoplasm; N, nucleus.
Previous studies have indicated that ATRA treatment is associated with degradation and localization of RARα fusion proteins.1,14,25 To investigate the effect of ATRA on expression and distribution of TBLR1-RARα, immunoblotting and immunofluorescence analyses were performed. The total amount of TBLR1-RARα was obviously decreased in cells after 1 μM ATRA treatment of 48 hours (Figure 4C), in keeping with the downregulation of TBLR1-RARα in nucleus and cytoplasm (Figure 4A). But the subcellular distribution pattern of TBLR1-RARα was unchanged; it was still diffusely distributed both in nucleus and cytoplasm (Figure 4D).
TBLR1-RARα functions as a transcriptional activator
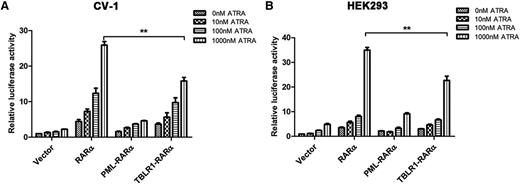
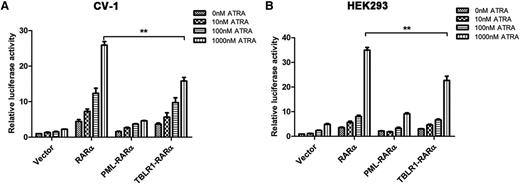
Different RARα fusion proteins have diverse transcriptional properties.1,14 For instance, PML-RARα, PLZF-RARα, and signal transducer and activator of transcription 5b (STAT5B)-RARα are dominant negative for retinoids, whereas NPM-RARα exhibits an ATRA-dependent transcriptional activation and has a dominant negative effect on wild-type RARα.26,27 To assess the ATRA-induced transcriptional activity of TBLR1-RARα, luciferase assays were conducted. The results indicated that TBLR1-RARα could increase the transcriptional activity of the reporter gene. In response to ATRA treatment, TBLR1-RARα exhibited an ATRA-induced transcriptional activation in a dose-dependent manner. Compared with wild-type RARα, TBLR1-RARα was a less efficient transcriptional activator, but its transcriptional capability was slightly higher than that of PML-RARα (Figure 5A-B).
TBLR1-RARα functions as a transcriptional activator. CV-1 (A) or HEK293 (B) cells were transfected with RARE Cignal reporter and expression plasmids containing vehicle, PML-RARα, RARα, and TBLR1-RARα, respectively. The transfected cells were then incubated with different dosages of ATRA or ethanol solvent for 48 hours and harvested for luciferase assay. TBLR1-RARα could increase the transcriptional activity of reporter gene. When treated with ATRA, TBLR1-RARα exhibited an ATRA-induced transcriptional activation in a concentration-dependent manner. The transcriptional capability of TBLR1-RARα was lower than that of wild-type RARα. Renilla luciferase activity was normalized to firefly luciferase activity. Ratios were normalized against the cells transfected with control plasmids. Bars represent mean values ± SD corresponding to at least 3 independent experiments executed in triplicate. Statistical significance was determined with the Student t test. **P < .01.
TBLR1-RARα functions as a transcriptional activator. CV-1 (A) or HEK293 (B) cells were transfected with RARE Cignal reporter and expression plasmids containing vehicle, PML-RARα, RARα, and TBLR1-RARα, respectively. The transfected cells were then incubated with different dosages of ATRA or ethanol solvent for 48 hours and harvested for luciferase assay. TBLR1-RARα could increase the transcriptional activity of reporter gene. When treated with ATRA, TBLR1-RARα exhibited an ATRA-induced transcriptional activation in a concentration-dependent manner. The transcriptional capability of TBLR1-RARα was lower than that of wild-type RARα. Renilla luciferase activity was normalized to firefly luciferase activity. Ratios were normalized against the cells transfected with control plasmids. Bars represent mean values ± SD corresponding to at least 3 independent experiments executed in triplicate. Statistical significance was determined with the Student t test. **P < .01.
TBLR1-RARα recruits more transcriptional corepressors resulting in diminished transcriptional activity compared with RARα
The RARα chimeric proteins exhibit an increased affinity for transcriptional corepressors, N-CoR/SMRT/SIN3A/HDAC, which are recruited to RARE resulting in repression of RARα transcriptional activity.1,13,14,25 TBLR1 was also reported to interact with N-CoR, stabilize the N-CoR/SMRT complex, bind to histones H2B and H4 in vitro, and finally mediate transcriptional repression.24,28-31 To assess the interactions of TBLR1-RARα with endogenous transcriptional corepressors, Co-IP assays were performed, and the interactions of RARα with endogenous transcriptional corepressors were examined as comparator Co-IPs. Consistent with other RARα fusion proteins, TBLR1-RARα coimmunoprecipitated with N-CoR, mSIN3A, HDAC3, H2B, and TBL1 but could not interact with RARα (Figure 6A, left). Further Co-IP assays showed that N-CoR also associated with TBLR1-RARα and HDAC3 (Figure 6B). The comparator Co-IPs showed the interaction of RARα with N-CoR, mSIN3A, HDAC3, and SMRT, but not with H2B (Figure 6A, right). In accordance with Co-IP results, immunofluorescence analysis revealed that Flag-tagged TBLR1-RARα colocalized with N-CoR, mSIN3A, and H2B in nucleus and overlapped with SMRT, HDAC3, and TBL1 both in nucleus and cytoplasm in 293T cells, but not with RARα (Figure 6C).
Recruitment of endogenous transcriptional corepressors by TBLR1-RARα in 293T cells. In Co-IP assays (A-B,D), 293T cells were transfected with Myc-tagged TBLR1-RARα (A[left],B,D[left]) or RARα (A[right],D[right]) expression plasmid, and Flag-tagged TBLR1-RARα expression plasmid in coimmunofluorescence analysis (C,E). (A) Co-IP between endogenous corepressors and TBLR1-RARα. TBLR1-RARα interacted with N-CoR, mSIN3A, HDAC3, H2B, and TBL1 but could not interact with RARα (left). Co-IP between endogenous corepressors and RARα. RARα could associate with N-CoR, SMRT, mSIN3A, and HDAC3, but not with H2B (right). (B) N-CoR was detected to associate with TBLR1-RARα and HDAC3 by Co-IP assays. (C) Coimmunofluorescence analysis of colocalization of Flag-tagged TBLR1-RARα and endogenous corepressors. Flag antibody and antibodies for corepressors were used as primary antibodies, and DAPI was used for nuclear staining. At least 100 cells were counted for colocalization analysis. (D) Co-IP assays between endogenous corepressors and TBLR1-RARα were conducted in the presence of 1 uM ATRA for 48 hours. TBLR1-RARα interacted with N-CoR, SMRT, and HDAC3, but not mSIN3A (left). Co-IP assays between endogenous corepressors and RARα were performed after ATRA treatment. RARα could only coimmunoprecipitate with HDAC3, but not N-CoR, SMRT, and mSIN3A (right). (E) Coimmunofluorescence analysis of colocalization of Flag-tagged TBLR1-RARα and endogenous corepressors was performed in the presence of ATRA. Bars (C,E) represent 8 μm.
Recruitment of endogenous transcriptional corepressors by TBLR1-RARα in 293T cells. In Co-IP assays (A-B,D), 293T cells were transfected with Myc-tagged TBLR1-RARα (A[left],B,D[left]) or RARα (A[right],D[right]) expression plasmid, and Flag-tagged TBLR1-RARα expression plasmid in coimmunofluorescence analysis (C,E). (A) Co-IP between endogenous corepressors and TBLR1-RARα. TBLR1-RARα interacted with N-CoR, mSIN3A, HDAC3, H2B, and TBL1 but could not interact with RARα (left). Co-IP between endogenous corepressors and RARα. RARα could associate with N-CoR, SMRT, mSIN3A, and HDAC3, but not with H2B (right). (B) N-CoR was detected to associate with TBLR1-RARα and HDAC3 by Co-IP assays. (C) Coimmunofluorescence analysis of colocalization of Flag-tagged TBLR1-RARα and endogenous corepressors. Flag antibody and antibodies for corepressors were used as primary antibodies, and DAPI was used for nuclear staining. At least 100 cells were counted for colocalization analysis. (D) Co-IP assays between endogenous corepressors and TBLR1-RARα were conducted in the presence of 1 uM ATRA for 48 hours. TBLR1-RARα interacted with N-CoR, SMRT, and HDAC3, but not mSIN3A (left). Co-IP assays between endogenous corepressors and RARα were performed after ATRA treatment. RARα could only coimmunoprecipitate with HDAC3, but not N-CoR, SMRT, and mSIN3A (right). (E) Coimmunofluorescence analysis of colocalization of Flag-tagged TBLR1-RARα and endogenous corepressors was performed in the presence of ATRA. Bars (C,E) represent 8 μm.
In the presence of pharmacologic doses of ATRA, some RARα fusion proteins, such as PML-RARα and NPM-RARα, release the transcriptional corepressors and recruit transcriptional activators to activate transcription.1,14,26 Moreover, TBLR1 can mediate an exchange of N-CoR/SMRT corepressors for coactivator complexes upon ligand binding.28,32,33 To gain insight into these effects of ATRA on TBLR1-RARα fusion protein, Co-IP and immunofluorescence analysis were performed. As shown in Figure 6D (left) and Figure 6E, TBLR1-RARα still interacted with N-CoR, SMRT, and HDAC3 when exposed to 1 μM ATRA for 48 hours but no longer associated with mSIN3A. However, in comparator Co-IPs, RARα could only interact with HDAC3; its interaction with N-CoR, SMRT, and mSIN3A could not be detected any more when exposed to 1 μM ATRA for 48 hours (Figure 6D, right). In addition, the distribution of colocalization between TBLR1-RARα and corepressors was also altered. TBLR1-RARα and corepressors, N-CoR, SMRT, and HDAC3, were colocalized mainly in cytoplasm, and only a small amount of them were found overlapping in nucleus, which was different from the dominant nuclear colocalization between TBLR1-RARα and N-CoR in the absence of ATRA (Figure 6C,E).
Overexpression of TBLR1-RARα induces ATRA-mediated cell differentiation
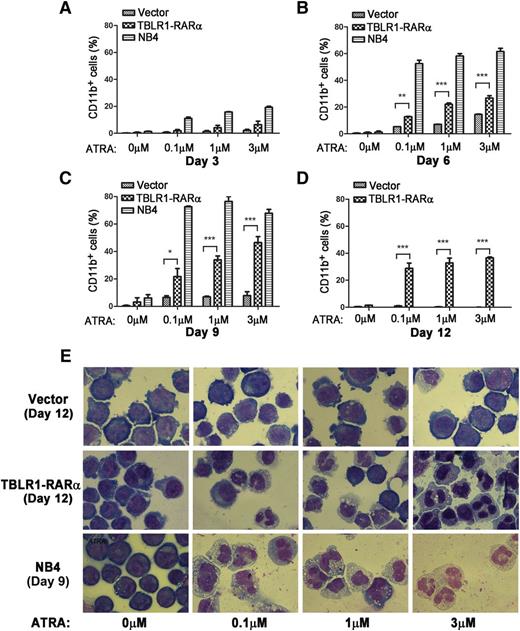
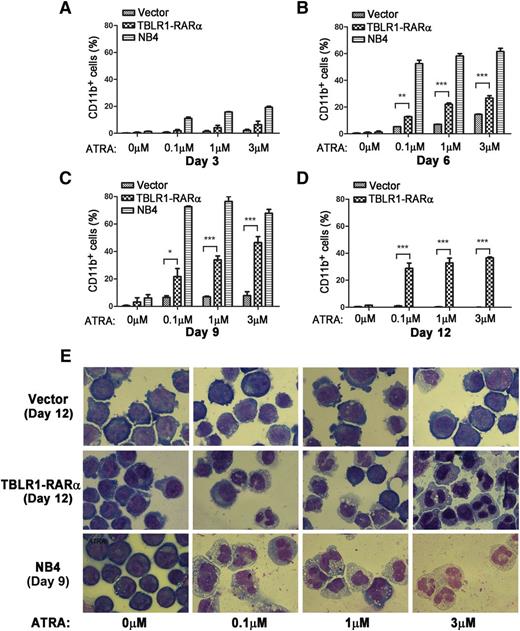
APL cells harboring different RARα fusion proteins exhibited diverse responses to ATRA-induced differentiation.1,14 Those harboring PLZF-RARα and STAT5B-RARα are ATRA resistant, whereas those with PML-RARα, NPM-RARα, nuclear mitotic apparatus–RARα, and so on are sensitive to ATRA.1,6,14,34 To examine whether expression of TBLR1-RARα could induce ATRA-mediated cell differentiation, TBLR1-RARα-transduced U937 and HL-60 cells were treated with ATRA, and the expression levels of cell surface marker CD11b were analyzed by flow cytometry. NB4 cells were used as a positive control. There is no statistical difference between the proportions of CD11b-positive cells in TBLR1-RARα- and vehicle-transduced cells before ATRA treatment. However, the proportion of CD11b-positive cells increased notably in TBLR1-RARα-transduced cells in a dose- and time-dependent manner of ATRA treatment (Figure 7A-D; supplemental Figure 1A-C, available on the Blood Web site). Nitroblue tetrazolium reduction assay in HL-60 cells also confirmed that TBLR1-RARα-expressed cells represented an ATRA-mediated cell differentiation in a dose- and time-dependent manner (supplemental Figure 1D-E). Cell morphology observation revealed that TBLR1-RARα-expressed cells presented certain differentiated characteristics in a dose-dependent manner of ATRA treatment compared with the control groups (Figure 7E and supplemental Figure 1G).
TBLR1-RARα induces ATRA-mediated cell differentiation of U937 cells. U937 cells were infected with lentivirus, which resulted in expression of TBLR1-RARα, or control lentivirus, respectively. The transduced cells were sorted for GFP-positive cell populations for further study. (A-D) Flow cytometry analysis of cell surface expression of CD11b for GFP-positive U937 cells and NB4 cells (positive control) incubated in different dosages of ATRA or ethanol solvent after 3, 6, 9, and 12 days, respectively. Because few live NB4 cells could be detected after 12 days, the data of NB4 cells in (D) were not shown. The expression levels of CD11b of the TBLR1-RARα-expressed group were increased significantly compared with the control group in a dose- and time-dependent manner in the presence of ATRA. In flow cytometry analysis, the results represented mean values ± SD corresponding to at least 3 independent experiments executed in triplicate. Statistical significance was determined with the Student t test. *P < .05;**P < .01; and ***P < .001 were accepted as statistically significant. (E) Morphologic changes of U937 and NB4 cells treated with different concentrations of ATRA or ethanol solvent for 12 and 9 days. The TBLR1-RARα-expressed group presented morphologic changes of differentiation in the presence of ATRA (Wright-Giemsa stain, original magnification ×1000).
TBLR1-RARα induces ATRA-mediated cell differentiation of U937 cells. U937 cells were infected with lentivirus, which resulted in expression of TBLR1-RARα, or control lentivirus, respectively. The transduced cells were sorted for GFP-positive cell populations for further study. (A-D) Flow cytometry analysis of cell surface expression of CD11b for GFP-positive U937 cells and NB4 cells (positive control) incubated in different dosages of ATRA or ethanol solvent after 3, 6, 9, and 12 days, respectively. Because few live NB4 cells could be detected after 12 days, the data of NB4 cells in (D) were not shown. The expression levels of CD11b of the TBLR1-RARα-expressed group were increased significantly compared with the control group in a dose- and time-dependent manner in the presence of ATRA. In flow cytometry analysis, the results represented mean values ± SD corresponding to at least 3 independent experiments executed in triplicate. Statistical significance was determined with the Student t test. *P < .05;**P < .01; and ***P < .001 were accepted as statistically significant. (E) Morphologic changes of U937 and NB4 cells treated with different concentrations of ATRA or ethanol solvent for 12 and 9 days. The TBLR1-RARα-expressed group presented morphologic changes of differentiation in the presence of ATRA (Wright-Giemsa stain, original magnification ×1000).
Discussion
In this study, we presented 3 case of APL with complex chromosomal rearrangement, t(3;17)(q26;q21),t(7;17)(q11.2;q21). Several groups have reported rare cases of APL involving t(3;17) chromosomal translocation,35,36 which is similar to what we found in our study. This suggests that t(3;17)(q26;q21) chromosomal translocation is recurrent. Case 1 failed to achieve CR with ATRA and MTZ. Subsequently treated with As2O3 and chemotherapeutic drugs, the patient achieved CR. Case 3 was initially treated with ATRA 25 mg/m2 from days 1 to 35 and daunorubicin 45 mg/m2 from days 4 to 6 and achieved CR on day 36. Combined with the outcome of case 1, it is currently difficult to evaluate the efficiency of ATRA on APL patients with TBLR1-RARα.
To clone the unknown RARα fusion transcript, 5′-RACE was conducted. We found that TBLR1 was a novel fusion partner of RARα chimeric gene by 5′-RACE and sequencing. TBLR1-RARα transcript was also identified in the other 2 APL patients harboring t(3;17) chromosomal translocation, which indicated that TBLR1-RARα was a bona fide RARα fusion transcript existing in the APL patient with t(3;17) chromosomal translocation. In case 1, t(7;17) (q11.2;q21) was detected by karyotype analysis; however, the partner gene in chromosome 7 fused to RARα was not identified by both 5′-RACE and 3′-RACE. Because the fusion gene derived from t(7;17) (q11.2;q21) was not cloned, the potential role of the fusion gene derived from t(7;17) cannot be ruled out. In case 3, karyotype analysis revealed t(15;17) (q22;q12) besides t(3;17) (q21;q25). PML-RARα transcript was also detected in this patient. These results suggest that t(3;17) may cooperate with other RARα rearrangements in pathogenesis.
TBLR1 encodes an F-box/WD-40 protein, which contains a LisH domain in the N-terminal region and 8 WD-40 repeats in the C-terminal region.15 LisH is a novel homodimerization domain and is required for homodimerization, heterodimerization, and oligomerization of TBLR1,24,37,38 which suggests that TBLR1-RARα may self-associate through its LisH domain. Co-IP assays in our study showed that TBLR1-RARα could self-associate and form homodimers, and the homodimerization of TBLR1-RARα homodimers could be interfered with by ATRA treatment. Similar to PML-RARα, TBLR1-RARα could also form transcriptional complexes with RXRα, which indicated that heterodimerization was still needed for TBLR1-RARα to bind RARE.
TBLR1 localized primarily in cytoplasm in 3T3 cells under normal culture conditions, and predominantly in nucleus in serum-starved 3T3 cells.15 Unlike TBLR1 and wild-type RARα, our findings demonstrated that TBLR1-RARα localized predominantly in nucleus and a small amount of it in cytoplasm, both in a diffused pattern, which is similar to that of B-cell CLL/lymphoma 6 corepressor–RARα.9 This disruption of TBLR1 and RARα distribution may account for transformation of APL cells. However, in the presence of ATRA, although TBLR1-RARα expression level was downregulated both in nucleus and cytoplasm, the subcellular distribution remained the same.
Wild-type RARα recruits transcriptional corepressors, including N-CoR, SMRT, SIN3A, and HDACs, to repress transcription.39,40 This is also verified in our work. Nevertheless, RARα associates with coactivators and triggers transcriptional activation at physiological doses of ATRA.41,42 Moreover, RARα fusion proteins can associate with other transcriptional corepressors, including polycomb-repressive complex, histone methyltransferases, and DNA methyltransferases.43-45 As a result, RARα fusion proteins repress transcription to a greater extent than RARα and block myeloid differentiation at the promyelocyte stage.46 Only pharmacologic doses of ATRA can release these transcriptional corepressors, recruit transcriptional coactivators to activate transcription, and finally induce differentiation of APL cells.1,14 But not all RARα fusion proteins can transactivate RARα target genes when treated with ATRA as a single agent. Our study showed that TBLR1-RARα functions as a transcriptional activator, although its transactivity was lower than that of wild-type RARα but was slightly higher than that of PML-RARα, which exhibits a similar transactivity to NPM-RARα.4
TBLR1 directly interacts with N-CoR and SMRT via its LisH domain in the N-terminal region and the first 3 WD-40 repeats in the C-terminal region and stabilizes the structure of the N-CoR/SMRT corepressor complex.29,30 Furthermore, TBLR1 directly binds to hypoacetylated histones H2B and H4 through its N-terminal domain. Interaction between TBLR1 and histones is necessary for transcriptional repression of the N-CoR/SMRT complex.30,31 In our study, we found that TBLR1-RARα associated with N-CoR, mSIN3A, HDAC3, H2B, and TBL1, but not RARα. The diminished transcriptional activation of TBLR1-RARα compared with RARα may also be attributable to recruitment of more transcriptional corepressors via both its N-terminal and C-terminal regions. We also found that TBLR1-RARα still interacted with N-CoR, SMRT, and HDAC3, but no longer with mSIN3A, when exposed to 1 μM of ATRA for 48 hours. This is reasonable, as previous studies and our results indicate that TBLR1-RARα binds to N-CoR, SMRT, and HDAC3 via its LisH domain and its RARα portion, but the association between TBLR1-RARα and mSIN3A is mainly through its RARα portion, which is ATRA responsive. When treated with ATRA, the transcriptional corepressors associated with RARα portion can be released, and the interaction between TBLR1-RARα remains. Although coimmunofluorescence results were consistent with Co-IP assays, the subcellular colocalization of TBLR1-RARα and corepressors was altered in the presence of ATRA. Recent studies demonstrated that TBLR1 could recruit ubiquitin conjugating/19S proteasome complex to nuclear receptor as an E3 ubiquitin ligase adaptor to degrade corepressors upon ligand binding.28,32,33 In the present study, the colocalization of corepressors and TBLR1-RARα primarily translocated into cytoplasm after ATRA treatment, which suggests that TBLR1-RARα may also function as a coactivator in the presence of ATRA, mediate dissociation and degradation of transcriptional corepressors via the ubiquitin/proteasome pathway, and finally induce transcriptional activation.
Different RARα fusion proteins exhibit diverse responses to ATRA. In our present study, TBLR1-RARα could increase the sensitivity of U937 and HL-60 cells to ATRA and induce a dose- and time-dependent cell differentiation, which was consistent with previous luciferase assays. Besides mediating the degradation of corepressors, TBLR1-RARα was found to be degraded as PML-RARα47 and homodimerization of TBLR1-RARα were interfered with in the presence of ATRA, which was also responsible for transactivation and cell differentiation.
In conclusion, we identified a novel RARα fusion partner, TBLR1, in a rare case of APL with complex chromosomal rearrangement. To our knowledge, TBLR1-RARα is the 10th RARα chimeric gene that has been reported up to now. TBLR1-RARα contained the B-F domains of RARα, exhibited a distinct subcellular localization, self-associated into homodimers, and associated with RXRα into heterodimers. In the presence of pharmacologic doses of ATRA, TBLR1-RARα could be degraded and the homodimerization was abrogated. Moreover, when treated with ATRA, TBLR1-RARα could mediate the dissociation and degradation of transcriptional corepressors and consequently transactivated transcription of RARα target genes and induced cell differentiation in a dose- and time- dependent manner.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Takeshi Kondo (Hokkaido University Graduate School of Medicine) for pFLAG-CMV4-RARα plasmid, Dr Timothy J. Ley (Washington University) for pcDNA3.1(+)-PML-RARα plasmid, and Dr Peter Tontonoz (Harvard Medical School) for pSV Sport RXRα plasmid.
This work was supported by grants from the National Natural Science Foundation of China (81270635 and 81370633), the National Basic Research Program of China (2011CB964801), and the Tianjin Applied Fundamental Research Planning Key Project (13JCZDJC29900).
Authorship
Contribution: Y.C. and S.L. performed all experimental validation and computational and statistical data analysis and wrote the manuscript; Q.R., H.X., Z.T., and K.T. helped with experimental validation and computational and statistical data analysis; C.L. and K.R. performed FISH, karyotype analysis, and morphologic analysis; Y.M., C.Z., and B.W. provided clinical samples and related data; and M.W. and J.W. conceived and designed this project, interpreted data, and revised and approved the manuscript.
Conflict-of-interest disclosure: Jianxiang Wang acts as consultant of Novartis and Bristol-Myers Squibb. The remaining authors declare no competing financial interests.
Correspondence: Jianxiang Wang, State Key Laboratory of Experimental Hematology, Institute of Hematology and Blood Diseases Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, 288 Nanjing Rd, Tianjin 300020, China; e-mail: wangjx@medmail.com.cn; and Min Wang, State Key Laboratory of Experimental Hematology, Institute of Hematology and Blood Diseases Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, 288 Nanjing Rd, Tianjin 300020, China; e-mail: wangjxm@ihcams.ac.cn.
References
Author notes
Y.C. and S.L. contributed equally to this study.

![Figure 1. Morphologic and cytogenetic analysis of BM sample of the APL case with t(3;17)(q26;q21),t(7;17)(q11.2;q21) translocation. (A) The May-Giemsa staining. BM cells were shown to be blocked at the hypergranular promyelocytic stage lacking Auer rods. (B) Myeloperoxidase staining. (C) Specific esterase staining. (D) Nonspecific esterase staining. (B-D) Cellular histochemical staining examination exhibited high positive rates of blast cells. (E) FISH analysis. PML probe (orange signal) and RARα probe (green signal) were used. No PML-RARα was detected. (F) Karyotype analysis. 46,XY,t(3;17)(q26;q21),t(7;17)(q11.2;q21) was found in 14 metaphases among 15 metaphases detected. (G) Karyotype analysis of relapsed BM cells. A more complex recurring chromosomal transcription, 46,XY,t(3;17)(q26;q21),add(5)(q32),del(6)(q22q24),t(7;17)(q11.2;q21),add(10)(q26),del(11)(p14)[13]/46,XY[1], was observed in 13 metaphases among 14 metaphases detected. The rearrangements and break points in panels F and G were indicated by arrows.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/6/10.1182_blood-2013-10-528596/4/m_936f1.jpeg?Expires=1765887528&Signature=xXSjjZ0eEGz6IjLuhC4iAS26H0yT6m-TCxeTl-Ukb9-AeMFIm6CjkeqGy5lzCp33y4NCPOPYRzBIfs~KkQu0rN7V017zhtRHph5vKvkYOQ01utq8DzkaW2SabsPhzdzJZps7xBMvSvtG9ia0x4XXmi9hF9YZMcPrfFex4jIU1yWPYXKoGHmkLBEWcciDkIH2FkQjEFrRYTXnkHYMwbLlJW~bRLyGAuPtvBmLYf9vjryKsNxRw6noljg2Ur4W0MxLIAe4FlM3QQU8ovR9-6sfv8-9iqCRsS0fq~lbYKZkC3bpIFn-beFh2pgsczNOLOGaogQyrHYeRjmP4zSOlQSQ7A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 6. Recruitment of endogenous transcriptional corepressors by TBLR1-RARα in 293T cells. In Co-IP assays (A-B,D), 293T cells were transfected with Myc-tagged TBLR1-RARα (A[left],B,D[left]) or RARα (A[right],D[right]) expression plasmid, and Flag-tagged TBLR1-RARα expression plasmid in coimmunofluorescence analysis (C,E). (A) Co-IP between endogenous corepressors and TBLR1-RARα. TBLR1-RARα interacted with N-CoR, mSIN3A, HDAC3, H2B, and TBL1 but could not interact with RARα (left). Co-IP between endogenous corepressors and RARα. RARα could associate with N-CoR, SMRT, mSIN3A, and HDAC3, but not with H2B (right). (B) N-CoR was detected to associate with TBLR1-RARα and HDAC3 by Co-IP assays. (C) Coimmunofluorescence analysis of colocalization of Flag-tagged TBLR1-RARα and endogenous corepressors. Flag antibody and antibodies for corepressors were used as primary antibodies, and DAPI was used for nuclear staining. At least 100 cells were counted for colocalization analysis. (D) Co-IP assays between endogenous corepressors and TBLR1-RARα were conducted in the presence of 1 uM ATRA for 48 hours. TBLR1-RARα interacted with N-CoR, SMRT, and HDAC3, but not mSIN3A (left). Co-IP assays between endogenous corepressors and RARα were performed after ATRA treatment. RARα could only coimmunoprecipitate with HDAC3, but not N-CoR, SMRT, and mSIN3A (right). (E) Coimmunofluorescence analysis of colocalization of Flag-tagged TBLR1-RARα and endogenous corepressors was performed in the presence of ATRA. Bars (C,E) represent 8 μm.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/6/10.1182_blood-2013-10-528596/4/m_936f6.jpeg?Expires=1765887528&Signature=csOX9SRW3WfprZ9vUA9HLJpkDh0KnvFY91PZFlpW-7jG7yxA1w4YSNHtMLrrJj93Zw6EZ-1B136lWNVBxr13Z166g3Wskf7GhNlDSXsU5SRq0bk1x25uTVtEPw4DL3U1BUEyEFd6d4DGWwyZDL8haDc~VdJXmsdq7w-EajSaffpni31FfdsbeaNvXcIkJhyo9wOhDa2UmHBzw83oLQI8yVGW-GXOaR4O4cMbBzEf~xPQshFHTVmmbsKZp~INOvEQlg4ED512byafOj1YXjcz4IyOckUK7OWTGwchCINWw72iJWO7g-7VZ7-SA3SWnkMGvoJcMttt7iPaY0r4W2wQ~w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 1. Morphologic and cytogenetic analysis of BM sample of the APL case with t(3;17)(q26;q21),t(7;17)(q11.2;q21) translocation. (A) The May-Giemsa staining. BM cells were shown to be blocked at the hypergranular promyelocytic stage lacking Auer rods. (B) Myeloperoxidase staining. (C) Specific esterase staining. (D) Nonspecific esterase staining. (B-D) Cellular histochemical staining examination exhibited high positive rates of blast cells. (E) FISH analysis. PML probe (orange signal) and RARα probe (green signal) were used. No PML-RARα was detected. (F) Karyotype analysis. 46,XY,t(3;17)(q26;q21),t(7;17)(q11.2;q21) was found in 14 metaphases among 15 metaphases detected. (G) Karyotype analysis of relapsed BM cells. A more complex recurring chromosomal transcription, 46,XY,t(3;17)(q26;q21),add(5)(q32),del(6)(q22q24),t(7;17)(q11.2;q21),add(10)(q26),del(11)(p14)[13]/46,XY[1], was observed in 13 metaphases among 14 metaphases detected. The rearrangements and break points in panels F and G were indicated by arrows.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/6/10.1182_blood-2013-10-528596/4/m_936f1.jpeg?Expires=1765906205&Signature=bUqOx8~AgJaU09EdKqOKrHZfkplLD9-I6uL1F7zpcHlqVkIbd9f28xNPNUfJl9AjzY7PavCHcH~jBw5Twt3VhKs1bp43sjMKPGWLJUcNmdl2j23p4603eAVBTfSmv2sElQi9AhqmRIhuPDFcJU0CmtmoUBYGA5nCjm-zr0YWHdLNn0dCatWMFWpz5bAX0zuDGOBe2nIAr1BiM3cV5VbhdFo1zurkd8yNoc4ljp7MkFCJwjNBpzGlqXLnIECK85bkbdhDt4zCWRO47~irfZN6dKiFdbDmfXJQ5jE2AjNTnhgZVXk98Xbei2RpzR5DMxjKlLKJ9FyVftxg0D5vkAEUzA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. Recruitment of endogenous transcriptional corepressors by TBLR1-RARα in 293T cells. In Co-IP assays (A-B,D), 293T cells were transfected with Myc-tagged TBLR1-RARα (A[left],B,D[left]) or RARα (A[right],D[right]) expression plasmid, and Flag-tagged TBLR1-RARα expression plasmid in coimmunofluorescence analysis (C,E). (A) Co-IP between endogenous corepressors and TBLR1-RARα. TBLR1-RARα interacted with N-CoR, mSIN3A, HDAC3, H2B, and TBL1 but could not interact with RARα (left). Co-IP between endogenous corepressors and RARα. RARα could associate with N-CoR, SMRT, mSIN3A, and HDAC3, but not with H2B (right). (B) N-CoR was detected to associate with TBLR1-RARα and HDAC3 by Co-IP assays. (C) Coimmunofluorescence analysis of colocalization of Flag-tagged TBLR1-RARα and endogenous corepressors. Flag antibody and antibodies for corepressors were used as primary antibodies, and DAPI was used for nuclear staining. At least 100 cells were counted for colocalization analysis. (D) Co-IP assays between endogenous corepressors and TBLR1-RARα were conducted in the presence of 1 uM ATRA for 48 hours. TBLR1-RARα interacted with N-CoR, SMRT, and HDAC3, but not mSIN3A (left). Co-IP assays between endogenous corepressors and RARα were performed after ATRA treatment. RARα could only coimmunoprecipitate with HDAC3, but not N-CoR, SMRT, and mSIN3A (right). (E) Coimmunofluorescence analysis of colocalization of Flag-tagged TBLR1-RARα and endogenous corepressors was performed in the presence of ATRA. Bars (C,E) represent 8 μm.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/6/10.1182_blood-2013-10-528596/4/m_936f6.jpeg?Expires=1765906205&Signature=07JrwY46WZXO2LhR943h-mFkkVAR35OaEeDb5Wc83PR91hO8CKZC3AZ~burO3wQEmbrGa2rFAloQs3MSu8vnvEdw4UJUt7u~jWKnaD-Q1HEe88LAaZTb-uHC88iaTPAjbnXvEcnZTLCqFKeXjY2R2bejBF0d1ynj-j3ATpEnzzEGHJGAZpicOBW7kEN4ZJUtlwVDYV97lo~-u7chzqL8TTwnxgAD2MZ-2ShS3fGygkq0nsIid7kXRIS4HJ0beP1zY6uvES1SpmyfP33L-mLVlJ01RBVGyOsnQUyWIV97MGcfXP00KXrTHq2E1UiIPQpopB8GUNOQsDsufGP1891JcA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)