Key Points
INPP4B promotes chemoresistance in AML independent of phosphoinositide phosphatase function.
Abstract
Phosphoinositide signaling regulates diverse cellular functions. Phosphoinositide-3 kinase (PI3K) generates PtdIns(3,4,5)P3 and PtdIns(3,4)P2, leading to the activation of proliferative and anti-apoptotic signaling pathways. Termination of phosphoinositide signaling requires hydrolysis of inositol ring phosphate groups through the actions of PtdIns(3,4,5)P3 3-phosphatase (PTEN), PtdIns(3,4,5)P3 5-phosphatases (eg, SHIP), and PtdIns(3,4)P2 4-phosphatases (eg, INPP4B). The biological relevance of most of these phosphoinositide phosphatases in acute myeloid leukemia (AML) remains poorly understood. Mass spectrometry–based gene expression profiling of 3-, 4- and 5-phosphatases in human AML revealed significant overexpression of INPP4B. Analysis of an expanded panel of 205 AML cases at diagnosis revealed INPP4B overexpression in association with reduced responses to chemotherapy, early relapse, and poor overall survival, independent of other risk factors. Ectopic overexpression of INPP4B conferred leukemic resistance to cytosine arabinoside (ara-C), daunorubicin, and etoposide. Expression of a phosphatase inert variant (INPP4B C842A) failed to abrogate resistance of AML cells to chemotherapy in vitro or in vivo. In contrast, targeted suppression of endogenously overexpressed INPP4B by RNA interference sensitized AML cell lines and primary AML to chemotherapy. These findings demonstrate a previously unsuspected and clinically relevant role for INPP4B gain of function as a mediator of chemoresistance and poor survival outcome in AML independent of its phosphoinositide phosphatase function.
Introduction
Acute myeloid leukemia (AML) is an aggressive bone marrow (BM) malignancy arising from somatic transformation of hematopoietic progenitor cells. Failure to achieve complete remission (CR) is associated with increased age, poor performance status, higher presenting white blood cell counts, secondary AML, and unfavorable risk karyotype.1-5 Molecular mechanisms of chemoresistance include constitutive activation of tyrosine kinase receptors and intracellular signal transducer molecules, such as phosphoinositide-3 kinase (PI3K) and protein kinase B (AKT).6-9 Phosphoinositide signaling second messengers such as PtdIns(3,4,5)P3 and PtdIns(3,4)P2 are phosphorylated by PI3K and negatively regulated by a tripartite family of 3-, 4-, and 5-phosphoinositide phosphatases.10 PtdIns(3,4,5)P3 and PtdIns(3,4)P2 function to recruit pleckstrin homology domain–containing proteins such as AKT to the plasma membrane.11-13 The 3-phosphatase PTEN (phosphatase and tensin homolog deleted on chromosome 10) removes the phosphate in the D-3 position from PtdIns(3,4,5)P3 to generate PtdIns(4,5)P2, whereas the 5-phosphatases hydrolyze the phosphate in the D-5 position from PtdIns(3,4,5)P3 to produce PtdIns(3,4)P2. The latter is further hydrolyzed by the 4-phosphatases at the phosphate in the D-4 position to produce PtdIns(3)P, which terminates PI3K/AKT-dependent signaling. Although the 3-phosphatase PTEN is uncommonly mutated in AML,14,15 loss of function has been ascribed to cytoplasmic mutated nucleophosmin 1 (NPM1MUT), which promotes PTEN ubiquitination and degradation.16 Constitutive phosphorylation and inactivation of PTEN also promotes AKT activation and poor survival outcome in AML.17 Expression of the inositol polyphosphate-5-phosphatase SH2 domain–containing inositol 5′-phosphatase (SHIP-1) is reduced in myelodysplastic syndrome progenitor cells, consistent with a potential tumor suppressor role.18 SHIP-1 coding mutations, however, are very rare, with isolated reports of potential dominant negative function linked to mutations in the phosphatase domain.19,20 The role of other phosphoinositides in AML has not been extensively studied.
We therefore undertook a quantitative screen of phosphoinositide phosphatase expression in human AML and we here report the aberrant overexpression of the phosphoinositide 4-phosphatase INPP4B in association with treatment failure, early relapse, and poor overall survival (OS) after intensive chemotherapy. Furthermore, we show that INPP4B confers resistance to common AML therapies by a mechanism that is independent of its phosphoinositide phosphatase activity, highlighting a novel gain-of-function role in AML.
Methods
Patients and characteristics
A retrospective analysis of 205 consecutive patients undergoing intensive induction chemotherapy for AML at the Alfred Hospital between November 2000 and July 2013 was conducted. Patients received induction with either 7 + 3 (ara-C 100 mg/m2 for 7 days plus idarubicin 12 mg/m2 for 3 days) or a similar regimen that included high-dose ara-C defined by the total use of 10 to 24 g/m2. Morphologic classification was by World Health Organization criteria,21 and cytogenetic risk was determined according to Medical Research Council criteria.22 The study was conducted with the approval of the Alfred Human Research Ethics Committee. Normal BM was obtained from patients in whom BM examinations were assessed as being morphologically uninvolved with pathology. Fms-like tyrosine kinase 3-internal tandem duplication (FLT3-ITD) detection was performed as previously published.23
Mononuclear cell isolation and Sequenom MassARRAY quantitative gene expression
For MassARRAY quantitative gene expression, BM samples with >70% blasts were chosen for expression analysis to reduce the confounding effects of contaminating non-leukemic cells. Mononuclear cells were isolated by using Ficoll-Hypaque (GE Healthcare, Amersham, UK) followed by NH4Cl lysis to remove red blood cells and resuspension in TRIzol reagent (Invitrogen, MA). Total RNA was purified by using a Qiagen miRNeasy kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Invitrogen Superscript RT III cDNA Synthesis kit (Invitrogen) was used to synthesize complementary DNA that was subjected to MassARRAY quantitative gene expression by Sequenom (San Diego, CA). Experiments were performed in quadruplicate at 4 dilutions for each transcript per the manufacturer’s protocols described on www.sequenom.com. The complementary DNA was mixed with a serially diluted synthetic competitor used as an internal standard on which quantification of the transcript was based. Normalized gene expression values (aM) were calculated by using GeNorm software.24 For each sample, the gene expression normalization factor was calculated on the basis of the geometric mean of housekeeping genes to which experimental raw data were normalized. Details of oligonucleotide primers are available on request.
Immunohistochemistry
Immunohistochemical analysis was performed on BM trephines fixed in 10% neutral buffered formalin followed by EDTA decalcification. Processing and wax infiltration was performed by using a Leica Peloris Automated Tissue Processor. Epitope retrieval was performed by using a Dako Pascal pressure cooker at 120°C for 1 minute with tris(hydroxymethyl)aminomethane-EDTA (pH 8.0) buffer. Peroxidase blockade was performed with 3% H2O2 for 10 minutes at room temperature. The primary antibody used for immunohistologic detection of INPP4B was clone 3D5 incubated at a dilution of 1:250 for 30 minutes at room temperature. This antibody was originally developed by Fedele et al,25 and it detects the N-terminus of the protein. Leica Max Polymer Detection kit was used with a Dako DAB+ chromogen for 10 minutes, and the cells were processed by using a LabVision 360 Autostainer. Each BM trephine was stained for INPP4B, and a representative field was photographed at low and high power. High-power images allowed the morphologic proportion of stained blasts to be assessed. The percentage of blasts expressing INPP4B was then scored by 3 qualified hematopathologists independently in a blinded manner, and the collective scores were averaged to obtain the final result.
Immunoblotting
AML mononuclear cells were derived as previously described. Normal CD34+ cells were isolated by using Dynabeads CD34 Positive Isolation Kit (Invitrogen) for granulocyte colony-stimulating factor–mobilized peripheral blood samples from healthy donors. Proteins were extracted by using ice-cold NP-40 detergent containing lysis buffer with protease inhibitors (Roche, IN) and subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis separation. Proteins were then transferred onto a nitrocellulose membrane (GE Biosciences, Bucks, UK) and the membrane was blocked by using Odyssey blocking buffer (LI-COR Biosciences, NE) prior to incubation with primary antibodies at 1 µg/mL dilutions. The following antibodies were used: pAkt (Ser473; D9E) XP rabbit monoclonal antibody (mAb), phospho-p44/42 MAPK (Erk1/2; Thr202/Tyr204) XP rabbit mAb, Akt rabbit mAb, Bcl-2 (50E3) rabbit mAb, BIMEL (C34C5) rabbit mAb, Bcl-xL (54H6) rabbit mAb (Cell Signaling, MA), Mcl-1 (S-19) rabbit polyclonal antibody (Santa Cruz, CA), β-tubulin mAb (Sigma-Aldrich, MO), and INPP4A (A-19) mAb (Santa Cruz). INPP4B (3D5 and 15E3) mouse mAb were obtained from Christina Mitchell and are available upon request.25 INPP4B 3D5 binds to the N-terminus of the protein, whereas 15E3 binds to the C-terminal end. The 15E3 antibody was used for immunoblotting a C2 domain–deleted version of INPP4B that lacks the 3D5 epitope. Membranes were incubated with a secondary antibody conjugated to IRDye 680 or 800 (LI-COR Biosciences) and scanned by using the Odyssey Infrared Imaging System (LI-COR Biosciences).
Malachite green phosphatase assay
Thirty micrograms of whole-cell lysates was mixed with 6 µL of INPP4A mAb at 200 µg/mL, and 50 µL of protein G sepharose beads (GE Healthcare) overnight to immunodeplete the supernatant of INPP4A protein for malachite green phosphatase assays. For the assay, the INPP4 substrate PtdIns(3,4)P2 (Echelon Research Laboratories, UT) was added to the cells for 60 minutes at 37°C in phosphatase assay buffer (50 mM 4-morpholinepropanesulfonic acid [pH 7.0]). Phosphatase activity was measured as the amount of free phosphate liberated by reacting with malachite green (Echelon Research Laboratories) and quantified by absorbance at 620 nm. Data were generated via comparison with a known standard curve for free phosphate, and any nonspecific background activity was accounted for. For more detail regarding methods, see supplemental Data (available at the Blood Web site).
Results
Overexpression of INPP4B in leukemic blasts is associated with poor survival outcome in AML
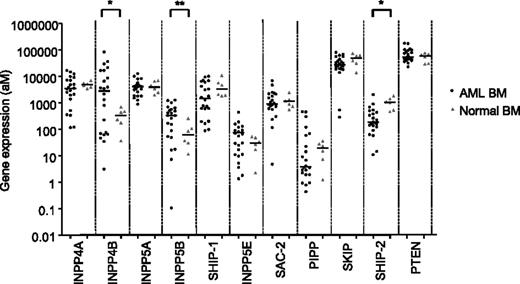
To quantitatively assess the expression of phosphoinositide phosphatases in human AML, a mass spectrometry detection system was used to detect absolute transcript number over a large dynamic range with attomolar sensitivity.26 Expression levels of the 3-phosphatase PTEN, the 4-phosphatases INPP4A and INPP4B, and the 5-phosphatases INPP5A, INPP5B, SHIP-1, INPP5E, PIPP, SKIP, SHIP-2, and SAC-2 were determined in a panel of AML BM samples taken at diagnosis and compared with normal control BM (Figure 1). The expression of the 5-phosphatase SHIP-2 was significantly reduced in AML (Figure 1). In contrast, two phosphoinositide phosphatases, INPP4B and INPP5B, were significantly overexpressed in AML compared with normal BM (P < .05) (Figure 1). By stratifying the AML patient cohort according to median phosphatase expression levels, high INPP4B expression emerged as a potential indicator of poor AML survival (supplemental Figure 1), although statistical significance was not attained in this pilot cohort of AML patient samples.
Gene expression profiling of phosphoinositide phosphatases in AML. Sequenom MassARRAY quantitative gene expression analysis of phosphoinositide phosphatases in human messenger RNA from 22 AML BM (black dots) samples at diagnosis with >70% blasts present were compared with messenger RNA from 6 normal BM samples (gray triangles). The mean expression level for both AML and normal patient cohorts for each gene is shown. *P < .05; **P < .01.
Gene expression profiling of phosphoinositide phosphatases in AML. Sequenom MassARRAY quantitative gene expression analysis of phosphoinositide phosphatases in human messenger RNA from 22 AML BM (black dots) samples at diagnosis with >70% blasts present were compared with messenger RNA from 6 normal BM samples (gray triangles). The mean expression level for both AML and normal patient cohorts for each gene is shown. *P < .05; **P < .01.
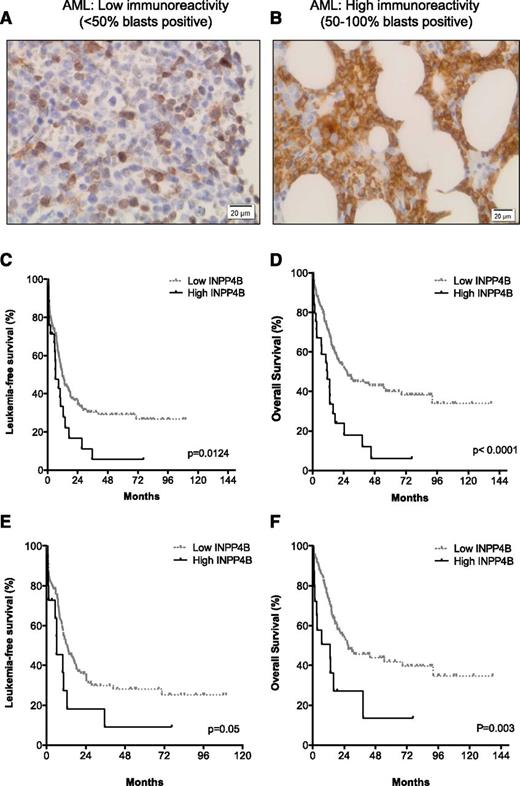
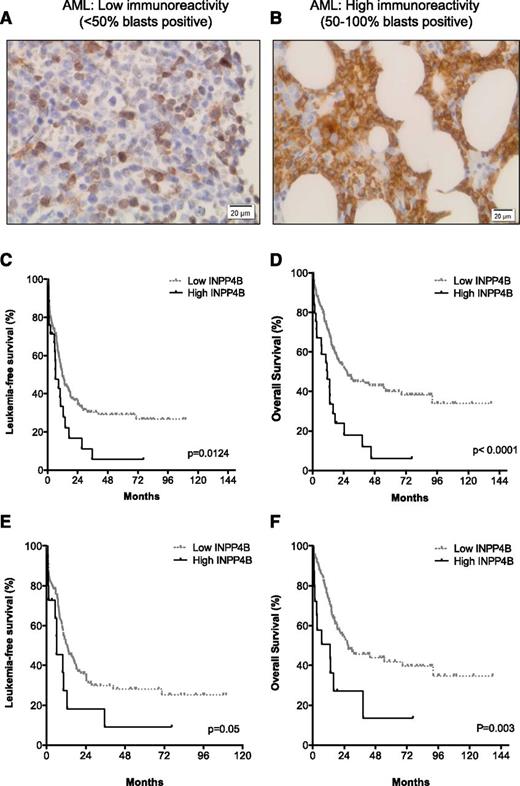
Good concordance between INPP4B gene expression and protein levels, as determined by immunoblotting with the 3D5 antibody (supplemental Figure 2), was demonstrated. We therefore expanded our analysis to examine INPP4B protein expression using the 3D5 clone in a panel of 205 BM trephine sections from patients with AML. The 3D5 monoclonal INPP4B antibody has been previously characterized and used to assess INPP4B expression in breast cancer.25 The INPP4B mAb immunostained normal megakaryocytes and occasional lymphocytes in non-leukemic BM (not shown). A Kaplan-Meier analysis of the AML cohort stratified into quartiles for INPP4B blast expression revealed separation of leukemia-free survival (LFS) and OS into two aggregated prognostic groups, namely low blast immunoreactivity (<50% blasts positive; Figure 2A) and high blast immunoreactivity (50% to 100% blasts positive; Figure 2B) for INPP4B (supplemental Figure 3A-B). The proportion of blasts positive for INPP4B was low in 88% (n = 180) and high in 12% (n = 25) of AML cases examined (Table 1). Patients with a high proportion of INPP4B-positive blasts had significantly inferior LFS (median, 6.2 vs 11.8 months; P = .01; Figure 2C) and OS (median, 11.5 vs 26.6 months; P < .01; Figure 2D) outcomes compared with patients with low INPP4B blast expression (Table 1). The inferior prognostic association of high INPP4B with clinical outcome in AML remained present despite censoring for the confounding impact of allogeneic stem cell transplantation (supplemental Figure 4). Examination of the prognostic impact of high INPP4B within cytogenetic risk groups revealed no impact among patients with adverse-risk karyotype (Table 1). Within intermediate-risk karyotype, however, high INPP4B was a significantly poor prognostic factor for both LFS (Figure 2E) and OS (Figure 2F and Table 1). Among patients with intermediate-risk karyotype AML, the majority (83%) with high INPP4B were FLT3 and NPM1 wild-type (Table 1). High INPP4B in intermediate-risk FLT3/NPM1 wild-type AML was also associated with significantly worse survival outcome (supplemental Figure 5 and Table 1). There was no association between high INPP4B and FLT3-ITD. The adverse OS impact of high INPP4B blast positivity was not curtailed by intensified chemotherapy approaches. There were 134 patients treated with standard-dose cytarabine-based induction and 71 patients treated with high-dose cytarabine-based chemotherapy. Median OS was worse for patients with high INPP4B if they were treated with either standard-dose cytarabine (11.1 vs 17.5 months; P = .001) or high-dose cytarabine-based induction regimens (14.5 months vs not reached; P = .007). Because increasing age is a well-known determinant of chemotherapy failure, we examined CR outcomes for patients up to the age of 65 years. Among patients with high INPP4B, the CR rate was only 39% compared with 71% for patients with low INPP4B blast reactivity (P = .01) (Table 1). These findings demonstrate that INPP4B is aberrantly overexpressed in a subset of AML and that a high proportion of INPP4B-positive blasts (50% to 100%) is associated with a lower initial response to induction chemotherapy and inferior long-term LFS and OS outcomes.
High INPP4B expression correlates with poor clinical outcome in AML. BM trephine sections from patients with AML (n = 205) undergoing intensive induction chemotherapy. In normal BM, only rare cells expressed INPP4B, such as megakaryocytes and occasional lymphocytes. In AML, representative cases displaying (A) low INPP4B expression (<50% blast immunoreactivity) or (B) high INPP4B expression (50% to 100% blast immunoreactivity) are shown. Kaplan-Meier survival analysis of patients with AML who received chemotherapy showed that patients with high INPP4B had significantly reduced (C) LFS and (D) OS outcomes compared with patients with low INPP4B. Worse AML (E) LFS and (F) OS outcomes were also evident for patients with intermediate cytogenetic risk.
High INPP4B expression correlates with poor clinical outcome in AML. BM trephine sections from patients with AML (n = 205) undergoing intensive induction chemotherapy. In normal BM, only rare cells expressed INPP4B, such as megakaryocytes and occasional lymphocytes. In AML, representative cases displaying (A) low INPP4B expression (<50% blast immunoreactivity) or (B) high INPP4B expression (50% to 100% blast immunoreactivity) are shown. Kaplan-Meier survival analysis of patients with AML who received chemotherapy showed that patients with high INPP4B had significantly reduced (C) LFS and (D) OS outcomes compared with patients with low INPP4B. Worse AML (E) LFS and (F) OS outcomes were also evident for patients with intermediate cytogenetic risk.
High INPP4B expression is an independent predictor of poor outcome in patients with AML undergoing intensive chemotherapy
Closer examination of characteristics typically associated with poor risk in AML revealed no significant association between patients with high INPP4B and age, secondary AML, hyperleukocytosis, adverse-risk karyotype, or FLT3-ITD (Table 1). A multivariate analysis revealed that high INPP4B was an independent predictor of poor OS outcome for patients with AML at diagnosis (hazard ratio [HR], 2.2), along with adverse-risk karyotype (HR, 1.7) and age >55 years (HR, 2.1) (Table 2). This effect as an independent predictor of survival still remained if patient outcome was censored for allograft (Table 2).
Endogenously overexpressed INPP4B in primary AML blasts is catalytically active
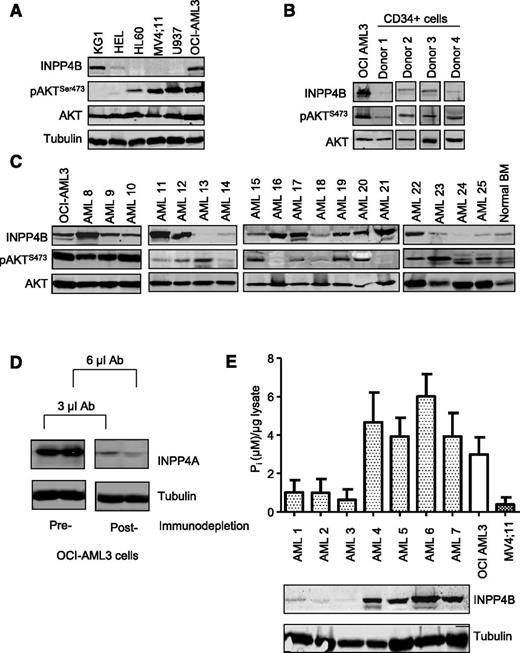
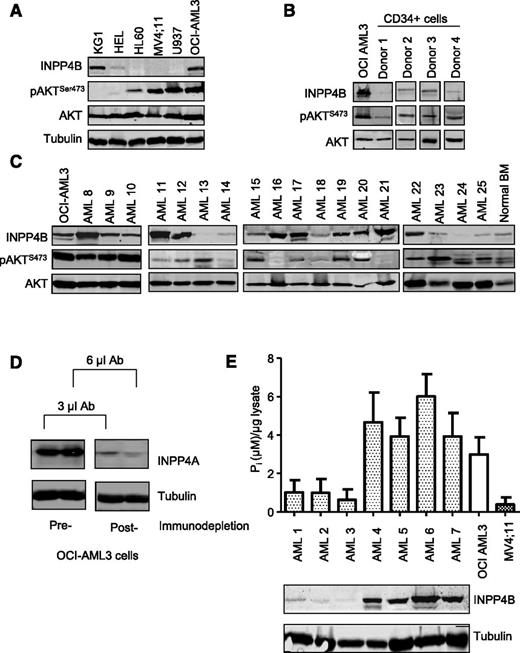
We next sought to determine whether the INPP4B expressed endogenously in human AML cells possessed lipid phosphatase activity. Immunoblot analysis of INPP4B across a panel of 6 human AML cell lines revealed high expression in OCI-AML3 and KG1 cells (Figure 3A). Furthermore, INPP4B was expressed strongly in 16 of 30 primary human AML patient samples when compared with BM samples from normal donors (Figure 3B-C and supplemental Figure 6A). To assess the catalytic function of INPP4B, a malachite green phosphatase assay was used to measure the ability of INPP4B to dephosphorylate PtdIns(3,4)P2 to PtdIns(3)P and liberate free phosphate molecules. Because both INPP4A and INPP4B can each dephosphorylate PtdIns(3,4)P2, INPP4A was first immunodepleted by using an mAb to INPP4A to ensure that the assay specifically measured INPP4B phosphoinositide phosphatase activity (Figure 3D). As shown in Figure 3E, patient samples with clearly detectable INPP4B protein (AML4-7) demonstrated robust INPP4B enzymatic activity, indicating that the phosphatase domain of endogenously overexpressed INPP4B in human AML was catalytically active. Although INPP4B has the potential to downregulate PI3K signaling, we did not observe any correlation between endogenous INPP4B protein levels and pAKTS473 among patient samples or in AML cell lines (Figure 3C and supplemental Figure 6). Thus, although INPP4B has been proposed in some cellular contexts to be a negative regulator of PI3K signaling,25 our results suggest that it is likely to have different roles in leukemia.
INPP4B is catalytically active but not coupled to pAKTS473 inhibition in AML. Immunoblotting analysis of INPP4B and pAKTS473 in (A) AML cell lines and (B) CD34+ sorted progenitors derived from normal donors relative to the INPP4B-expressing OCI-AML3 cell line. (C) Analysis of INPP4B and pAKTS473 in patient-derived AML samples relative to OCI-AML3 cells and normal BM. (D) Cell lysates from OCI-AML3 cells were immunodepleted with INPP4A antibody (Ab) to demonstrate removal of INPP4A protein in lysates prior to performing INPP4B enzymatic assays. (E) Endogenous INPP4B expression in primary AML samples relative to PtdIns(3,4)P2 4-phosphatase activity. Lysates were immunodepleted of INPP4A and the capacity for INPP4B to process PtdIns(3,4)P2 and release free phosphate ions (Pi) quantified by a malachite green reporter assay. Results represent the mean ± standard deviation (SD) of 2 independent experiments.
INPP4B is catalytically active but not coupled to pAKTS473 inhibition in AML. Immunoblotting analysis of INPP4B and pAKTS473 in (A) AML cell lines and (B) CD34+ sorted progenitors derived from normal donors relative to the INPP4B-expressing OCI-AML3 cell line. (C) Analysis of INPP4B and pAKTS473 in patient-derived AML samples relative to OCI-AML3 cells and normal BM. (D) Cell lysates from OCI-AML3 cells were immunodepleted with INPP4A antibody (Ab) to demonstrate removal of INPP4A protein in lysates prior to performing INPP4B enzymatic assays. (E) Endogenous INPP4B expression in primary AML samples relative to PtdIns(3,4)P2 4-phosphatase activity. Lysates were immunodepleted of INPP4A and the capacity for INPP4B to process PtdIns(3,4)P2 and release free phosphate ions (Pi) quantified by a malachite green reporter assay. Results represent the mean ± standard deviation (SD) of 2 independent experiments.
INPP4B overexpression is associated with resistance to leukemia therapy independent of its phosphoinositide phosphatase function
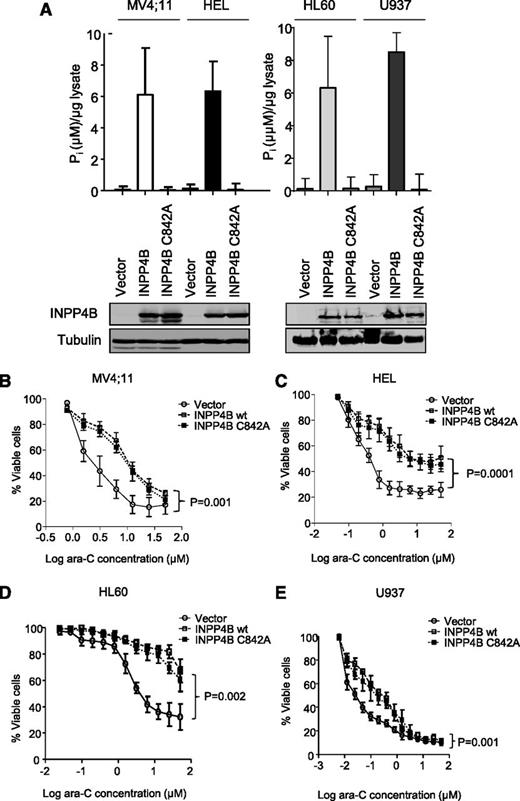
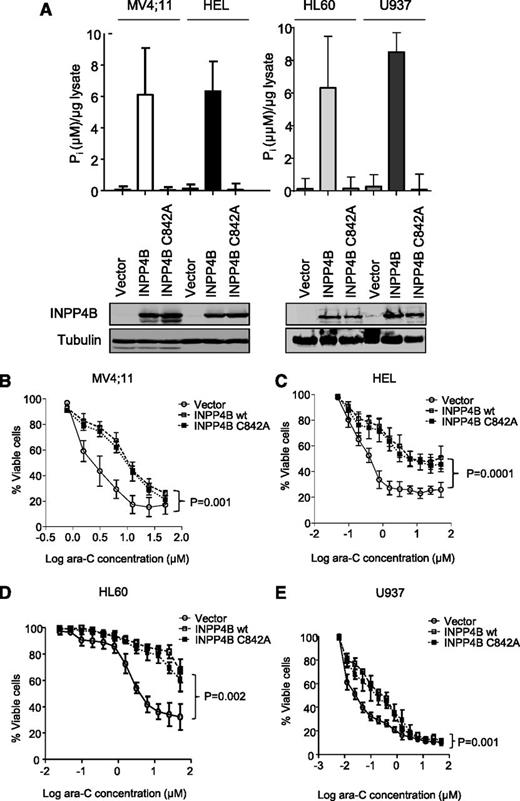
Because multivariate analysis indicated that INPP4B overexpression in AML was significantly associated with poorer patient outcomes, we then investigated the possibility that high INPP4B levels may have a functional role in promoting chemoresistance in AML cells. To examine this possibility further, wild-type INPP4B was overexpressed in 4 independent AML cell lines (MV4;11, HEL, HL60, and U937), each of which had low INPP4B expression (Figure 3A). Increased INPP4B catalytic activity in association with overexpressed INPP4B was verified in each cell line (Figure 4A). Importantly, we also expressed a catalytically inactive mutant of INPP4B (INPP4B C842A) that lacked catalytic activity in the malachite green lipid phosphatase assay (Figure 4A). Compared with vector control cells, MV4;11, HEL, HL60, and U937 AML cells overexpressing wild-type INPP4B were significantly more resistant to the induction of cell death by ara-C (Figure 4B-E). MV4;11 and HEL cell lines were also resistant to increasing doses of daunorubicin (supplemental Figure 7A-B) and etoposide (supplemental Figure 7C-D), which represent standard cytotoxic drugs used in the clinical management of AML. INPP4B overexpression also conferred resistance to ara-C in long-term clonogenic assays (supplemental Figure 8). Unexpectedly, the catalytically inactive INPP4B C842A mutant also mediated chemoresistance in short-term cell survival assays as well as longer-term clonogenic assays, indicating that the phosphatase function of INPP4B was not required for mediating drug resistance (Figure 4B-E and supplemental Figure 8). Although there was no clear association between high INPP4B and pAKT in cell lines or primary AML samples, the effect of transgenic overexpression of INPP4B and INPP4B C842A on signaling pathways associated with chemoresistance was explored. Despite overexpression of INPP4B in MV4;11 and HEL in cell lines, no changes in the phosphorylation status of AKTS473 or in the expression levels of Bcl-2 family members Bcl-2, Bcl-x, Mcl-1, or Bim were observed (supplemental Figure 9A). Although a minor increase in pERK was noted in MV4;11 cells, this was not evident in HEL cells (supplemental Figure 9A). Furthermore, the marginal increase in pERK was also present in MV4;11 cells overexpressing the phosphatase inactive form of INPP4B, thereby weakening any potential link between INPP4B phosphatase function and pERK signaling.
INPP4B overexpression confers resistance to ara-C independent of its phosphoinositide phosphatase function. (A) Ectopic overexpression of wild-type (wt) INPP4B and phosphatase inactive INPP4B C842A in AML cell lines showing expression by western blot (bottom panels) and the concordant phosphatase activity (top panels). Overexpression of both INPP4B and the INPP4B C842A variant resulted in resistance to cytarabine in (B) MV4;11, (C) HEL, (D) HL60, and (E) U937 AML cells compared with vector control. Cells were subjected to drugs for 48 hours, and viability was measured by using flow cytometry. Results represent the mean ± SD of 3 independent experiments. P < .05 for survival curve comparisons between vector control and wild-type INPP4B (as indicated) and INPP4B C842A (not shown).
INPP4B overexpression confers resistance to ara-C independent of its phosphoinositide phosphatase function. (A) Ectopic overexpression of wild-type (wt) INPP4B and phosphatase inactive INPP4B C842A in AML cell lines showing expression by western blot (bottom panels) and the concordant phosphatase activity (top panels). Overexpression of both INPP4B and the INPP4B C842A variant resulted in resistance to cytarabine in (B) MV4;11, (C) HEL, (D) HL60, and (E) U937 AML cells compared with vector control. Cells were subjected to drugs for 48 hours, and viability was measured by using flow cytometry. Results represent the mean ± SD of 3 independent experiments. P < .05 for survival curve comparisons between vector control and wild-type INPP4B (as indicated) and INPP4B C842A (not shown).
To determine whether transgenic INPP4B was expressed at physiologically relevant levels, a comparison was made with INPP4B expressed in a range of primary AML cells (supplemental Figure 9B). This confirmed that transgenic levels of INPP4B and INPP4B C842A were comparable to levels observed in AML cells manifesting high levels of constitutive INPP4B. To determine whether chemoresistance could be explained by a non-phosphatase–dependent role for INPP4B, we next examined whether small interfering RNA (siRNA) –mediated knockdown of INPP4B could sensitize AML cell lines and primary AML cells overexpressing INPP4B to chemotherapy. We identified two si-RNAs (INPP4B siRNA-1 and siRNA-2), which efficiently suppressed INPP4B protein in OCI-AML3 cells (supplemental Figure 10A-B). Both si-RNAs directed at INPP4B sensitized KG-1 and OCI-AML3 cells to the cytotoxic effects of ara-C (Figure 5A-B). Transduction of primary AML blasts with INPP4B si-RNA (supplemental Figure 10C) also led to ara-C sensitization (Figure 5C-D) in examples of primary AML shown to have INPP4B expressed in a high proportion of blasts (supplemental Figure 10D-E). Thus, targeting INPP4B in a variety of AML cell contexts supports the hypothesis that INPP4B is an important determinant of chemoresistance in human AML.
INPP4B siRNA sensitizes AML to chemotherapy. (A) KG-1 or (B) OCI-AML3 cells were cultured for 48 hours prior to being treated with 5 μM or 25 μM ara-C for 24 hours, and viability was measured by using flow cytometric enumeration of propidium iodide–negative cells. Results represent the mean ± SD of 2 independent experiments. (A) KG-1 or (B) OCI-AML3 cells were transduced with siRNAs targeting either INPP4B (INPP4B siRNA-1 or siRNA-2) or a scrambled control for 12 hours and were cultured for 48 hours prior to cytotoxic treatment as above. Similar experiments were performed in primary AML samples (C) AML 31 and (D) AML 32 shown to have high INPP4B expression (AML 31 [supplemental Figure 10D] and AML 32 [supplemental Figure 10E], confirming sensitization of primary AML blasts to ara-C after INPP4B siRNA targeting.
INPP4B siRNA sensitizes AML to chemotherapy. (A) KG-1 or (B) OCI-AML3 cells were cultured for 48 hours prior to being treated with 5 μM or 25 μM ara-C for 24 hours, and viability was measured by using flow cytometric enumeration of propidium iodide–negative cells. Results represent the mean ± SD of 2 independent experiments. (A) KG-1 or (B) OCI-AML3 cells were transduced with siRNAs targeting either INPP4B (INPP4B siRNA-1 or siRNA-2) or a scrambled control for 12 hours and were cultured for 48 hours prior to cytotoxic treatment as above. Similar experiments were performed in primary AML samples (C) AML 31 and (D) AML 32 shown to have high INPP4B expression (AML 31 [supplemental Figure 10D] and AML 32 [supplemental Figure 10E], confirming sensitization of primary AML blasts to ara-C after INPP4B siRNA targeting.
INPP4B overexpression confers resistance to chemotherapy in vivo
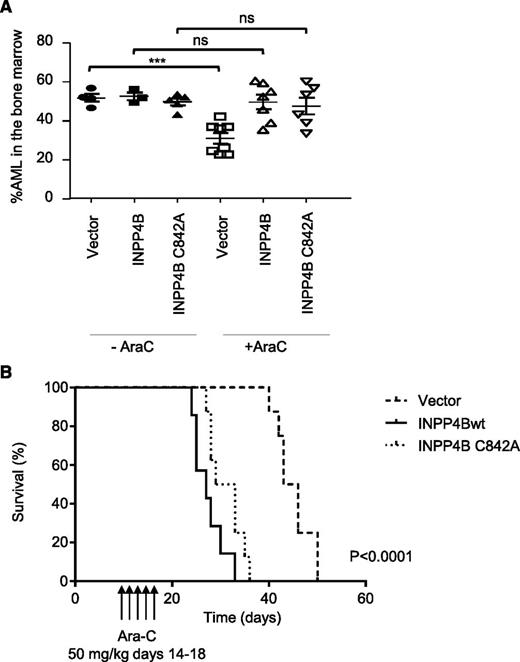
To analyze the relevance of INPP4B as a mediator of drug resistance and to determine the importance of the phosphoinositide 4-phosphatase catalytic activity on leukemia outcomes in vivo, we examined the impact of INPP4B on human AML growth in a mouse xenograft model. MV4;11 cells overexpressing wild-type or the catalytically inactive INPP4B C842A mutant were engrafted into NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice. Once disease was established, mice were treated with a 5-day course of chemotherapy (ara-C at 75 mg/kg by intraperitoneal injection once per day). After completing cytotoxic treatment, mice were culled, and the effect of chemotherapy on leukemic BM infiltration (percentage of human CD45+ cells and cells expressing ZsGreen1 reporter encoded within the transgene) was examined (Figure 6A). As expected, treating mice with ara-C significantly reduced the burden of vector control MV4;11 cells in BM compared with untreated mice (P = .0006; Figure 6A). However, chemotherapy was unable to reduce the leukemic burden in mice engrafted with MV4;11 cells expressing either INPP4B or the catalytically inactive INPP4B C842A variant, verifying in vivo resistance to cytotoxic killing linked to both forms of overexpressed INPP4B (Figure 6A). Consistent with the ability of INPP4B and INPP4B C842A to promote chemoresistance in vivo, mice engrafted with MV4;11 cells expressing either INPP4B or INPP4B C842A had significantly poorer survival outcomes following ara-C treatment (Figure 6B). These findings confirm that INPP4B is a potent mediator of resistance to standard AML therapies in vitro and in vivo and that this effect is independent of INPP4B PtdIns(3,4)P2 4-phosphatase function.
Ectopic overexpression of INPP4B confers resistance to ara-C and reduces OS in vivo. (A) Cohorts of 3 to 8 NSG mice were given 105 MV4;11 cells transduced with wild-type INPP4B, phosphatase inactive INPP4B C842A mutant, or vector control intravenously after irradiation. After confirmation of engraftment on day 14, mice were left untreated (– araC) or treated with ara-C at 75 mg/kg intraperitoneally for 5 days, after which leukemia infiltration in the BM was quantified by flow cytometric assessment of MV4;11 cells expressing the Zsgreen1 reporter protein as well as human CD45 staining of BM cells. (A) BM analysis revealed that MV4;11 cells expressing both INPP4B and INPP4B C842A were resistant to ara-C administration compared with vector control. ***P < .0001; ns, not significant. (B) For survival analysis, cohorts of 7 to 8 NSG mice were engrafted with MV4;11 cells transduced with wild-type INPP4B, phosphatase inactive INPP4B C842A mutant, or vector control and treated with ara-C at 50 mg/kg intraperitoneally for 5 days and monitored until ethical euthanasia according to signs of morbidity. Survival was plotted by using the Kaplan-Meier method. P < .0001 for comparisons between vector control ands wild-type INPP4B (as indicated) and INPP4B C842A (not shown).
Ectopic overexpression of INPP4B confers resistance to ara-C and reduces OS in vivo. (A) Cohorts of 3 to 8 NSG mice were given 105 MV4;11 cells transduced with wild-type INPP4B, phosphatase inactive INPP4B C842A mutant, or vector control intravenously after irradiation. After confirmation of engraftment on day 14, mice were left untreated (– araC) or treated with ara-C at 75 mg/kg intraperitoneally for 5 days, after which leukemia infiltration in the BM was quantified by flow cytometric assessment of MV4;11 cells expressing the Zsgreen1 reporter protein as well as human CD45 staining of BM cells. (A) BM analysis revealed that MV4;11 cells expressing both INPP4B and INPP4B C842A were resistant to ara-C administration compared with vector control. ***P < .0001; ns, not significant. (B) For survival analysis, cohorts of 7 to 8 NSG mice were engrafted with MV4;11 cells transduced with wild-type INPP4B, phosphatase inactive INPP4B C842A mutant, or vector control and treated with ara-C at 50 mg/kg intraperitoneally for 5 days and monitored until ethical euthanasia according to signs of morbidity. Survival was plotted by using the Kaplan-Meier method. P < .0001 for comparisons between vector control ands wild-type INPP4B (as indicated) and INPP4B C842A (not shown).
In addition to its consensus phosphatase domain, INPP4B also possesses a C2 domain, which may play an adaptor role to anchor proteins to the plasma membrane.27-32 A plausible hypothesis is that overexpressed INPP4B could promote chemoresistance via an increased adapter function. Our attempts to express an INPP4B C2 deletion mutant in AML, however, were not successful, because the INPP4B ΔC2 variant protein was inherently unstable, with cellular expression levels much lower than those observed for transgenic INPP4B or INPP4B C842A (supplemental Figure 11). Although overexpressed INPP4B ΔC2, unlike full-length INPP4B, did not cause ara-C chemoresistance (not shown), our inability to express both constructs at comparable levels made interpretation of these results difficult. Similar problems with protein stability in the absence of the C2 domain have also been reported for PTEN.31,33,34
Discussion
Constitutive activation of kinase-mediated proliferation and survival pathways is a frequent finding in AML.9,35 Exploration of the role played by phosphoinositide phosphatases, which are negative regulators of PI3K-AKT signaling, has been limited to a small number of phosphatases in AML. In this study, we sought to identify aberrant patterns of phosphoinositide phosphatase expression in AML by using a highly sensitive mass spectrometry–based platform. This survey revealed significant overexpression of the phosphoinositide 4-phosphatase INPP4B, which on further evaluation in a panel of 205 primary AML samples from patients undergoing intensive chemotherapy was found to be associated with lower rates of CR and inferior LFS and OS. We also showed that overexpressed INPP4B had independent prognostic relevance for overall AML survival in multivariate analysis.
INPP4B has been shown in some cellular settings to downregulate PI3K signaling through its role in dephosphorylating PtdIns(3,4)P2 and PtdIns(4,5)P2. We and others have previously shown that INPP4B knockdown promoted Akt activation, proliferation, and xenograft growth in models of breast cancer.25 In breast cancer, INPP4B loss was associated with higher clinical grade and more aggressive histologic subtypes.25,36 In AML, we surprisingly found no correlation between INPP4B expression levels and AKT phosphorylation status in a panel of 30 primary samples. To rule out the possibility of an aberrant form of INPP4B, we confirmed that endogenously overexpressed INPP4B in primary AML samples was able to process PtdIns(3,4)P2 (Figure 3). In addition, coding mutations affecting INPP4B were not identified by using next-generation sequencing (not shown). Further experiments revealed that overexpressed INPP4B promoted resistance to drugs commonly used to treat AML, including ara-C, daunorubicin, and etoposide (Figures 4 and 6). To determine whether the phosphoinositide phosphatase domain of INPP4B was important for mediating chemoresistance, we compared in vitro and in vivo responses to chemotherapy in AML cells overexpressing wild-type or the phosphatase inactive INPP4B C842A variant. These studies indicated that the mechanism by which INPP4B promoted chemoresistance was not linked to its phosphoinositide phosphatase function (Figures 4 and 6). Suppression of high endogenous levels of INPP4B in AML cells by RNA interference, however, sensitized responses to chemotherapy in contrast to the minimal effect observed from ablating lipid phosphatase activity (Figure 5). These findings implicate a novel phosphoinositide phosphatase–independent gain-of-function role for INPP4B as a mediator of chemoresistance in AML in contrast to the phosphoinositide phosphatase–dependent loss-of-function role for INPP4B described in some solid cancers.
Although INPP4B overexpression has been linked to chemoresistance in laryngeal, lung, and gastric cancer cells,37,38 our study is the first to show relevance in a large cohort of primary patient AML samples and the first to demonstrate that chemoresistance may be mediated by a mechanism independent of the phosphoinositide phosphatase domain of INPP4B. Previous studies have revealed missense mutations affecting SHIP-1 (INPP5D) in 3% of AML in association with reduced 5-phosphatase function.19,39 Although some SHIP-1 mutations (eg, SH2 [F28L] and SH3 [P1039S]) have been reported, which do not affect 5-phosphatase function, such variants were linked to loss-of-adaptor functions associated with the molecule.19,39 Although INPP4B has been linked to a tumor suppressor role in solid cancers, the functional consequences of INPP4B expression may be tissue-specific. Our finding that INPP4B may also have a non-phosphatase–dependent role may provide some insight into how such diverse functions may be orchestrated as well as providing the stimulus for future studies focused on elucidating the precise mechanisms by which INPP4B promotes chemoresistance in AML.
Therapeutic resistance manifests as either failure to achieve CR or early relapse, with increasing age and cytogenetic risk being the most important predictors.5 Forecasting chemoresistance has important implications for patients and physicians when making treatment decisions. To date, few molecular targets linked to treatment response have been clinically validated in AML. Selected examples include FLT3-ITD and mutations affecting p53.5,40 INPP4B overexpression has previously been linked to poor OS as part of a leukemic stem cell gene expression signature in AML.41 In our current study, we substantially extend these observations by showing that high INPP4B, defined as protein positivity in at least 50% of AML blasts, delineates a high-risk patient population characterized by reduced response to induction chemotherapy, earlier leukemic relapse, and poorer OS outcome. For patients age 65 years and younger, the CR rate was only 39%, substantially lower than CR rates reported in other studies (typically 63% to 77%).42,43 Importantly, the poor prognostic value of high INPP4B was independent of other established prognostic markers, such as FLT3-ITD, age, white blood cell count, and adverse-risk karyotype. Furthermore, the prognostic value was most apparent in patients with intermediate-risk karyotype, especially those with wild-type FLT3 and NPM1 AML, thus enhancing the clinical value of this potential prognostic marker. If validated, patients with high INPP4B could be considered candidates for experimental treatment options. Although the frequency of overexpressed INPP4B in AML in our series was approximately 12%, this is similar to the frequency of mutated isocitrate dehydrogenase-2, for which targeted therapies are being actively pursued in the clinic.44
In conclusion, this study identifies overexpressed INPP4B as a potential marker of treatment failure in patients with AML. Contrary to other studies, we find that INPP4B has phosphoinositide phosphatase–independent functions that contribute to chemoresistance. A better understanding of how INPP4B gain of function enhances drug resistance could stimulate the development of INPP4B-directed targeted therapies in the future.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Darryl Irwin from Sequenom for assistance with MassArray quantitative gene expression, David Huang from Walter and Eliza Hall Institute for Medical Research for input and guidance with experiments, and Monash Micro Imaging for assistance and guidance.
This work was supported by a Leukemia Foundation Grant in Aid, the Victorian Cancer Agency, Arrow Bone Marrow Transplant Foundation, and the National Health and Medical Research Council.
Authorship
Contribution: S.R., A.H.W., and C.A.M conceived and designed the research; S.R., N.-Y.N.N., N.C., J.F.M., C.M., and T.-C.T. performed the research; S.A., N.K.R., and L.M.O. provided reagents and database information; S.R., S.F., A.T.P., C.M., C.A.M., and A.H.W. analyzed and interpreted the data; S.R., S.F., C.M., C.A.M., and A.H.W. wrote the paper; and all authors approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andrew H. Wei, Department of Clinical Haematology, Alfred Hospital and Monash University, Melbourne, VIC 3004, Australia; e-mail: a.wei@alfred.org.au; Christina A. Mitchell, Department of Biochemistry and Molecular Biology, Monash University, Clayton, VIC 3800, Australia; e-mail: christina.mitchell@monash.edu.

![Figure 5. INPP4B siRNA sensitizes AML to chemotherapy. (A) KG-1 or (B) OCI-AML3 cells were cultured for 48 hours prior to being treated with 5 μM or 25 μM ara-C for 24 hours, and viability was measured by using flow cytometric enumeration of propidium iodide–negative cells. Results represent the mean ± SD of 2 independent experiments. (A) KG-1 or (B) OCI-AML3 cells were transduced with siRNAs targeting either INPP4B (INPP4B siRNA-1 or siRNA-2) or a scrambled control for 12 hours and were cultured for 48 hours prior to cytotoxic treatment as above. Similar experiments were performed in primary AML samples (C) AML 31 and (D) AML 32 shown to have high INPP4B expression (AML 31 [supplemental Figure 10D] and AML 32 [supplemental Figure 10E], confirming sensitization of primary AML blasts to ara-C after INPP4B siRNA targeting.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/18/10.1182_blood-2014-09-603555/4/m_2815f5.jpeg?Expires=1769592840&Signature=1zCoZ3wbYUOdu1~hFF7BdDHrCcxLL9QXcPmzwHAyXAVozjat5r7OVdkGuvhgQCsbjvCbRfXBSNIK1KJXSkEogSZzJ0z1lRQM8udUhMxQ1H5N7ThAFpGAKxXo007-d62aoGIMgv1j3o0nlKWJYWlRU5KgS-oldfxhisZanmAyNC~yMgwiU94wj7nitLg29NlPjm7WnKAu4lDapfz-P5XuvosQVhvjI9ljK~-gTIDVbeYog5-yfUew9Aqkgfbt8t0byCRGAcNs62YiBk719QKXxRMoxPICTHZZRbhwJwn8u9CIa0JvIdYHe~adLIuaMFx~zmaKbCWoXPwsUPiC4X8iGg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 5. INPP4B siRNA sensitizes AML to chemotherapy. (A) KG-1 or (B) OCI-AML3 cells were cultured for 48 hours prior to being treated with 5 μM or 25 μM ara-C for 24 hours, and viability was measured by using flow cytometric enumeration of propidium iodide–negative cells. Results represent the mean ± SD of 2 independent experiments. (A) KG-1 or (B) OCI-AML3 cells were transduced with siRNAs targeting either INPP4B (INPP4B siRNA-1 or siRNA-2) or a scrambled control for 12 hours and were cultured for 48 hours prior to cytotoxic treatment as above. Similar experiments were performed in primary AML samples (C) AML 31 and (D) AML 32 shown to have high INPP4B expression (AML 31 [supplemental Figure 10D] and AML 32 [supplemental Figure 10E], confirming sensitization of primary AML blasts to ara-C after INPP4B siRNA targeting.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/18/10.1182_blood-2014-09-603555/4/m_2815f5.jpeg?Expires=1770006114&Signature=mzDbdbjX5AGPTS4q3~djjy2a7Wer6QclDinvpKmfrJHJcDIMjCBWFY~t03GPLSJ0UBNmu38lEqUNMZ4liNwDlA~E0tMiglfQGasKuO4fKZuDRtxBAkK9XtnuiAavXk1znNXKdbERR-ua2yPcYsIsMb-Ei8wHXEHok3Ten~z2C-zwRcVdtRRaGJFw1BDxZUkwkK2rniZ9PQpi8Sds48PWD9uovhW1DnzGvEccueCP9x4K56NDARvNXpDlmmi8pazcrSg-I6ETcEGyqABakMTi7PLKh7~jCwhjpBOYCzPV-r6UUilWLBhaN6yuyH9WWK5E53aJ67kKB7Kt0lVdNhyktA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
