Key Points
High-risk copy number gains of 1q21 originate in part by the hypomethylation of 1q12 pericentromeric heterochromatin.
Novel CNAs can result from juxtaposition of chromosomal regions to hypomethylated 1q12.
Abstract
Multiple myeloma is a B-cell malignancy stratified in part by cytogenetic abnormalities, including the high-risk copy number aberrations (CNAs) of +1q21 and 17p−. To investigate the relationship between 1q21 CNAs and DNA hypomethylation of the 1q12 pericentromeric heterochromatin, we treated in vitro peripheral blood cultures of 5 patients with balanced constitutional rearrangements of 1q12 and 5 controls with the hypomethylating agent 5-azacytidine. Using G-banding, fluorescence in situ hybridization, and spectral karyotyping, we identified structural aberrations and copy number gains of 1q21 in the treated cells similar to those found in patients with cytogenetically defined high-risk disease. Aberrations included 1q12 triradials, amplifications of regions juxtaposed to 1q12, and jumping translocations 1q12. Strikingly, all 5 patients with constitutional 1q12 rearrangements showed amplifications on the derivative chromosomes distal to the inverted or translocated 1q12 region, including MYCN in 1 case. At the same time, no amplification of the 1q21 region was found when the 1q12 region was inverted or absent. These findings provide evidence that the hypomethylation of the 1q12 region can potentially amplify any genomic region juxtaposed to it and mimic CNAs found in the bone marrow of patients with high-risk disease.
Introduction
The copy number aberrations (CNAs) deletion of 17p and gain of 1q21 are cytogenetic markers for high-risk multiple myeloma.1,2 Of these, the gain of the 1q21 region occurs most frequently, is found in about 40% of cases, and is often found in proliferative relapsed and/or refractory disease.3-7 Gains of 1q21 are most often associated with secondary aberrations involving jumping translocations of 1q12 (JT1q12). In patients with JT1q12s, we have shown that extra copies of 1q21 result from the formation of triradial chromosomes involving the 1q12 region.8 We and others have previously speculated that hypomethylation of the highly repetitive elements that make up the 1q12 pericentromeric region may be related to the origin and progression of CNAs of 1q21,8-10 suggesting epigenetic factors are involved. In fact, methylation studies in multiple myeloma (MM) have shown a gradual progression from overall global hypomethylation to DNA hypermethylation of specific genes11-13 and that hypomethylation of repetitive elements may contribute to tumor progression and genomic instability.14,15
It has previously been demonstrated that hypomethylation of the pericentromeric regions of metaphase chromosomes by 5-azacytidine can induce 1q12 chromosome decondensation16,17 and somatic associations in chromosome regions 1, 9, and 16 in human lymphocytes.18-20 The aim of this study was to test the hypothesis that hypomethylation of 1q12 pericentromeric heterochromatin by 5-azacytidine can initiate copy number (CN) gains of 1q21. Here we present evidence that the hypomethylation of the 1q12 region not only induces CN gains of 1q21, but also can induce novel CN gains of chromosome regions inverted or translocated next to hypomethylated 1q12 pericentromeric heterochromatin.
Patients and methods
The Institutional Review Board of the University of Arkansas for Medical Sciences approved the research studies, and all subjects provided written informed consent approving the use of their samples for research purposes. Research was conducted in accordance with the Declaration of Helsinki.
Patients were selected for the study based on the diagnosis of MM and the presence of balanced 1q12 rearrangements, which changed the orientation or chromosomal location of the 1q12 region. Constitutional rearrangements are characterized in supplemental Figure 1 on the Blood Web site. Patients 1 and 2 showed pericentromeric inversions of chromosome 1 involving breakpoints on both sides of the centromere and a reversal of the 1q12 region. Patient 3 demonstrated a balanced constitutional translocation in which most of the short arm of chromosome 2 is juxtaposed to the 1q12 region of the long arm of chromosome 1. Patient 4 showed a translocation between chromosomes 1 and 2 in which the 1q12 region is translocated to the distal end of the long arm of chromosome 2. Patient 5 showed a balanced whole-arm translocation of the entire 1q to the long arm of the der(9). Five volunteer control subjects with normal constitutional karyotypes were also studied.
In vitro phytohemagglutinin-stimulated peripheral blood cultures were initiated under identical conditions on patients and controls as previously described.21 Briefly, cultures were grown for 96 hours and a low dose of 5-azacytidine (10 μM final concentration) was added for the final 72 hours of culture.17
All results are presented as percentages of metaphases expressing a particular lesion or rearrangement based on the examination of 100 metaphases (supplemental Table 1). CN data were tabulated for the presence or absence of the 1q21 (CKS1B) signal. Probes for chromosome loci 1q21(CKS1B), 1p13 (AHCYL1), and 2p24 (MYCN) were prepared as previously described.22 Probes for pericentromeric regions including 1q12, sat III DNA (red), 9q11 (α sat, aqua), and 16q11 (sat II, aqua) were acquired from Vysis and used according to manufacturer’s instructions (Vysis, Downers Grove, IL). The SKY probe mixture and hybridization reagents were prepared by Applied Spectral Imaging (Carlsbad, CA) and applied as previously described.22 Original magnification was ×1000 for all G-band and fluorescence in situ hybridization (FISH) images and ×630 for SKY.
Results and discussion
Tabulations of CN aberrations and structural aberrations are presented in supplemental Table 1. CN gains of 3 for 1q21 were identified in patients in the 1% to 10% range. Four copies of 1q21 were identified in patients 3, 4, 5, and 5 copies in patients 2, 3, 4, and 5. Triradials of regions distal to 1q12 occurred in 1% to 11% of cells in patients. In the control samples, 3 copies of 1q21 occurred in 7% to 14%, 4 copies in 1% to 6%, 5 copies in 1% to 5%, and six copies in 1% to 6% of cells (supplemental Table 1). In control samples, triradials distal to 1q12 occurred in 2% to 10% of cells. Whole-arm JT1q12s to 16q11 occurred in 3 patients and 3 controls (supplemental Table 1).
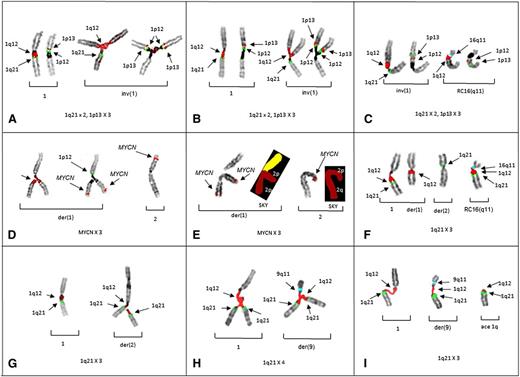
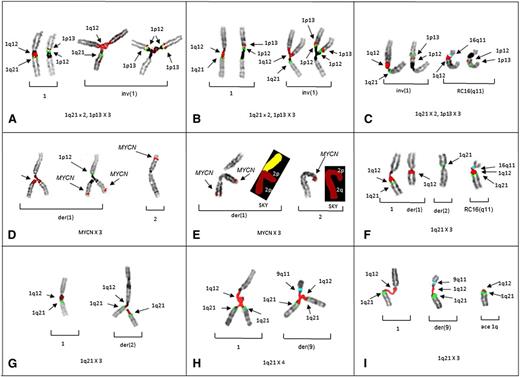
The triradials of 1p in patients 1 and 2, demonstrate both the origin of the gain for 1p13 (Figure 1A-B) and a novel whole-arm JT1q12 translocation of 1p to 16q11 (Figure 1C). In patient 3, the CN gain of MYCN resulted from triradials of 2p. Importantly, this case demonstrates that novel CN gains of regions harboring genes such as MYCN can occur by juxtaposition to hypomethylated 1q12 pericentromeric heterochromatin (Figure 1D-E). A whole-arm JT1q12 to 16q11 was also found in patient 3 (Figure 1F). In patients 4 and 5, CN gains of 1q21 resulted from 1q12 triradials occurring on nonhomologous chromosomes involving a der(2) (Figure 1G) and a der(9), respectively (Figure 1H-I).
Partial karyotypes from patients 1-5 demonstrate the origin of CN aberrations of regions juxtaposed to 1q12. Successive hybridizations of chromosome pairs with different probe sets for 1p and 1q illustrate the CN changes or translocations in the respective chromosome arms. (A) Left bracket shows a normal pattern of probes on 1q (left) and 1p (right) on the normal chromosome 1 (patient 1). Right bracket demonstrates chromosome 1p triradial on the inv(1) showing branching of 1q12 (red) and 1 copy of 1q21 (green) to the left, whereas the same triradial chromosome on the right shows 2 copies of probes for 1p12 (green) and 1p13 (red). Triradials for 1p demonstrate the origin of a CN of 3 for 1p13. (B) Partial karyotype of patient 2 showing the same probe pattern as patient 1, with a normal pattern shown in the left bracket and gains of 1p in the right bracket. (C) Partial karyotype of patient 2 showing a novel whole-arm translocation of 1p to RC16q11 (aqua) with a CN gain of 1p. The left bracket shows a pattern of probes on inv(1), whereas the right bracket shows 1p translocation to RC16q11(aqua). Normal chromosome 1 is not shown. (D) Partial karyotype of patient 3 demonstrating the origin of CN gains of 2p and MYCN. The left bracket shows successive hybridization of a triradial of 2p. The triradial of 2p (left) originates on the der(1) showing the branching 1q12 probe (red). Rehybridization of same triradial (right) with a probe for MYCN (red) demonstrates an extra copy of MYCN and 1 copy of 1p12 (green) for the short arm. The right bracket shows normal 2 with probe for MYCN (red). Normal 1 and der(2) not shown. (E) Cell demonstrating triradial of 2p with FISH for MYCN and subsequently rehybridized with SKY (left bracket). The right bracket shows normal 2 by FISH and SKY. (F) FISH probes for 1q and 16q show whole-arm translocation of 1q12 to RC16(q11) (aqua). (G) Partial karyotype from patient 4 demonstrating CN gains of 1q21 originating on nonhomologous chromosome. A normal FISH pattern on chromosome 1 in left bracket. In the right bracket, a triradial of 1q12 (red) and 2 signals for 1q21 (green) on the der(2) demonstrate the origin of the 1q21 CN gain is the der(2). (H) Patient 5 demonstrates triradial of 1q21 on the normal 1 (left bracket) and a triradial of 1q12 on the der(9)(q11) (aqua) (right bracket), resulting in a CN of 4 for 1q21. (I) Patient 5 showing 1q12 decondensation in both the normal 1 (left bracket), and der(9) (middle bracket). The right bracket shows acentric (ace) copy of JT1q12 which will subsequently result in the formation of a micronucleus. FISH hybridizations to metaphase chromosomes are shown in inverse 4,6 diamidino-2-phenylindole to delineate G-banding patterns.
Partial karyotypes from patients 1-5 demonstrate the origin of CN aberrations of regions juxtaposed to 1q12. Successive hybridizations of chromosome pairs with different probe sets for 1p and 1q illustrate the CN changes or translocations in the respective chromosome arms. (A) Left bracket shows a normal pattern of probes on 1q (left) and 1p (right) on the normal chromosome 1 (patient 1). Right bracket demonstrates chromosome 1p triradial on the inv(1) showing branching of 1q12 (red) and 1 copy of 1q21 (green) to the left, whereas the same triradial chromosome on the right shows 2 copies of probes for 1p12 (green) and 1p13 (red). Triradials for 1p demonstrate the origin of a CN of 3 for 1p13. (B) Partial karyotype of patient 2 showing the same probe pattern as patient 1, with a normal pattern shown in the left bracket and gains of 1p in the right bracket. (C) Partial karyotype of patient 2 showing a novel whole-arm translocation of 1p to RC16q11 (aqua) with a CN gain of 1p. The left bracket shows a pattern of probes on inv(1), whereas the right bracket shows 1p translocation to RC16q11(aqua). Normal chromosome 1 is not shown. (D) Partial karyotype of patient 3 demonstrating the origin of CN gains of 2p and MYCN. The left bracket shows successive hybridization of a triradial of 2p. The triradial of 2p (left) originates on the der(1) showing the branching 1q12 probe (red). Rehybridization of same triradial (right) with a probe for MYCN (red) demonstrates an extra copy of MYCN and 1 copy of 1p12 (green) for the short arm. The right bracket shows normal 2 with probe for MYCN (red). Normal 1 and der(2) not shown. (E) Cell demonstrating triradial of 2p with FISH for MYCN and subsequently rehybridized with SKY (left bracket). The right bracket shows normal 2 by FISH and SKY. (F) FISH probes for 1q and 16q show whole-arm translocation of 1q12 to RC16(q11) (aqua). (G) Partial karyotype from patient 4 demonstrating CN gains of 1q21 originating on nonhomologous chromosome. A normal FISH pattern on chromosome 1 in left bracket. In the right bracket, a triradial of 1q12 (red) and 2 signals for 1q21 (green) on the der(2) demonstrate the origin of the 1q21 CN gain is the der(2). (H) Patient 5 demonstrates triradial of 1q21 on the normal 1 (left bracket) and a triradial of 1q12 on the der(9)(q11) (aqua) (right bracket), resulting in a CN of 4 for 1q21. (I) Patient 5 showing 1q12 decondensation in both the normal 1 (left bracket), and der(9) (middle bracket). The right bracket shows acentric (ace) copy of JT1q12 which will subsequently result in the formation of a micronucleus. FISH hybridizations to metaphase chromosomes are shown in inverse 4,6 diamidino-2-phenylindole to delineate G-banding patterns.
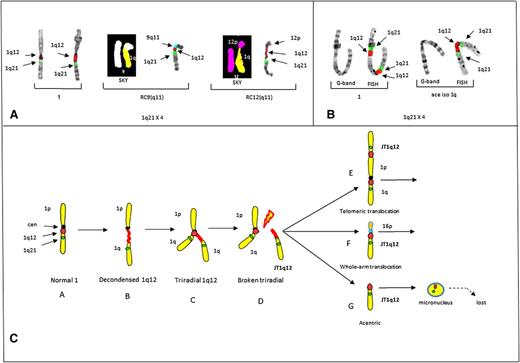
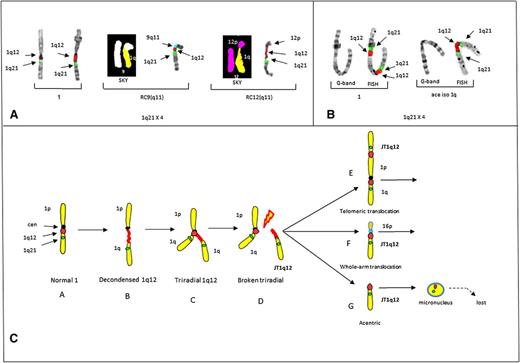
In this study, the most striking example of a JT1q12 that mimics the clonal progression found in the bone marrow of patients was identified in control 2.23 In this case, a JT1q12 to 9(q11) was found in 2 cells, increasing the CN of 1q21 to 3. Remarkably, in a third cell, additional progression was noted as a second jump of JT1q12 to the receptor chromosome (RC) RC12(q11) occurred, increasing the CN of 1q21 to 4 (Figure 2A). Finally, in control 3 a telomeric intrachromosomal JT1q12 to 1p was identified (Figure 2B). These same aberrations, including the whole-arm JT1q12 to 16q11, have been reported in the bone marrow of high-risk patients.8,9,22,23
Partial karyotypes from controls 2 and 3 demonstrating the origin of JT1q12 and a model for the origin of 1q12 aberrations. (A) Partial karyotypes of control # 2 demonstrating CN gains of 1q21 and CN losses in RCs. Normal chromosomes 1 (left bracket), RC9(q11) by both SKY (chromosome 1 yellow, chromosome 9 white) and FISH (9q11) (aqua) (middle bracket), and progression of CN aberrations resulting from an additional jump of JT1q12 to the RC12(q11) (SKY, chromosome 12 pink) and FISH (right bracket). CN of 1q21 is 4 with the loss of both 9q and 12q in the RCs. (B) Control 3 shows a chromosome 1 by both G-banding and FISH (left bracket) with a 1q21 CN of 2 resulting from a JT1q12 to the telomere of 1p and an isochromosome 1q (right bracket). (C) Model for the origin of CN gains of 1q21. Characterization of aberrations is identified following hypomethylation of 1q12 pericentromeric heterochromatin. Normal chromosome 1 (A) is depicted with a centromere (black) and FISH probes 1q12 (red) and 1q21 (green) (far left). Transient aberrations include cells with decondensation of the 1q12 region (B), triradials of 1q12 resulting in a CN gain of 1q21 (C), and cells with breakage in the 1q12 (red) pericentromeric heterochromatin (D). Extra copies of the JT1q12 originating from a triradial usually either translocate to the telomeric region of an RC (E) or alternatively to the pericentromeric region of an RC (F). Copies of JT1q12s that do not successfully translocate to an RC (G) result in acentric copies of chromosome 1q21 and are subsequently encapsulated into micronuclei and lost from the cell. FISH hybridizations to metaphase chromosomes are shown in inverse 4,6 diamidino-2-phenylindole to delineate G-banding patterns.
Partial karyotypes from controls 2 and 3 demonstrating the origin of JT1q12 and a model for the origin of 1q12 aberrations. (A) Partial karyotypes of control # 2 demonstrating CN gains of 1q21 and CN losses in RCs. Normal chromosomes 1 (left bracket), RC9(q11) by both SKY (chromosome 1 yellow, chromosome 9 white) and FISH (9q11) (aqua) (middle bracket), and progression of CN aberrations resulting from an additional jump of JT1q12 to the RC12(q11) (SKY, chromosome 12 pink) and FISH (right bracket). CN of 1q21 is 4 with the loss of both 9q and 12q in the RCs. (B) Control 3 shows a chromosome 1 by both G-banding and FISH (left bracket) with a 1q21 CN of 2 resulting from a JT1q12 to the telomere of 1p and an isochromosome 1q (right bracket). (C) Model for the origin of CN gains of 1q21. Characterization of aberrations is identified following hypomethylation of 1q12 pericentromeric heterochromatin. Normal chromosome 1 (A) is depicted with a centromere (black) and FISH probes 1q12 (red) and 1q21 (green) (far left). Transient aberrations include cells with decondensation of the 1q12 region (B), triradials of 1q12 resulting in a CN gain of 1q21 (C), and cells with breakage in the 1q12 (red) pericentromeric heterochromatin (D). Extra copies of the JT1q12 originating from a triradial usually either translocate to the telomeric region of an RC (E) or alternatively to the pericentromeric region of an RC (F). Copies of JT1q12s that do not successfully translocate to an RC (G) result in acentric copies of chromosome 1q21 and are subsequently encapsulated into micronuclei and lost from the cell. FISH hybridizations to metaphase chromosomes are shown in inverse 4,6 diamidino-2-phenylindole to delineate G-banding patterns.
Epigenetic modification of the 1q12 pericentromeric heterochromatin is characterized by transient decondensations and triradials of the 1q12 pericentromeric region (Figure 2C). Triradials of 1q12 originate CN gains of the entire 1q or potentially any other region distal to it. JT1q12s result in clonal copy number gains of 1q21 if the 1q12 region successfully translocates to either the telomere of an RC, or the pericentromeric region of an RC. Telomeric JT1q12s result in only a CN gain for 1q21, whereas in pericentromeric JT1q12s both a gain of 1q21 and a whole-arm loss of the RC16q occur (Figure 2C). The loss of 16q results in the deletion of both WWOX and CYLD1, which have been associated with a worse prognosis in MM.24 Unsuccessful JT1q12s lead to acentric copies of 1q that are subsequently lost as micronuclei (Figure 2C).
Hypomethylation of 1q12 pericentromeric heterochromatin appears to be at least 1 aspect of 1q21 CNAs in MM; however, clearly other epigenetic factors must contribute to the clonal aberrations found in the bone marrow.25 Here we provide evidence for the origin of 1q21 CN gains by showing that 5-azacytidine can induce morphologically identical aberrations to those in high-risk patients. We also demonstrate for the first time that hypomethylation of the 1q12 region can potentially amplify any genomic region juxtaposed distal to it and can account for novel CN gains of nonhomologous chromosome regions associated with JT1q12 translocations.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and staff of the Myeloma Institute for Research and Therapy. The authors also thank Diane Schrantz for editorial assistance.
This work was supported in part by the National Institutes of Health National Cancer Institute (research program project grant CA 0055819).
Authorship
Contribution: J.R.S. conceived hypothesis, analyzed and interpreted data, and wrote the manuscript; C.J.H., D.J.J., and J.E. analyzed the data; C.M.S., M.J., and G.S. performed chromosome analysis; E.T., J.L.L., and R.L.B. performed FISH and SKY studies; M.Z. and F.v.R. provided patient samples; and F.E.D., G.J.M., and B.B. wrote, reviewed, or revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jeffrey R. Sawyer, Cytogenetics Laboratory, Suite 200, Freeway Medical Tower, 5800 West 10th St, Little Rock, AR 72204; sawyerjeffreyr@uams.edu.