Abstract
The use of unrelated donors matched in all alleles of HLA-A, -B, -C, and -DRB1 loci has been associated with superior outcomes compared with those having 1 or more mismatches. Recent studies showed increased transplant-related mortality (TRM) with the use of HLA-DPB1 mismatched donors supporting the notion that the ideal volunteer unrelated donor should fully match at HLA-A, -B, -C, and -DRB1 and lack -DPB1 mismatches. The issue of the effect of HLA-DPB1 mismatch on the disease progression rate is still controversial and we aimed to investigate the impact of HLA-DPB1 mismatch in the graft versus host direction on transplant outcomes in patients categorized according to the recently defined disease risk index (DRI) for disease risk classification.
Our study cohort included 1,211 transplant patients with hematological malignancies whohave received an hematopoietic stem cell transplant (HSCT) from an unrelated HLA-A, -B, -C,-DRB1 matched donor by high resolution typing (8/8 matched) after 2005 through 2014. The study cohort had a median age of 55 (range, 19-77); the hematopoietic stem cell source was peripheral blood (PB) in 698 and bone marrow (BM) in 513 patients. Disease risk index (DRI) at HSCT was high or very high in 382 (33%), intermediate in 598 (51%), low in 185 (16%) patients.
Of the pairs, 1,154 (95%) were matched atHLA-DQB1 and 1,116 (92%) at HLA- DRB3/4/5 by high resolution testing. However, 633 (52%) had mismatch at one of the DPB1 alleles and 208 (17%) had two mismatches. There was association between matching for DPB1and matching for DRB3/4/5 (p=0.002) but not with DQB1.
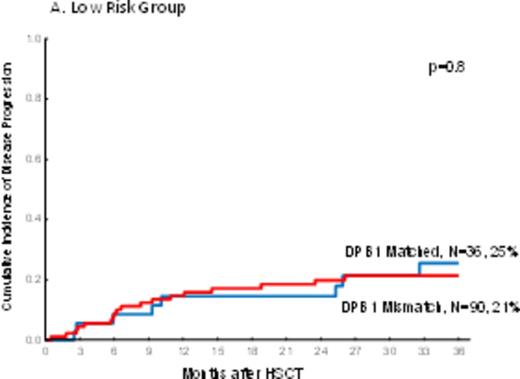
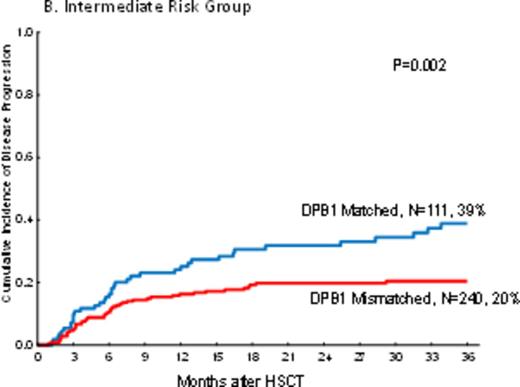
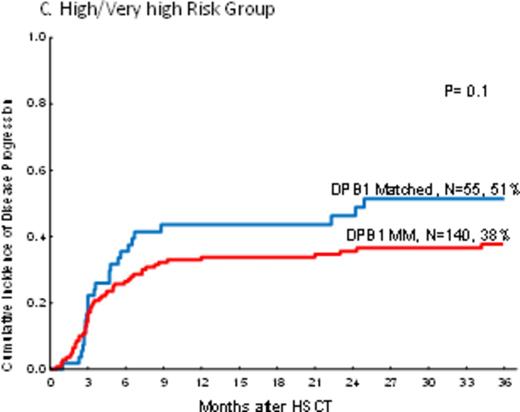
In PB recipients, there was a highly significant decreaseof disease progression in DPB1 mismatched pairs (one and two allele; HR=0.7, p=0.01 and HR=0.6, p=0.01 respectively) as compared tothose pairs with DPB1 matched. The impact of mismatches at one or two alleles were not different on disease progression (HR=1.2, p=0.4). However, the impact of DPB1 mismatch on disease progression was not uniform in different disease risk groups by DRI. Mismatch at DPB1 significantly decreased disease progression only in the intermediate risk group (HR=0.5, p=0.002) but not in low risk and high/very high disease groups by DRI (HR=0.9, p=0.8 and HR=0.7, p=0.1 respectively) (Figure 1a-c).
In BM recipients, increasing number of DPB1 incompatibilities decreased disease progression (HR=0.9, p=0.4 and HR=0.6, p=0.1 for 1 and 2 allele mismatches respectively) but did not reach significance. Mismatches at HLA-DQB1 and -DRB3/4/5 had no impact on disease progression in both PB and BM recipients.
Pairs with one or two allele-level DPB1 mismatches increased TRM compared with DPB1 matched pairs in PB (HR=1.5, p=0.04 and HR=1.9, p=0.006 respectively) and BM recipients (HR=1.8, p=0.03 and HR=1.9, p=0.05). There was no difference between two and one allele DPB1 mismatched for TRM in PB and BM recipients. Multivariate analyses revealed that the negative impact of DPB1 mismatch on TRM was not uniform in younger or (?) older patients. Interestingly, DPB1 mismatches increased TRM only in younger (aged<55) patients (HR=2.3, p=0.02) if they were PB recipients but only in older patients (HR=2.03, p=0.046) if they were BM recipients.
We next analyzed the impact of DPB1 matching on progression free survival (PFS) and did not observe any impact of DPB1 mismatches on PFS in PB (HR=0.9, p=0.9) and BM (HR=1.12, p=0.6) recipients. Subgroup analyses by DRI to identify a specific risk group that the use of HLA-A, -B, -C and -DRB1 matched but DPB1 mismatched unrelated donor might lead to improved PFS did not reveal any particular risk group in both PB and BM recipients.
Thus, in recipients of HLA-A, -B-C and DRB1 allele-level matched unrelated donors a mismatch for DPB1 is associated with a significantlydecreased risk of disease progression with no impact on PFS in intermediate risk group by DRI. Further analysis permissive vs. non-permissive DPB1 mismatches would be warranted.
The cumulative incidence of disease progression by DPB1 mismatch and Disease Risk Index in peripheral blood recipients.
The cumulative incidence of disease progression by DPB1 mismatch and Disease Risk Index in peripheral blood recipients.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.