Abstract
Introduction
Induction chemotherapy results in complete remission in 80% of children with acute myeloid leukemia (AML). However, many patients either fail to achieve a remission, or relapse after an initial response and subsequently die of their disease. Although large numbers of somatic karyotypic and molecular alterations have been identified, the majority of them do not indicate a specific target or distinct pathway that can be readily exploited for therapeutic intervention.
Materials & Methods
As part of a genome-scale approach to identify prognostic markers and therapeutic targets, we provide a comprehensive characterization of the pediatric AML transcriptome, detailing miRNA & mRNA expression patterns and miRNA:mRNA interactions that are characteristic of the disease. A total of 676 patients were considered for this study. Our discovery cohort consisted of miRNA-seq from 259 primary, 22 refractory and 38 relapse samples, and mRNA-seq from 158 primary, 12 refractory and 47 relapse samples. We confirmed our survival analyses on a validation cohort that consisted of miRNA-seq and mRNA-seq from 378 and 87 primary samples, respectively.
Unsupervised non-negative matrix factorization (NMF) was used to identify patient subgroups based on miRNA/mRNA expression. To identify miRNA/mRNA expression associated with patient survival, Cox proportional hazards analysis was performed. Wilcoxon tests were performed to identify differentially expressed miRNAs/mRNAs between samples. To screen for functional miRNA:mRNA interactions, we identified miRNA and mRNA pairs with anti-correlated expression profiles and miRNA binding site predictions consistent with miRNA:mRNA interaction.
Results
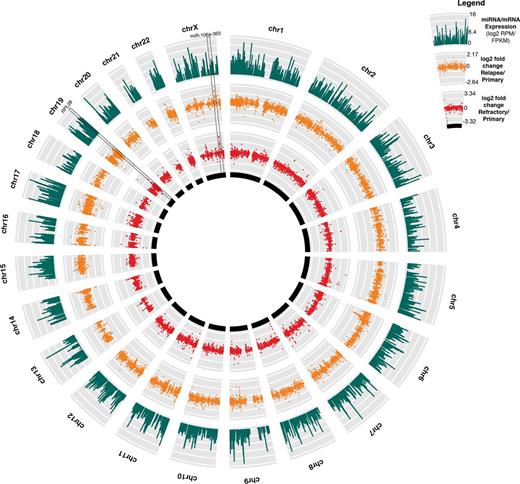
Survival analysis of both the discovery and validation cohorts revealed that 6 miRNAs were associated with overall survival (OS) and event free survival (EFS) (p-val<0.05, q-val<0.1): miR-181c-3p and miR-378c were associated with superior OS and EFS (Hazard Ratio (HR): 0.79-0.88), while miR-106a-3p, miR-106a-5p, miR-363-3p and miR-20b-5p were associated with inferior OS and EFS (HR: 1.14-1.36). All 4 of the miRNAs that were associated with inferior survival are members of the polycistronic miR-106a-363 cluster. Differential expression analysis revealed that miR-106a-363 was abundantly expressed in relapse and refractory samples and in primary samples of refractory patients (q-val<0.05). Integrative miRNA:mRNA expression analysis and luciferase reporter assays further revealed that targets of miR-106a-5p include NDUFC2, NDUFA10, UQCRB, ATP5J2-PTCD1 and ATP5S. Interestingly, these genes are involved in oxidative phosphorylation, a process that is suppressed in treatment-resistant leukemic cells[1].
NMF clustering of miRNA expression profiles revealed 2 groups of patients, with each group characterized by particular genomic alterations: Group 1 cases were enriched for NPM1 mutation and FLT3 -ITD, while Group 2 cases were enriched for t(8;21), inv(16), MLL rearrangements and CEBPA mutation (Fisher's exact test p-val<0.05).
NMF clustering of mRNA expression revealed 5 groups of patients, in which the group with abundant expression of ribosomal genes was further distinguished by superior OS and EFS (log-rank p-val<0.05). Analysis of the mRNA data showed a decrease in expression of one mRNA isoform of ribosomal protein L28 (RPL28) in relapse samples (q-val<0.05). In addition, survival analyses revealed that abundant expression of ribosomal protein L10 (RPL10) is associated with superior OS in both the discovery and validation cohorts (p-val<0.05, q-val<0.1, HR: 0.78 & 0.47).
Conclusions
Through a detailed analysis of the transcriptome (Figure A), we identified miRNAs whose expression levels were significantly associated with clinical outcome. In addition, we showed that abundant expression of miR-106a-363 might contribute to treatment resistance by modulating genes involved in energy metabolism. We also demonstrated that reduced expression of ribosomal genes is associated with inferior outcomes, suggesting a dysregulation of protein translation in treatment resistance. Overall, our transcriptome profiles provide clinically meaningful data for risk and response identification and define novel pathways that may be amenable to therapeutic targeting.
Summary of the pediatric AML transcriptome
Lagadinou ED, et al. Cell Stem Cell. 2013.
Summary of the pediatric AML transcriptome
Lagadinou ED, et al. Cell Stem Cell. 2013.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.