Key Points
More than 75% of primary diagnostic BCP-ALL samples engraft in the CNS in xenograft models.
We find no evidence for selective trafficking to the CNS but show that CNS entry is a generic property of BCP-ALL cells.
Abstract
Prevention of central nervous system (CNS) relapse is critical for cure of childhood B-cell precursor acute lymphoblastic leukemia (BCP-ALL). Despite this, mechanisms of CNS infiltration are poorly understood, and the timing, frequency, and properties of BCP-ALL blasts entering the CNS compartment are unknown. We investigated the CNS-engrafting potential of BCP-ALL cells xenotransplanted into immunodeficient NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ mice. CNS engraftment was seen in 23 of 29 diagnostic samples (79%): 2 of 2 from patients with overt CNS disease and 21 of 27 from patients thought to be CNS negative by diagnostic lumbar puncture. Histologic findings mimic human pathology and demonstrate that leukemic cells transit the blood–cerebrospinal fluid barrier situated close to the dural sinuses, the site of recently discovered CNS lymphatics. Retrieval of blasts from the CNS showed no evidence for chemokine receptor–mediated selective trafficking. The high frequency of infiltration and lack of selective trafficking led us to postulate that CNS tropism is a generic property of leukemic cells. To test this, we performed serial dilution experiments which showed CNS engraftment in 5 of 6 mice after transplant of as few as 10 leukemic cells. Clonal tracking techniques confirmed the polyclonal nature of CNS-infiltrating cells, with multiple clones engrafting in both the CNS and periphery. Overall, these findings suggest that subclinical seeding of the CNS is likely to be present in most BCP-ALL patients at original diagnosis, and efforts to prevent CNS relapse should concentrate on effective eradication of disease from this site rather than targeting entry mechanisms.
Introduction
One of the earliest advances in curative treatment of childhood acute lymphoblastic leukemia (ALL) came with the recognition that without central nervous system (CNS)-directed therapy, up to 75% of children relapse within the CNS.1 Introduction of universal CNS-directed treatment resulted in a dramatic reduction in overt CNS relapses. However, disease in the CNS still poses many clinical challenges.2 CNS-directed therapy is potentially toxic to the developing brain,3 and efforts to risk stratify and devise less-toxic therapy are hampered by a lack of knowledge regarding mechanisms of CNS disease and the absence of biomarkers predictive of CNS relapse.
CNS involvement is classified by the identification of lymphoblasts in cytospin preparations of cerebrospinal fluid (CSF): CNS-1 (CSF white cell count [WCC] <5/μL, no blasts), CNS-2 (WCC <5/μL, visible blasts), and CNS-3 (WCC >5/μL). It is important to appreciate that CNS-1 status does not equate with absence of leukemia in the CNS; early postmortem studies on children succumbing to leukemia frequently showed leptomeningeal involvement despite negative CSF cytology.4 Cytologic classification is insensitive5-7 and clearly inadequate for risk stratification because the majority of relapses occur in CNS-1 children.8,9 In addition, the CNS is one of the major sites of relapse in children with otherwise excellent prognosis as determined by low-risk bone marrow (BM) minimal residual disease measurements,10 suggesting that factors influencing leukemic kill in the periphery may not apply to the CNS. It is clear that a better understanding of CNS disease is required in order to develop rational risk-stratified treatment.
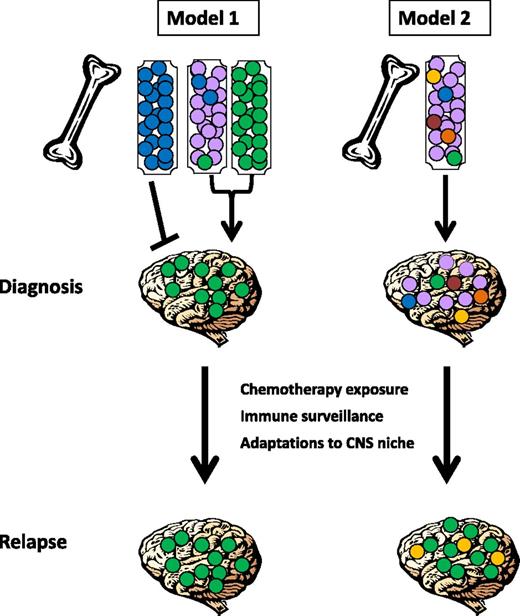
Two possible models for CNS relapse can be postulated (see Figure 1). First, it is possible that only some leukemic cells acquire the ability to enter the CNS, and the risk of CNS relapse depends on the presence or absence of a clone with the capacity to leave the BM and enter the CNS; this may occur at diagnosis or later during the disease course (model 1). Alternatively, all leukemic cells may have the ability to seed this compartment, and subclinical CNS involvement at diagnosis may be universal. In this case, CNS relapse is determined by whether cells can adapt to the foreign microenvironment of the CNS and evade elimination by ALL-directed therapy and/or immunologic surveillance (model 2). Distinguishing between these two models is critical in order to determine the optimal approaches for risk stratification and the development of biomarkers and novel therapeutics to prevent CNS relapse.
Schematic representation of proposed mechanisms underlying CNS infiltration and subsequent relapse. In model 1, only some leukemic cells acquire the ability to enter the CNS, and the risk of CNS relapse depends on the presence or absence of a clone (shown in green) with the capacity to leave the BM and enter the CNS. Different leukemia subtypes may vary in this capacity, with some (shown as blue) unable to enter the CNS compartment, others avidly trafficking to the CNS (shown as green), and some acquiring this capacity in rare subclones (shown as mixed purple, blue, and green). In model 2, all leukemic cells may have the ability to seed this compartment, and subclinical CNS involvement at diagnosis may be universal and show little or no subclonal selection. In both cases, CNS relapse may also be determined by whether cells can adapt to the foreign microenvironment of the CNS and evade elimination by ALL-directed therapy and/or immunologic surveillance (in this example, green and/or yellow subclones have been selected for at relapse).
Schematic representation of proposed mechanisms underlying CNS infiltration and subsequent relapse. In model 1, only some leukemic cells acquire the ability to enter the CNS, and the risk of CNS relapse depends on the presence or absence of a clone (shown in green) with the capacity to leave the BM and enter the CNS. Different leukemia subtypes may vary in this capacity, with some (shown as blue) unable to enter the CNS compartment, others avidly trafficking to the CNS (shown as green), and some acquiring this capacity in rare subclones (shown as mixed purple, blue, and green). In model 2, all leukemic cells may have the ability to seed this compartment, and subclinical CNS involvement at diagnosis may be universal and show little or no subclonal selection. In both cases, CNS relapse may also be determined by whether cells can adapt to the foreign microenvironment of the CNS and evade elimination by ALL-directed therapy and/or immunologic surveillance (in this example, green and/or yellow subclones have been selected for at relapse).
Here, we describe experiments that test these alternative models by addressing the qualitative question of whether every leukemic blast and/or every individual patient/subtype of leukemia has the intrinsic capacity to enter the CNS. We demonstrate that primary B-cell precursor (BCP)-ALL blasts, even from low-risk CNS-1 patients, frequently infiltrate the CNS in xenograft models by transiting the blood-CSF barrier. We find no evidence for selective trafficking of subclones to the CNS but show that CNS infiltration is a generic and ubiquitous property of BCP-ALL cells. These findings support the current dogma that all children require CNS-directed therapy and suggest that novel therapies to reduce the risk of CNS relapse and/or to provide safer and less-toxic CNS-directed therapy should concentrate on effective eradication of cells from this site rather than targeting selected entry mechanisms.
Materials and methods
Cell culture and primary cells
SD1 and REH (DSMZ, Braunschweig, Germany), as well as Sup B15 (American Type Culture Collection, LGC Standards, Middlesex, United Kingdom) cell lines were grown at 37°C in 5% carbon dioxide in complete RPMI 1640, 10% fetal bovine serum, and 1% penicillin/streptomycin (Invitrogen, Paisley, United Kingdom). Human primary meningeal cells (catalog #1400) and choroid plexus epithelial cells (catalog #1310) were obtained from ScienCell Research Laboratories (Carlsbad, CA) and cultured according to supplier’s instructions.
After informed consent, diagnostic BM samples from children with BCP-ALL underwent mononuclear cell enrichment using density-gradient centrifugation (Ficoll-Paque; GE Healthcare, Amersham, United Kingdom), cryopreservation in 10% dimethylsulfoxide/90% fetal bovine serum, and storage in liquid nitrogen until use. Samples originated from local institutions and the Leukaemia & Lymphoma Research Childhood Leukaemia Cell Bank. Patient details are listed in Table 1, as well as in supplemental Table 1, available on the Blood Web site. The use of human samples was approved by the West of Scotland Research Ethics Committee. ScienCell Research Laboratories primary tissues were obtained after informed consent (www.sciencellonline.com/site/ethics.php).
Xenotransplants
JAX NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG; Charles River Laboratories, Harlow, United Kingdom) or NOD.Cg-PrkdcscidIL2rgtm1Sug/JicTac (Taconic Biosciences, Ry, Denmark) mice were kept in sterile isolators with autoclaved food, bedding, and water. Xenotransplant was performed at 6 to 10 weeks of age using IV (tail vein) or intrafemoral injections of up to 1 × 107 leukemic cells, as previously described.11 Supplemental Tables 2 and 3 give details of individual experiments. Methods for serial dilution and sorting of leukemic subpopulations have been published.11 Mice were euthanized after they became unwell, had clinical evidence of leukemia, or had significant weight loss. All animal experiments were approved by Institutional Ethical Review Process Committees and were performed under UK Home Office licenses.
Histology
Murine heads were stripped of soft tissues, fixed in 10% neutral buffered formalin (CellPath, Powys, United Kingdom), and decalcified in Hilleman and Lee EDTA solution (5.5% EDTA in 10% formalin) for 2 to 3 weeks. Samples were processed as described previously.12
Anti-CD45 immunohistochemistry on paraffin-embedded sections was performed as previously described.13
Imaging used Axiostar Plus or Axio Imager M2 microscopes with AxioVision and ZEN software (Carl Zeiss, Cambridge, United Kingdom).
Cell retrieval
After terminal carbon dioxide asphyxiation, mice were perfused with phosphate-buffered saline (PBS) to eliminate peripheral blood contamination. Pilot experiments were performed comparing retrieval of leukemic cells from the meninges alone vs performing whole-brain extracts of meninges and parenchyma. The former produced excellent yields of pure leukemic cells, whereas the latter did not add to the yield and substantially reduced the viability and purity of the retrieved cells. Therefore, all experiments used direct retrieval of leukemic cells from the leptomeninges by gentle scraping of the skull vault and by vortex mixing the intact brain in PBS for 5 minutes. Femoral BM cells were retrieved by flushing with PBS. Spleen samples were collected by homogenizing material through a cell strainer with PBS.
Quantitative polymerase chain reaction (PCR)
RNA extraction, on-column DNase digestion, complementary DNA synthesis, and custom-designed TaqMan low-density arrays were performed as described previously.14 Two reference endogenous control genes were included on the plate: TATA binding protein and 18sRNA. Gene assay identification numbers are given in supplemental Table 4. Both reference genes were validated according to “Minimum Information for Publication of Quantitative Real-Time PCR Experiments” guidelines.15 Data were analyzed using the ΔΔCT method using RQ Manager 1.2.4 software (Applied Biosystems, Paisley, United Kingdom). For gene expression arrays, arbitrary expression values were derived from the CT value as described previously.16 Gene expression assay identification numbers are given in supplemental Table 4.
Flow cytometry
Cells were washed twice in 0.5% fetal calf serum plus 0.5 mM EDTA in PBS, incubated with anti-human Fc-receptor binding inhibitor (eBioscience, Hatfield, United Kingdom), and then with directly conjugated antibodies (supplemental Table 5). Viability staining used Via-Probe (BD Biosciences, Oxford, United Kingdom) or DRAQ7 (Biostatus, Shepshed, United Kingdom). Data were acquired on a MACSQuant flow cytometer (Miltenyi Biotec, Bergisch Gladbach, Germany) and analyzed using FlowJo 7.2.4 software (Tree Star, Ashland, OR).
Clonal tracking
Primograft ALL blasts were lentivirally transduced and transplanted intrafemorally into NSG mice as described previously.17 Genomic DNA was extracted from splenic and leptomeningeal leukemic blasts using a DNeasy kit (Qiagen, Hilden, Germany). Analysis of lentiviral integration sites using nonrestriction-based linear amplification–mediated PCR was described previously.18 The linear PCR step was performed using a biotinylated primer (Btn-GCACTGACAATTCCGTGGTGTTGTC) for 99 cycles (98°C for 10 seconds, 64°C for 45 seconds, and 72°C for 15 seconds). The final amplification used 2 successive 30-cycle PCR reactions (98°C for 10 seconds, 62°C for 30 seconds, and 72°C for 2 minutes) with the following primers: round 1, forward GACCCGGGAGATCTGAATTC and reverse GCTACGTAACTCCCAACGAAG; and round 2, forward AGTGGCACAGCAGTTAGG and reverse GTGTGGAAAATCTCTAGCA. Illumina adaptors were added by PCR, and samples were sequenced using an Illumina MiSeq. Reads were considered valid if they perfectly matched the expected vector flanking sequence. Sequences composing <0.1% of the total were removed, and reads with truncations or mismatches within the remaining sequences were merged to create the final list.
Statistics
Student t tests were used to analyze 2 parametric groups, and χ2 tests were used to examine frequency of CNS infiltration. A P value ≤.05 was considered significant. All analyses were performed using GraphPad Prism (La Jolla, CA).
Results
CNS engraftment in NSG mice is common and involves passage across the blood-CSF barrier
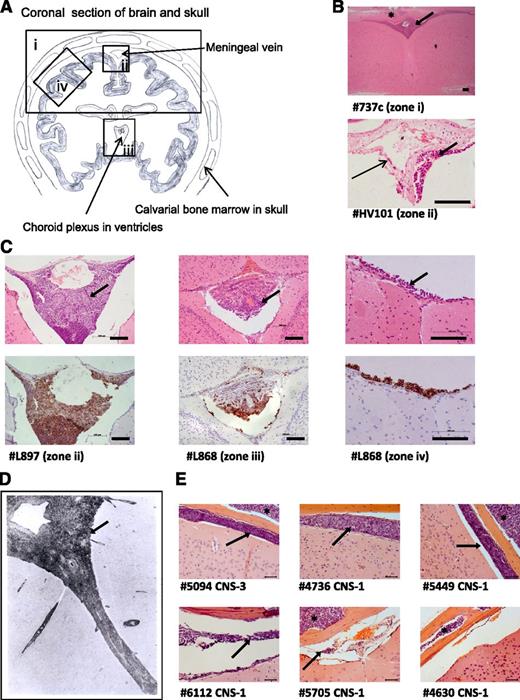
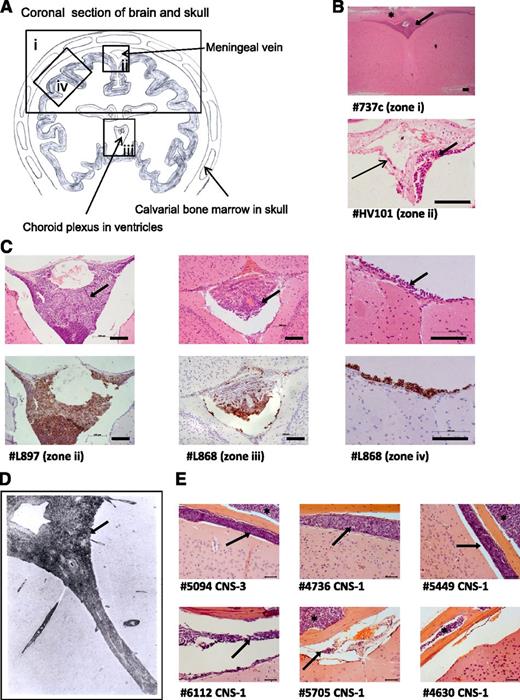
To investigate the frequency and distribution of CNS engraftment, brains were examined from NSG mice xenografted with 29 different ALL samples (diagnostic BM samples and primary cells previously passaged through mice [primografts]; see supplemental Tables 1-3 for clinical and experimental details). CNS engraftment was observed in 23 of 29 patient samples (79%) across 13 different cytogenetic subtypes (Table 1). There was no difference in the frequency of CNS engraftment in primary BM samples vs primografts (P = .303, χ2 test). CNS engraftment was seen in samples with both high- and low-risk clinical features (Table 2), and semiquantitative scoring of the degree of CNS infiltration did not show clear differences between high- and low-risk samples (supplemental Table 2). Histopathology was consistent, showing early infiltration around the dural venous sinuses, plaques of disease in the leptomeninges, relative sparing of the ventricles, and absence of gross parenchymal involvement (Figure 2A-C). This histology suggests that ALL cells primarily transit the blood-CSF barrier (comprising the choroid plexus and the meningeal postcapillary venules) rather than the blood-brain barrier. Importantly, this histopathology closely resembles that observed in patients (Figure 2D).4,19
Histologic analysis of brains from xenografted mice. (A) Line drawing of a coronal section of murine brain showing approximate locations (zones i-iv) of images shown in panels B-D. (B) Photomicrographs of hematoxylin and eosin (H&E)-stained brain sections. Top: low-power view of cerebral cortex and leptomeninges (original magnification ×5); bottom: close-up of a central meningeal vessel (original magnification ×40). Long thin arrow indicates leukemic cells in the vessel wall. Bar represents 100 μm. (C) H&E (top) and corresponding anti-human CD45 (bottom) staining of leukemic deposits. Left: Cells surrounding the dural venous sinus (original magnification ×20); center: cells within choroid plexus (original magnification ×20); right: high-power view of meninges (original magnification ×40). Bar represents 100 μm. (D) Postmortem image of grade 2 arachnoid leukemia (zone ii) in a child with ALL (H&E, original magnification ×33), reproduced with permission from Price and Johnson.4 (E) Representative coronal sections (all zone iv) from mice engrafted with a CNS-3 sample and 5 matched CNS-1 controls (H&E, original magnification ×40). Bar represents 50 μm. Thick arrows (panels B-E) mark the leukemic infiltrate within the leptomeninges and asterisks (panels B and E) mark leukemic infiltrates within the calvarial BM cavity. Numbers preceded by the # symbol (panels B, C, and E) denote the sample identifiers in Table 1.
Histologic analysis of brains from xenografted mice. (A) Line drawing of a coronal section of murine brain showing approximate locations (zones i-iv) of images shown in panels B-D. (B) Photomicrographs of hematoxylin and eosin (H&E)-stained brain sections. Top: low-power view of cerebral cortex and leptomeninges (original magnification ×5); bottom: close-up of a central meningeal vessel (original magnification ×40). Long thin arrow indicates leukemic cells in the vessel wall. Bar represents 100 μm. (C) H&E (top) and corresponding anti-human CD45 (bottom) staining of leukemic deposits. Left: Cells surrounding the dural venous sinus (original magnification ×20); center: cells within choroid plexus (original magnification ×20); right: high-power view of meninges (original magnification ×40). Bar represents 100 μm. (D) Postmortem image of grade 2 arachnoid leukemia (zone ii) in a child with ALL (H&E, original magnification ×33), reproduced with permission from Price and Johnson.4 (E) Representative coronal sections (all zone iv) from mice engrafted with a CNS-3 sample and 5 matched CNS-1 controls (H&E, original magnification ×40). Bar represents 50 μm. Thick arrows (panels B-E) mark the leukemic infiltrate within the leptomeninges and asterisks (panels B and E) mark leukemic infiltrates within the calvarial BM cavity. Numbers preceded by the # symbol (panels B, C, and E) denote the sample identifiers in Table 1.
Our original panel of primografts and primary samples comprised mainly CNS-1 patients (Table 1). Therefore, CNS-engrafting capacity appears to be more prevalent than suggested by CSF-cytospin status. However, because primograft samples have a priori shown successful engraftment in mice, there may have been a selection bias for aggressive leukemias. To address this, we prospectively investigated CNS engraftment of BM-derived leukemic cells from a CNS-3 patient (#5094) and 5 matched CNS-1 controls (#4736, #5449, #6112, #5705, and #4630). These samples had unknown xenografting capability and all carried the good-prognosis translocation t(12;21). All samples engrafted in the BM, whereas CNS infiltration was seen in the CNS-3 sample in addition to 4 of 5 CNS-1 samples (Figure 2E).
Together, these observations indicate that the majority of diagnostic BCP-ALL BM samples contain cells capable of entering the CNS compartment, irrespective of initial CSF-cytospin findings.
Chemokine receptors do not drive CNS entry in BCP-ALL
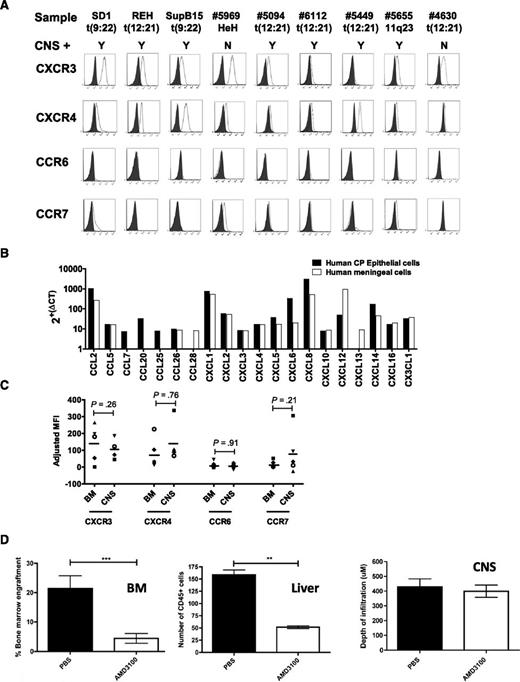
In a murine model of T-cell ALL (T-ALL), expression of the chemokine receptor CCR7 determines CNS engraftment.20 In addition, a recent report has highlighted a possible association between CXCR3 expression and ALL migration to the CNS.21 Therefore, we investigated the role of chemokine receptors in directing leukemic cells to the CNS compartment in our model. As shown in Figure 3 and supplemental Figures 1 and 2, only CXCR3 and CXCR4 were consistently expressed on the cell surface, with variable expression of CCR7 and CCR6 (Figure 3A) and CXCR7 (supplemental Figure 2). Comparison of chemokine receptor expression in 2 non-CNS-homing primary samples (#4630 and #5969) showed the same pattern of chemokine receptor expression as 7 CNS homing primary samples (Figure 3A; supplemental Table 6). Therefore, there was no apparent chemokine receptor expression signature that marked the ability to enter the CNS compartment. Next, we examined the repertoire of chemokine ligands expressed by human blood-CSF barrier tissues. Of importance, the CXCR3 ligand CXCL10, the CXCR4/CXCR7 ligand CXCL12, and the CCR6 ligand CCL20 were detected (Figure 3B), suggesting that these pairings could be functionally important in ALL transit across this barrier. To investigate whether cells expressing high levels of any particular chemokine receptor were being positively selected for in the CNS compartment in vivo, we compared expression profiles of leukemic cells retrieved from the meninges and BM of engrafted mice. As seen in Figure 3C, there was no evidence of positive selection for high-expressing subclones in the CNS, with leukemic blasts showing similar chemokine receptor expression profiles at the 2 sites. Lastly, given the importance of the chemokine receptor CXCR4 in BM engraftment, we went on to specifically interrogate the role of CXCR4 in CNS engraftment by utilizing the CXCR4 inhibitor AMD3100 in vivo. Interestingly, mice in the AMD3100-treated group showed a significant reduction in leukemic burden in the liver and BM but no reduction of CNS disease (Figure 3D; supplemental Figures 3 and 4).
Chemokine receptors and CNS engraftment. (A) Flow cytometry for chemokine receptor expression in BCP-ALL. Shaded histogram represents isotype control; open histogram represents specific staining. Sample names and associated translocations are indicated. CNS+ denotes CNS engraftment, with the results of xenografting this sample into mice categorized as Y (evidence of CNS engraftment) or N (no evidence of CNS engraftment). (B) Quantitative PCR for chemokine ligand expression by cultured human primary meningeal cells (white bars) and choroid plexus (CP) epithelial cells (black bars) (both passage 3). Arbitrary expression values were derived from ΔCT. (C) Primary ALL cells from 1 CNS-3 patient (open symbols) and 4 CNS-1 matched controls (closed symbols) were retrieved from BM and meninges of xenografted mice and analyzed by flow cytometry. Contaminating murine cells were excluded by gating on human CD45. A total of 105 events were analyzed when possible. Data represent adjusted mean fluorescence intensity (MFIspecific − MFIisotype) of live leukemic cells (huCD45+/Draq7−). Bars represent means of adjusted MFIs. Differences between CNS and BM expression were analyzed using 2-tailed paired Student t tests. (D) Leukemic infiltration in NSG mice engrafted with cells expressing REH–luciferase–green fluorescent protein (GFP) and treated with the CXCR4 inhibitor AMD3100 or PBS control. Left: Leukemic engraftment in the BM as measured by numbers of GFP-positive cells (REH-luciferase-GFP) on flow cytometry. Data show mean ± standard error of the mean (SEM) in n = 7 and n = 6 mice for PBS and AMD3100 groups, respectively, analyzed by an unpaired Student t test, ***P < .001. Center: Liver infiltration quantified by counting human-CD45+ cells (brown diaminobenzidine-stained cells) in 8 random fields of view per section. Data show mean ± SEM for n = 4 and n = 5 mice in PBS and AMD3100 groups, respectively, analyzed by an unpaired Student t test, **P < .01. Right: Histologic analysis of murine brains from xenografts (n = 5 mice in each group). Each brain was divided into 5 segments, sections were cut from each segment, and the maximal depth of meningeal infiltrates was recorded for each section using AxioVision Rel 4.3 software (Carl Zeiss). Data show mean ± SEM. Representative histology and bioluminescent in vivo imaging from these mice, along with full experimental details are provided in supplemental Figure 3.
Chemokine receptors and CNS engraftment. (A) Flow cytometry for chemokine receptor expression in BCP-ALL. Shaded histogram represents isotype control; open histogram represents specific staining. Sample names and associated translocations are indicated. CNS+ denotes CNS engraftment, with the results of xenografting this sample into mice categorized as Y (evidence of CNS engraftment) or N (no evidence of CNS engraftment). (B) Quantitative PCR for chemokine ligand expression by cultured human primary meningeal cells (white bars) and choroid plexus (CP) epithelial cells (black bars) (both passage 3). Arbitrary expression values were derived from ΔCT. (C) Primary ALL cells from 1 CNS-3 patient (open symbols) and 4 CNS-1 matched controls (closed symbols) were retrieved from BM and meninges of xenografted mice and analyzed by flow cytometry. Contaminating murine cells were excluded by gating on human CD45. A total of 105 events were analyzed when possible. Data represent adjusted mean fluorescence intensity (MFIspecific − MFIisotype) of live leukemic cells (huCD45+/Draq7−). Bars represent means of adjusted MFIs. Differences between CNS and BM expression were analyzed using 2-tailed paired Student t tests. (D) Leukemic infiltration in NSG mice engrafted with cells expressing REH–luciferase–green fluorescent protein (GFP) and treated with the CXCR4 inhibitor AMD3100 or PBS control. Left: Leukemic engraftment in the BM as measured by numbers of GFP-positive cells (REH-luciferase-GFP) on flow cytometry. Data show mean ± standard error of the mean (SEM) in n = 7 and n = 6 mice for PBS and AMD3100 groups, respectively, analyzed by an unpaired Student t test, ***P < .001. Center: Liver infiltration quantified by counting human-CD45+ cells (brown diaminobenzidine-stained cells) in 8 random fields of view per section. Data show mean ± SEM for n = 4 and n = 5 mice in PBS and AMD3100 groups, respectively, analyzed by an unpaired Student t test, **P < .01. Right: Histologic analysis of murine brains from xenografts (n = 5 mice in each group). Each brain was divided into 5 segments, sections were cut from each segment, and the maximal depth of meningeal infiltrates was recorded for each section using AxioVision Rel 4.3 software (Carl Zeiss). Data show mean ± SEM. Representative histology and bioluminescent in vivo imaging from these mice, along with full experimental details are provided in supplemental Figure 3.
Therefore, although chemokine receptors may play a permissive role in ALL transit across the blood-CSF barrier, single members do not appear to play an instructive role in determining localization of leukemic blasts in the CNS compartment.
CNS leukemia-initiating cells are frequent and not related to blast maturation status
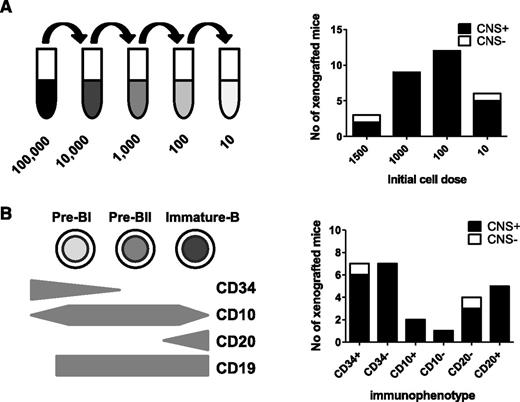
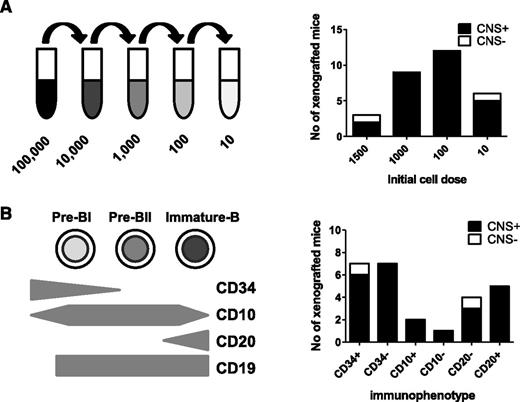
Overall, the evidence of frequent CNS involvement and lack of chemokine receptor–mediated selective trafficking to the CNS suggested to us that CNS-engrafting capability may be a generic property of BCP-ALL lymphoblasts rather than an acquired property of a rare subclone. To test this, we examined the frequency of cells within an individual leukemia sample capable of CNS engraftment. Using a cohort of BCR-ABL and BCR-ABL-like (defined as an activated kinase gene expression profile that clusters with BCR-ABL on microarray but without a classical Philadelphia chromosome22 ) samples, cell suspensions were prepared containing 10, 100, 1000, or 1500 cells for intrafemoral transplant (Figure 4A). The results of BM engraftment in this cohort of mice are already published.11 Figure 4A shows no relationship between cell number and likelihood of CNS engraftment, and as few as 10 cells produced CNS disease in 5 of 6 cases (Figure 4A; supplemental Table 3).
CNS engraftment of sorted subpopulations of leukemic blasts. Sorted and unsorted leukemic blasts from 6 primografts (#4540, #M120, #WB51, #HV101, #737c, and #758b) were injected intrafemorally, at limiting dilutions, into the femurs of 1 to 4 mice each. CNS involvement was assessed histologically after transplant of 10 to 1500 cells (A) or different immunophenotypic subpopulations of leukemic blasts (B). Black bars represent the number of mice in each experimental group with evidence of CNS involvement (CNS+) on histology. White bars represent number of mice without any visible CNS infiltration (CNS−). Individual results of mice injected with each cell number/immunophenotypic subpopulation for the different primografts are given in supplemental Table 3.
CNS engraftment of sorted subpopulations of leukemic blasts. Sorted and unsorted leukemic blasts from 6 primografts (#4540, #M120, #WB51, #HV101, #737c, and #758b) were injected intrafemorally, at limiting dilutions, into the femurs of 1 to 4 mice each. CNS involvement was assessed histologically after transplant of 10 to 1500 cells (A) or different immunophenotypic subpopulations of leukemic blasts (B). Black bars represent the number of mice in each experimental group with evidence of CNS involvement (CNS+) on histology. White bars represent number of mice without any visible CNS infiltration (CNS−). Individual results of mice injected with each cell number/immunophenotypic subpopulation for the different primografts are given in supplemental Table 3.
Because the ability to egress from the BM to the periphery is acquired during normal B-cell development, we next investigated whether CNS engraftment rates differed in mature vs immature ALL subpopulations. Leukemia-propagating ability in childhood ALL does not appear to follow a hierarchical stem cell model,11,23 and indeed, a stem cell–like transcriptional signature is seen in both CD34+ and CD34− ALL blasts.11 However, downregulation of CD34 expression in ALL is associated with increased expression of B-cell differentiation genes,23 and diagnostic BCP-ALL samples contain blasts at different maturation stages.11 B-cell precursors were identified by gating on CD19 and then flow sorted into CD34high (immature), CD34low (more mature), CD10low (immature), CD10high (more mature), CD20low (immature), and CD20high (more mature) populations. Sorted cells underwent limiting dilution, and 10 to 1500 cells were injected intrafemorally (supplemental Table 3). As seen in Figure 4B, all fractions showed CNS-engrafting capability, and both immature and more-mature B-cell subpopulations appeared to engraft in the CNS with equal competence.
These experiments support our hypothesis that the ability to enter the CNS is not restricted to rare subclones but is instead a generic property of the majority of leukemic cells present at original diagnosis.
Clonal tracking experiments identify very similar clonal composition in CNS and periphery
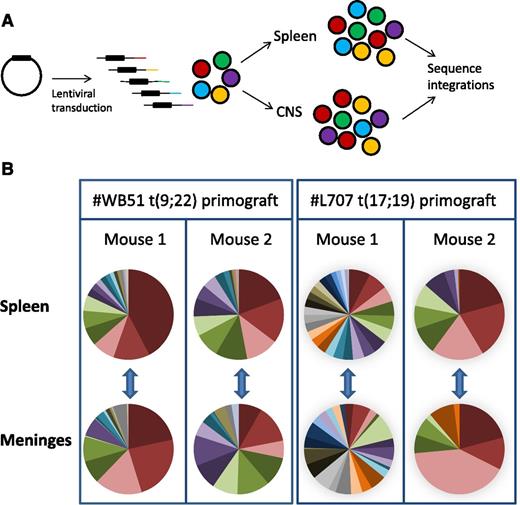
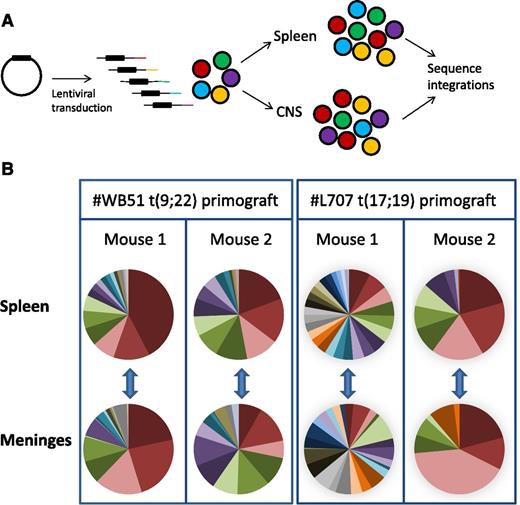
To lend further support to our hypothesis, we performed clonal tracking of lentivirally marked #WB51 primograft cells, which carry the Philadelphia chromosome, and #L707 cells, which have a t(17;19) translocation. This clonal tracking allowed us to investigate both the clonal architecture of leukemic subpopulations in the CNS and their relationship to cells in the periphery. We used a modified linear amplification–mediated PCR protocol18 to detect lentiviral integration sites in samples from the spleen and leptomeninges. Each integration site is unique, so it can be used as a heritable marker to track the spread of individual clones (Figure 5A). Up to 10 000 cells were transplanted per mouse, with a maximum lentiviral transduction rate of 10% to limit the risk of multiple integrations in a single cell.24 Analysis of the most prevalent integrations in 2 mice per sample demonstrated that the CNS disease was polyclonal and very similar in clonal composition to the splenic disease (Figure 5B; supplemental Figure 5). All integrations present in the spleen at >0.5% of the total population were also detectable in the meninges, demonstrating that all major clones had CNS engrafting capability. In the case of #WB51, femoral leukemic cells were also analyzed, and again, all detectable clones in the femur were also present in the CNS (supplemental Figure 5).
Subclonal composition of CNS and splenic compartments. (A) Schematic of experimental design. (B) Clonal composition of CNS (leptomeninges) and splenic compartments of mice transplanted with primograft samples #WB51 and #L707. Pie charts show frequencies of most prevalent integrations in paired CNS and spleen samples from 2 mice per sample. Pie chart colors are unique to each mouse spleen-meninges pair and do not represent the same clones between different mice. Corresponding tables of integration site frequencies for each mouse are given in supplemental Figure 5.
Subclonal composition of CNS and splenic compartments. (A) Schematic of experimental design. (B) Clonal composition of CNS (leptomeninges) and splenic compartments of mice transplanted with primograft samples #WB51 and #L707. Pie charts show frequencies of most prevalent integrations in paired CNS and spleen samples from 2 mice per sample. Pie chart colors are unique to each mouse spleen-meninges pair and do not represent the same clones between different mice. Corresponding tables of integration site frequencies for each mouse are given in supplemental Figure 5.
Together, these experiments confirm our hypothesis that the ability to engraft in the CNS is a generic property of the bulk leukemic population at initial diagnosis rather than being due to acquisition of a metastatic phenotype by a rare subclone.
Discussion
We have shown that CNS involvement is detectable in more than three-fourths of xenografts from diagnostic primary BCP-ALL samples and is seen with equal frequency in samples from patients with high- and low-risk cytogenetic and clinical features. These observations support a model whereby CNS relapse is determined by whether cells can adapt to the foreign microenvironment of the CNS and evade elimination by ALL-directed therapy and/or immunologic surveillance (model 2) rather than whether individual clones present within a patient’s leukemia acquire the ability to enter the CNS compartment (model 1) (illustrated in Figure 1). The importance of this observation lies in the concept that CNS infiltration is likely to be present in the majority of patients at the time of diagnosis. Current cytologic classification may lead to the erroneous impression that CNS-1 patients do not have any leukemia in the CNS and, consequently, that these children are at very low risk of CNS relapse.
We acknowledge some limitations in the clinical interpretation of our work. Xenograft models are unlikely to completely faithfully recapitulate all the receptor-ligand interactions governing leukemic engraftment in humans, although most trafficking molecules are highly conserved25 and this limitation would be predicted to result in reduced rather than enhanced engraftment in xenografts. In addition, our limiting-dilution and clonal-tracking experiments were performed using high-risk leukemias, which may more readily engraft in mice.26-28 However, our original cohort of mice showed frequent CNS involvement in both high- and low-risk primary samples, suggesting that this is a universal property of BCP-ALL cells. Additional support for high rates of subclinical seeding of the CNS at the time of diagnosis comes from clinical observations. The use of more-sensitive detection methods such as flow cytometry7 and PCR5,6 is able to detect occult CNS involvement in up to 40% of patients. In addition, before the era of routine CNS “prophylaxis,” 50% to 75% of children relapsed in the CNS,1 usually within a couple of months of original diagnosis, suggesting that occult CNS leukemia was present from the outset. This rate of early CNS relapse in patients mirrors the rate of CNS infiltration in our xenograft model.
Postmortem histopathology from children with ALL demonstrates a pattern of CNS infiltration closely resembling our xenograft model.4 The earliest leukemic infiltrates appear in the walls of superficial arachnoid veins, with progressive infiltration of the leptomeninges and subsequent extension into the deep arachnoid following the course of penetrating vessels. Parenchymal infiltration is seen only in late-stage disease, always accompanied by a breach of the pia-glial membrane.4 Therefore our results, along with these historical data, provide evidence for ALL cells primarily transiting the blood-CSF barrier rather than the blood-brain barrier. This is important when considering cellular trafficking, microenvironmental influences, and drug pharmacokinetics because the leptomeninges and brain parenchyma are distinct physiological compartments.29 In addition, it highlights that commonly used in vitro models of the blood-brain barrier are inappropriate to study mechanisms of CNS entry of leukemic blasts. The close relationship of leukemic cells to the dural sinuses is particularly intriguing, given the recent description of lymphatic vessels in this area in mice.30 Of note, our histology and the human postmortem data4 also show ALL blasts adherent to meningeal stroma rather than free floating in the CSF. This may explain why sampling a small volume of CSF from the lumbar spine may significantly underestimate CNS infiltration.
We went on to examine whether chemokine receptors directed this migration across the blood-CSF barrier. In common with previous reports,21,31-33 we show that BCP-ALL cells express CXCR3 and CXCR4, with some samples expressing CCR7, CCR6, and CXCR7. Corresponding chemokine ligands were expressed by blood-CSF barrier tissues. However, there was no evidence for positive selection of cells bearing these receptors within the CNS compartment. Previous work has shown that CXCR3 inhibitors cause a global reduction in leukemic engraftment in BM, spleen, and CNS.21 In contrast, we show that treatment with the CXCR4 inhibitor AMD3100 leads to a reduced disease burden in the liver and BM but no reduction in CNS infiltration, suggesting that CNS engraftment may be a CXCR4-independent niche.
Our findings differ from results in T-ALL, in which a single chemokine receptor–ligand pairing (CCR7-CCL19) determined the ability of lymphoblasts to enter the CNS in a humanized murine model.20 Our findings may be due to differences in experimental models34 or to the intrinsic biology of T and B cells; CCR7-expressing memory T cells are the most abundant leukocyte to be found in normal CSF,35 and so it is perhaps not surprising that CCR7 expression enhances T-ALL entry into this site.
Lastly, we investigated the subclonal composition of the CNS compartment. We found that CNS-engrafting ability was a generic ability of leukemic cells and that the clonal composition of CNS and splenic engrafting cells was remarkably similar. These findings suggest that CNS-infiltrating ability is ubiquitous in BCP-ALL and strongly argues against our proposed model 1 and supports model 2, which is that CNS relapse originates from inadequate eradication of cells from this sanctuary site rather than from selective entry (Figure 1).
Of note, our studies do not exclude that individual cells/clones differ in their biological fitness to invade, proliferate, and survive in the CNS but do indicate that CNS invasion is a generic ability of the leukemic blasts. Differences in biological fitness may determine leukemic load in the CNS and thus explain why some children have visible disease on CSF cytology and others do not. These differences may also play a role in the likelihood of survival of cells in this compartment and the attendant risk of CNS relapse. Future efforts to investigate risk factors for CNS relapse should focus on advantageous leukemic adaptations to this new niche using approaches such as transcriptomics, metabolomics, and proteomics. We and others have reported on the potential role of the cytokine interleukin-15 in promoting leukemic cell survival in the hostile environment in the CSF.13,36 A recent report has highlighted a critical role for MER tyrosine kinase in promoting survival of t(1:19)-positive ALL in the CNS,37 and increased intercellular adhesion molecule 1,38 stearoyl-CoA desaturase, and osteopontin39 expression have also been associated with CNS disease. Additional mechanisms of CNS relapse may relate to evasion of chemotherapy40 or immune surveillance34,41 at this site.
Our findings have important implications for the design of risk-adapted CNS therapy. First, our studies indicate that the current dogma of CNS-directed therapy for all patients appears to have a rational scientific basis. Second, it is unlikely that chemokine receptor expression profiling will be a useful biomarker for CNS disease in BCP-ALL. Third, identifying factors that enable long-term survival of cells in the CNS (which may also enhance long-term survival in the BM) may be a better therapeutic strategy than attempts to block cell entry.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Leukaemia & Lymphoma Research Childhood Leukaemia Cell Bank, the ALL2003 trial coordinators, the Clinical Trial Services Unit at Oxford, and all contributing centers and patients; Lynn Stevenson, Alasdair Fraser, Saeeda Bhatti, Antony Cousins, and Michelle Le Brocq (University of Glasgow) for technical assistance; and Monique den Boer (Rotterdam) for the original bcr-abl-like samples.
This work was supported by the Kay Kendall Leukaemia Fund (KKL454, KKL515) (C.H. and L.J.R.), with additional funding from Cancer Research UK (C27943/A12788) (J.V. and O.H.), Leukaemia & Lymphoma Research (D.M., T.P., K.D., and P.K.), the European Research Council (P.S.), the William Lawrence and Blanche Hughes Foundation, the German Israel Foundation, and an Israeli Ministry of Health Chief Scientist European Research Area Network grant (S.I.), the Chief Scientist Office (SCD/08) and the Scottish Funding Council (C.H.), a Wellcome Trust Senior Investigator award (G.J.G.), and the Medical Research Council (G0901113, G0802259) (G.J.G. and S.B.).
Authorship
Contribution: C.H., J.V., O.H., S.I., and G.J.G. designed the research, analyzed the data, and wrote the paper; Y.M.Y., M.T.S.W., and A.E. performed the experiments and analyzed the data; K.R., S.T., L.F.-L., and S.B. performed the experiments; P.S., K.D., D.M., T.P., V.J.W., P.K., H.B., L.J.R., and J.A.E.I. performed the xenografting experiments and provided essential data. All authors contributed to the writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christina Halsey, Institute of Cancer Sciences, College of Medical, Veterinary, and Life Sciences, University of Glasgow, Glasgow G61 1QH, United Kingdom; e-mail: chris.halsey@glasgow.ac.uk.
References
Author notes
M.T.S.W., Y.M.Y., and A.E. contributed equally to this study.