Abstract
Even though two-thirds of acute myeloid leukemia (AML) patients respond to induction chemotherapy and achieve complete remission (CR), the majority of these patients will eventually relapse. The time from CR to relapse is an important clinical indicator of disease aggressiveness, as patients relapsing within the first 6 months after initial diagnosis have a poorer prognosis in terms of response to salvage therapy and overall survival compared to patients with a later relapse. To learn about the evolution during the course of disease, we analyzed the somatic mutation patterns from initial diagnosis to relapse in 50 cytogenetically normal (CN) AML patients. Based on the ELN classification, 38% of the patients (n=19) were assigned as "favorable" at diagnosis, all other patients were classified as "intermediate-I". ELN classification was associated with time to relapse as "intermediate-I" patients relapsed earlier than "favorable" patients (median 9.3 months vs. 16.1 months, p=0.008, log-rank test).
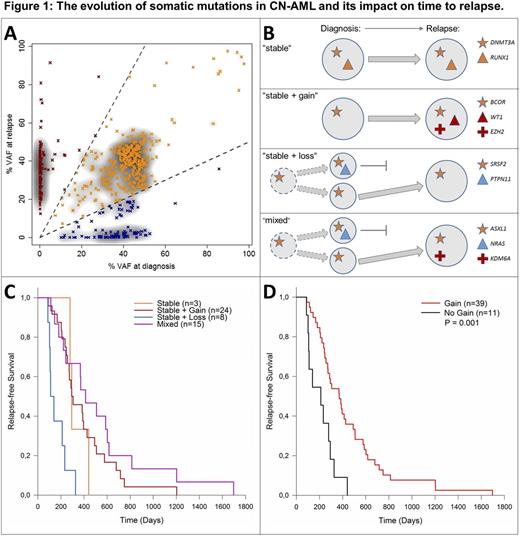
Somatic alterations were detected by exome sequencing and confirmed by targeted amplicon sequencing of matched diagnostic, remission and relapse samples. FLT3-ITD and NPM1 mutation status were obtained from routine diagnostic tests as the reliable detection of these markers by NGS remains challenging. The vast majority of somatic alterations were present both at diagnosis and at relapse, hereafter referred to as stable mutations (70%, Fig. 1A). All patients in our cohort had ≥1 stable mutation with DNMT3A being the most stably altered gene. In 47 out of 50 patients (94%), we observed mutations that were only found at diagnosis or only at relapse. Based on the mutation patterns, four distinct 'evolutionary' subgroups of patients were defined (Fig. 1B): (I) patients with an identical mutation profile at diagnosis and at relapse ("stable", n=3, 6%), (II) patients who gained mutations at relapse ("stable + gain", n=24, 48%), (III) patients that lost mutations at relapse ("stable + loss", n=8, 16%), and (IV) patients with both loss and gain of mutations at relapse ("mixed", n=15, 30%). Mutations that were lost during the course of the disease were detected in e.g. PTPN11 or NRAS. Relapse-specific mutations were identified in e.g. IDH1/2, WT1, KPNB1 or KDM6A. Evolutionary subgroups showed differences in time to relapse (Fig. 1C). Patients with "stable + loss" relapsed earlier (median 4.1 months) than patients with gain of mutation at relapse (groups "stable + gain" and "mixed", median 12.2 months). All patients in the category "stable + loss" developed relapse within the first year after complete remission. The "stable" group of 3 patients showed an intermediate time to relapse (median 9.6 months), but was too small for a statistically valid comparison. Ultimately, the genetic evolution of CN-AML patients without gain of new mutations at relapse (categories "stable" and "stable + loss") was associated with significantly earlier relapse compared to patients that gained mutations at relapse (categories "stable + gain" and "mixed", Fig. 1D, p=0.001, log-rank test). Distinct predominant patterns of clonal evolution were observed in the ELN genetic groups, as only one patient of the "stable + loss" group was initially classified as "favorable". Interestingly, applying the ELN classification on relapse samples revealed a switch from "favorable" to "intermediate-I" in six patients, all with gain of mutations at relapse. This points towards more aggressive genetic profiles at relapse in these patients.
The acquisition of mutations and/or the outgrowth of a resistant clone during/after chemotherapy might require a longer time or is per se associated with a longer time to relapse and a more favorable prognosis. Loss of mutations at relapse suggest the presence of two clones at diagnosis, with a chemotherapy resistant clone expanding after the eradication of a chemotherapy sensitive clone. As both clones share mutations and only the sensitive clone contains specific alterations, the resistant clone might be an ancestor of the sensitive clone.
Taken together, in some patients the AML cells may require additional genetic alterations to become chemotherapy resistant, whereas in other patients the selective eradication of a sensitive clone is a potential mechanism underlying disease progression. Understanding the evolution of AML under selective pressure of chemotherapy is essential to cure or prevent AML relapse.
Hiddemann:Roche: Other: Grants; Genentech: Other: Grants; Roche: Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.