Abstract
Epigenetic modifiers and signaling factors are frequently mutated and often co-occur in various myeloid malignancies. However, precisely how these mutations cooperate to cause myeloid leukemia is not fully understood. Here, we show that cells with concurrent Ten-eleven-translocation 2 (Tet2) loss and Nras mutation can cause lethal chronic myelomonocytic leukemia (CMML) like disease in vivo and synergistically activate Ras signaling through epigenetic silencing of Sprouty2 (Spry2), thereby making cells with both disease alleles dependent on MAPK signaling and highly sensitive to MEK inhibition.
To assess if Tet2 loss and Nras mutation cooperate in myeloid transformation, we crossed Tet2 conditional knockout mice (Mx1-Cre+Tet2f/f) and Nras mutant mice (Mx1-Cre+Nras+/G12D) to generate Mx1-Cre+Tet2f/fNras+/G12D mice (Tet2Δ/ΔNras+/G12D). These mice, compared to single mutant mice with either allele alone, had more significant monocytosis, expansion of Lineage- Sca-1+ c-Kit+ (LSK) and myeloid progenitors in both bone marrow (BM) and spleen and development of lethal CMML-like disease (median survival 264 days). Moreover, serial transplantation of splenic cells derived from leukemic Tet2Δ/ΔNras+/G12D mice caused similar CMML-like disease in recipients, which emanates from LSK-positive stem/progenitor cells as the disease propagating population.
To delineate how Tet2 loss and Nras mutation synergize in leukemic transformation, we next performed western blot and phospho-flow analysis of MAPK and PI3K signaling in primary hematopoietic cells. Interestingly, pErk, pAkt and pS6 expression were significantly higher in Tet2Δ/ΔNras+/G12D cells compared to WT or single mutant cells, indicating that Tet2 loss and Nras mutation cooperates to further activate Ras signaling (Figure 1). Consistent with our murine model, TET2 silencing in NRAS mutant human leukemia cells increased MAPK output, consistent with augmentation of signaling by concurrent TET2/NRAS alterations in human leukemia cells.
To unravel the molecular mechanism of Ras signaling activation, we assessed mRNA / protein expression and performed bisulfite sequencing of known regulators of MAPK signaling, including Sprouty family members. We observed significant decrease of Spry2 expression, stepwise and specific hyper-methylation of CpG islands in the Spry2 promoter region in Tet2Δ/ΔNras+/G12D cells compared to WT or single mutant cells, consistent with progressive epigenetic remodeling in these leukemia cells in vivo (Figure 2). Genome wide methylation profiling of WT, single mutant and Tet2Δ/ΔNras+/G12D LSK cells using enhanced reduced representation bisulfite sequencing demonstrated clear separation of leukemic Tet2Δ/ΔNras+/G12D LSKs from WT or single mutant LSKs. Most importantly, restoration of Spry2 expression in Tet2Δ/ΔNras+/G12D cells led to decrease in pErk / pAkt level and significantly reduced colony formation, which functionally validates Spry2 as a key epigenetic target in Tet2/Nras mutant leukemia cells.
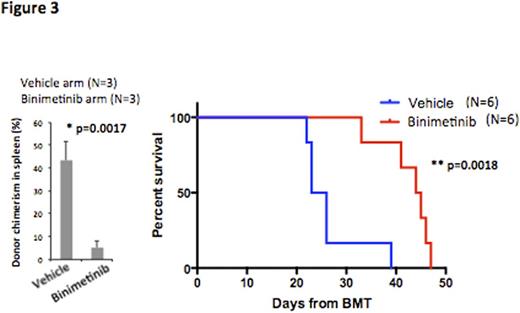
We next assessed whether the increased MAPK signaling seen in Tet2Δ/ΔNras+/G12D cells leads to differential sensitivity to MEK inhibition by performing studies with the clinical MEK inhibitor binimetinib (ARRY162). Tet2Δ/ΔNras+/G12D cells showed significantly higher sensitivity to binimetinib compared to Nras+/G12D cells in vitro (IC50, 6.948nM vs. 690.4nM). Moreover, in vivo treatment of Tet2Δ/ΔNras+/G12D leukemic recipients with binimetinib restored splenomegaly, significantly reduced disease burden in BM and spleen and improved overall survival compared to vehicle treatment (median survival 24.5 days vs. 44.5 days, p=0.0018, Figure 3). Of note, knockdown of human TET2 in NRAS mutant human leukemia cells sensitized to MEK inhibition in a similar manner demonstrating this approach may have value in leukemia patients with concurrent TET2 and NRAS mutations.
These data clearly indicate that Tet2 loss and Nras mutation synergize in myeloid transformation and cooperatively remodel DNA methylation, which leads to epigenetic silencing of Spry2 and synergistic activation of MAPK signaling, which can be leveraged through therapeutic MEK inhibition. Our studies provide novel insights into how signaling and epigenetic mutations cooperate in leukemic transformation and provide a rationale for mechanism based therapy in CMML patients with these high risk genetic lesions.
Melnick:Janssen: Research Funding. Levine:Qiagen: Membership on an entity's Board of Directors or advisory committees; Novartis: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.