Key Points
Ex vivo drug profiling captures disease-relevant features and relevant sensitivity to therapeutic agents in ALL.
A subset of drug-resistant T-ALL without mutations in ABL1 is highly responsive to dasatinib, which provides a rationale for drug repurposing.
Abstract
Drug sensitivity and resistance testing on diagnostic leukemia samples should provide important functional information to guide actionable target and biomarker discovery. We provide proof of concept data by profiling 60 drugs on 68 acute lymphoblastic leukemia (ALL) samples mostly from resistant disease in cocultures of bone marrow stromal cells. Patient-derived xenografts retained the original pattern of mutations found in the matched patient material. Stromal coculture did not prevent leukemia cell cycle activity, but a specific sensitivity profile to cell cycle–related drugs identified samples with higher cell proliferation both in vitro and in vivo as leukemia xenografts. In patients with refractory relapses, individual patterns of marked drug resistance and exceptional responses to new agents of immediate clinical relevance were detected. The BCL2-inhibitor venetoclax was highly active below 10 nM in B-cell precursor ALL (BCP-ALL) subsets, including MLL-AF4 and TCF3-HLF ALL, and in some T-cell ALLs (T-ALLs), predicting in vivo activity as a single agent and in combination with dexamethasone and vincristine. Unexpected sensitivity to dasatinib with half maximal inhibitory concentration values below 20 nM was detected in 2 independent T-ALL cohorts, which correlated with similar cytotoxic activity of the SRC inhibitor KX2-391 and inhibition of SRC phosphorylation. A patient with refractory T-ALL was treated with dasatinib on the basis of drug profiling information and achieved a 5-month remission. Thus, drug profiling captures disease-relevant features and unexpected sensitivity to relevant drugs, which warrants further exploration of this functional assay in the context of clinical trials to develop drug repurposing strategies for patients with urgent medical needs.
Introduction
The treatment of relapsed and refractory acute lymphoblastic leukemia (ALL) remains challenging.1 Progress in ALL genomics2 provides unprecedented insight into potentially actionable targets, such as activating mutations in tyrosine kinases,3 RAS,4 or interleukin-7R.5 Recurrent features such as MLL-AF4 rearrangements, the TCF3-HLF fusion,6 and hypodiploid karyotypes4 define rare subgroups with highly drug-resistant disease. However, a majority of patients who may benefit from innovative therapies are still identified on the basis of the persistence of minimal residual disease (MRD)1,7,8 or failure of remission induction therapy.9
Large integrative studies on cell line panels illustrate the difficulty of extrapolating drug responses on the basis of genomic data,10,11 even when indicative lesions in druggable pathways occur. Moreover, such alterations may be over- or underrepresented in cell lines, whereas patient-derived xenografts (PDXs) seem to reproduce the genetic driver mutation landscape in leukemia more closely.12-14 To obtain insight into interpatient drug response heterogeneity, we developed an in vitro platform directly using patient-derived leukemia cells. We hypothesized that drug response profiling of ALL, even without a priori information on genetic lesions or activated pathways, will detect sensitivity that may otherwise be overlooked. This approach will complement in vivo PDX models, which have obvious limitations in compound coverage and flexibility.15 Drug sensitivity testing revealed individual drug response phenotypes in acute myeloid leukemia (AML)16 and identified new strategies to bypass resistance to tyrosine kinase inhibitors (TKIs) in patients with deleterious BCR-ABL mutations.17 Conversely, characteristic tyrosine kinase mutations could be predicted on the basis of drug activity.18 Drug activity patterns can also identify an ALL subtype with tonic pre-BCR signaling.19
To establish a drug profiling platform, we took advantage of PDXs from clinically relevant ALL subgroups20-24 and, on the basis of previous reports,16,18,21-23,25,26 adapted a serum-free ALL coculture system on hTERT immortalized human bone marrow–derived mesenchymal stromal cells (MSCs)27,28 to an automated microscopic image readout for drug testing. This population-based approach reveals relevant activity clusters of therapeutic agents, thus identifying actionable targets that have not yet been exploited in conventional ALL treatment.
Patients and methods
Human samples
Primary human ALL cells were recovered from cryopreserved bone marrow aspirates of patients enrolled in the ALL-BFM 2000, ALL-BFM 2009, and ALL-REZ-BFM 2002 studies. Informed consent was given in accordance with the Declaration of Helsinki and the ethics commission of the Kanton Zurich (approval number 2014-0383). Samples were classified as standard risk, medium risk, high risk, or very high risk according to the ALL-BFM 2000 stratification,22 or as relapse (R) or refractory relapse (RR). Patients from a second cohort consented to protocols reviewed by the institutional review boards at Oregon Health & Science University and University of Texas Southwestern Medical Center.
Xenograft model
PDXs were generated as described23 by intrafemoral injection of 1 × 105 to 5 × 106 viable primary ALL cells in NSG mice. Leukemia progression was monitored in the peripheral blood by flow cytometry using anti-mouse-CD45 (anti-mCD45), anti-human-CD45 (anti-hCD45), anti-hCD19, or anti-hCD7 antibodies. Xenograft identity was verified by DNA fingerprinting using the commercial AmpFLSTR NGM SElect polymerase chain reaction amplification kit.
Genomic characterization of leukemia samples
Primary patient material and matched xenografts were analyzed by targeted sequencing and multiplex ligation-dependent probe amplification. In 19 BCP-ALL patients without an established abnormality (B-other)29 or targetable kinase-activating lesions,3 fluorescence in situ hybridization was performed (probes from Cytocell, Cambridge, United Kingdom). Detailed protocols are provided in the supplemental Data (available on the Blood Web site).
In vitro drug profiling platform
Drug responses were assessed in ALL cell cocultures on hTERT-immortalized primary bone marrow MSCs20 as described22 in 384-well plates (Greiner, REF781090). MSCs at 2.5 × 103 per well were plated in 30 µL AIM-V medium 24 hours before adding 2 × 104 to 3 × 104 ALL cells in 27.5 µL of medium recovered from cryopreserved samples. Compounds were reconstituted in dimethyl sulfoxide (10 mM stock concentrations) and stored at −80°C. Serially diluted drugs were prepared by using epMotion 5070 and Tecan D300 robots. An independent T-cell ALL (T-ALL) cohort was tested as described in Tyner et al.18
Drug response quantification and statistical analysis
We used a fitting routine based on the 4-parameter log-logistic function (R package drc, version 2.3-96) on data normalized against samples treated with dimethyl sulfoxide. Outliers were detected and removed before curve fitting by detecting local slope changes with a linear fit. Nonconvergent cases were identified on the basis of linear fit parameters. R codes are available at https://github.com/pampernickel/drTools. Hierarchical clustering was performed to group patients according to their drug responses (R package gplots). Differential drug responses of patient groups of interest were evaluated by using the nonparametric Mann-Whitney U test.
In vivo drug treatment
For venetoclax and combinations, 5 to 8 mice were transplanted intravenously with 1 × 106 ALL cells per treatment arm. Randomized cohorts were treated with vehicle, 100 mg/kg/day venetoclax (Activebiochem30 ) orally, 10.5 mg/kg dexamethasone (Mepha) intraperitoneally daily ×5 days per week for 2 weeks, and 0.5 mg/kg vincristine (Teva) intraperitoneally once per week. For cytarabine, docetaxel, and dasatinib, animals (1 per condition) were intravenously transplanted with 7 × 106 ALL cells. After 5 days, animals were treated with 50 mg/kg cytarabine (Sandoz) daily intraperitoneally for 5 days, 5 mg/kg docetaxel (Taxotere) intravenously on day 1 and day 5, or 50 mg/kg dasatinib (Selleck; dissolved as described31 ) orally for 5 days. Leukemic burden was determined posttreatment by flow cytometry.
Cell assays
Viability of 2.5 × 104 ALL cells in suspension or coculture with 2.5 × 103 MSCs in AIM-V medium was measured by flow cytometry at 1, 4, and 7 days (7-aminoactinomycin D; reported as day 4 mean ± standard deviation [SD]). Regarding proliferation and apoptosis, 1 × 105 ALL cells were seeded with 1 × 104 MSCs. Proliferating and apoptotic cells were labeled by using the Click-iT EdU Imaging Kit and Cell Event Caspase-3/7 Green, respectively. Proliferating and nonproliferating groups were identified with an expectation-maximization mixture model (R package mixtools).
Intracellular flow cytometry and western blot
ALL cells (10 × 106) were fixed in 2% paraformaldehyde, permeabilized with ice-cold methanol, and indirectly tagged with fluorescein isothiocyanate–labeled antibodies. Whole-cell extracts from 3 × 106 to 5 × 106 cells were used for western blots (Bio-Rad Criterion). Detailed protocols are provided in the supplemental Data.
Results
Drug response profiling reveals distinct clusters of activity in ALL
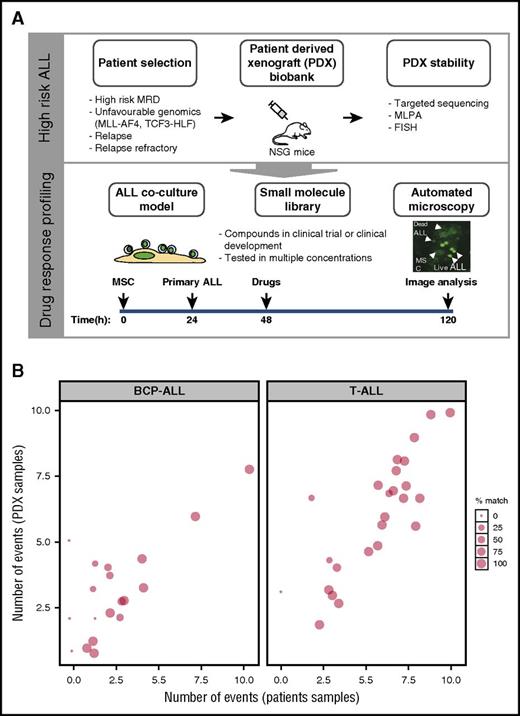
Cocultures on hTERT-immortalized MSCs28 facilitate survival of most BCP-ALL and T-ALL cells.21 This protective effect may even increase the stringency of drug testing.32 We tested 60 preclinical and clinical compounds (supplemental Table 1) by using an imaging-based cell viability readout21 (Figure 1) on ALL xenografts derived from patients with standard-risk or high-risk ALL based on MRD persistence and relapsed and refractory ALL. Patient and PDX samples were characterized by using established diagnostic workflows, including tests for most recurrent translocations that activate tyrosine kinase pathways (supplemental Table 2A-B) and by targeted sequencing of 52 frequently mutated genes in ALL. We retrieved the expected pattern of mutations (supplemental Figure 1; supplemental Table 3) with frequent events in KRAS (13 of 25) and TP53 (10 of 25), consistent with previous reports.33,34 On average, 74% of single nucleotide variants and insertions/deletions were conserved between the primary diagnostic samples and PDXs (Figure 1B; supplemental Figure 1). Oncogenic translocations were always maintained. We also included samples from TCF3-HLF– and TCF3-PBX1–positive ALL subtypes for which we recently reported a strong conservation of the genomic landscape in PDXs.35
Setup of the drug response profiling platform. (A, top panel) Patient material, notably from high-risk patients, including relapse patients and patients with translocations linked to poor survival, were prioritized for PDX and drug response profiling; PDX stability was evaluated against primary material by comparing targeted deep-sequenced leukemogenesis markers. (A, bottom panel) Drug profiling was performed on primary ALL cells in coculture with bone marrow–derived MSCs. Automated microscopy-based image analysis was used to quantify living ALL cells and generate dose-response curves. Imaging results were analyzed with a toolkit that performed dose-response normalization, outlier removal, rapid curve fitting, and extraction of response parameters (IC50, area under the curve, Emax, which corresponds to the percentage of viable cells at the maximum dose of the drug). Selected single compounds and combinations were validated in the xenograft model. This platform enabled the identification of drug-response phenotypes in individual ALL patients, providing an additional layer of information to facilitate individual treatment approaches. (B) Our PDX model preserves an average of 74% of the mutations and insertions/deletions initially detected in patients, making it an ideal source of material for drug-response testing in multicenter, co-clinical settings. MLPA, multiplex ligation-dependent probe amplification; FISH, fluorescent in situ hybridization.
Setup of the drug response profiling platform. (A, top panel) Patient material, notably from high-risk patients, including relapse patients and patients with translocations linked to poor survival, were prioritized for PDX and drug response profiling; PDX stability was evaluated against primary material by comparing targeted deep-sequenced leukemogenesis markers. (A, bottom panel) Drug profiling was performed on primary ALL cells in coculture with bone marrow–derived MSCs. Automated microscopy-based image analysis was used to quantify living ALL cells and generate dose-response curves. Imaging results were analyzed with a toolkit that performed dose-response normalization, outlier removal, rapid curve fitting, and extraction of response parameters (IC50, area under the curve, Emax, which corresponds to the percentage of viable cells at the maximum dose of the drug). Selected single compounds and combinations were validated in the xenograft model. This platform enabled the identification of drug-response phenotypes in individual ALL patients, providing an additional layer of information to facilitate individual treatment approaches. (B) Our PDX model preserves an average of 74% of the mutations and insertions/deletions initially detected in patients, making it an ideal source of material for drug-response testing in multicenter, co-clinical settings. MLPA, multiplex ligation-dependent probe amplification; FISH, fluorescent in situ hybridization.
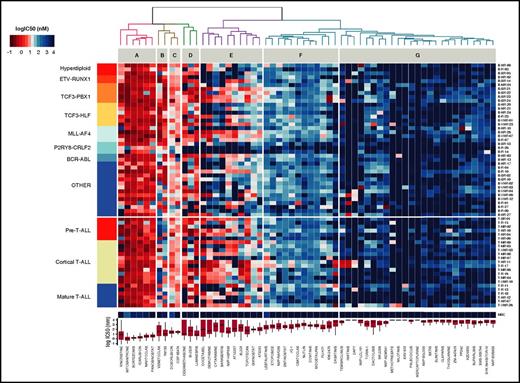
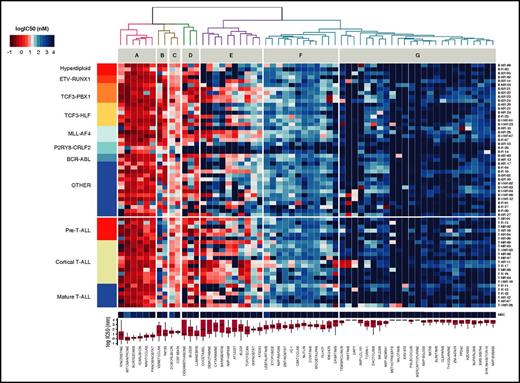
To evaluate the potential of this ex vivo platform, we profiled 24 T-ALL and 44 BCP-ALL PDXs derived from pretreatment diagnostic samples (ALL-BFM 2000 study36 ; Figure 2). For each drug, we used 8 doses optimized from an initial 5-point screen (supplemental Table 4). None of the tested compounds affected MSC viability at concentrations lethal to ALL cells, indicating selective drug activity (Figure 2). Unsupervised clustering of drug responses (half maximal inhibitory concentration [IC50] values) identified various patterns of response. Compounds including anthracyclines, the BH3 mimetic navitoclax (ABT-263), and the proteasome inhibitor bortezomib were effective at low (IC50 <10 nM) and narrow IC50 range in most cases (cluster A). A second group of agents, including the BCL2-specific BH3 mimetic venetoclax,30 TKIs, and conventional cytotoxic agents such as glucocorticoids, topoisomerase inhibitors, and nucleotide analogs (gemcitabine, cytarabine), showed responses distributed over a wider concentration range, with high activity in the nanomolar range in some cases and low activity in others (clusters B, C, and E). Venetoclax (cluster C) was generally more active in BCP-ALL but showed similar activity in a T-ALL subset. In cluster E, we identified 2 groups of agents whose separation was driven by differences in response to antimetabolites (cytarabine, gemcitabine), antimitotic drugs (vincristine, docetaxel), the Aurora kinase inhibitors AT9283 and barasertib, and the Polo-like kinase inhibitor BI-2536. Finally, very strong sensitivity was detected in a few ALL patients for drugs that were otherwise generally not active in ALL on this platform (cluster G). These included second mitochondrial activator of caspases (SMAC) mimetics (eg, LCL161), an observation that led us to show that a BCP-ALL subset was extremely responsive to SMAC mimetics through RIP1 kinase–dependent necroptosis and apoptosis.37 The ABL/SRC inhibitor dasatinib is also highly active in a T-ALL subset discussed later in this article. High peak plasma concentrations (Cmax) were reported for most clinical compounds in our panel (supplemental Figure 2), suggesting that effective concentrations may be achieved in vivo. Our platform provides reproducible drug activity profiles that identify functional phenotypes and give new insights for therapeutic targeting. No correlations between drug responses and genetic lesions were found (supplemental Table 5).
Drug response profiles of BCP-ALL and T-ALL. Heatmap indicating the response of BCP-ALL (n = 44) and T-ALL (n = 24) to 60 compounds and represented by IC50 values. Samples (rows) were ordered according to clinical classification, and compounds (columns) were ordered according to activity. The IC50 distribution range for each compound is shown on the lower panel forming drug clusters. (A) Generally active drugs, mean IC50 values <10 nM. (B) Drugs more active in BCP-ALLs than T-ALLs. (C) Generally active drugs with IC50 values <100 nM. (D) Drugs with variable activity. (E) Drugs with activity linked to cycling activity. (F) Generally active drugs with high nanomolar range. (G) Generally inactive drugs, with sporadic exceptions. Heatmap of MSCs and drug IC50 distribution box plot are shown on the lower part of the graph.
Drug response profiles of BCP-ALL and T-ALL. Heatmap indicating the response of BCP-ALL (n = 44) and T-ALL (n = 24) to 60 compounds and represented by IC50 values. Samples (rows) were ordered according to clinical classification, and compounds (columns) were ordered according to activity. The IC50 distribution range for each compound is shown on the lower panel forming drug clusters. (A) Generally active drugs, mean IC50 values <10 nM. (B) Drugs more active in BCP-ALLs than T-ALLs. (C) Generally active drugs with IC50 values <100 nM. (D) Drugs with variable activity. (E) Drugs with activity linked to cycling activity. (F) Generally active drugs with high nanomolar range. (G) Generally inactive drugs, with sporadic exceptions. Heatmap of MSCs and drug IC50 distribution box plot are shown on the lower part of the graph.
Drug profiling captures leukemia-intrinsic differences in cell proliferation and survival
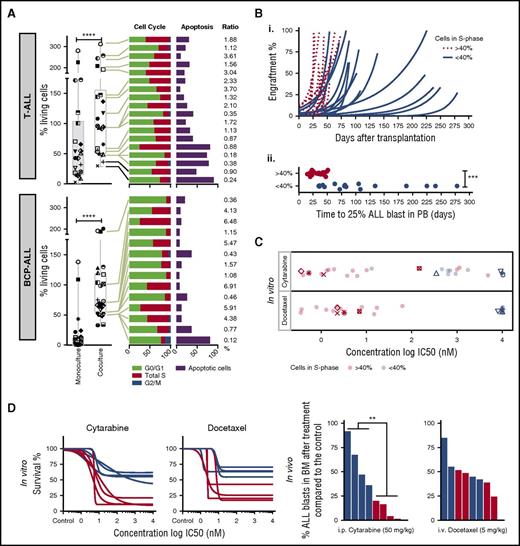
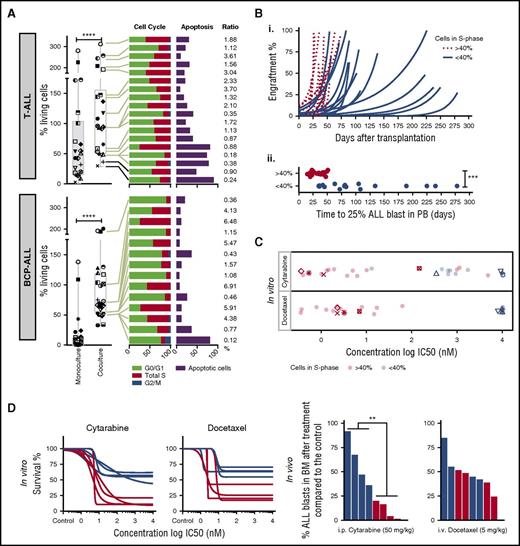
Although most ALL samples tested in coculture survive on MSCs, we noticed relative cell survival heterogeneity, suggesting differences in cell proliferation and spontaneous cell death rates across samples (Figure 3A). We did not detect ALL migration beneath stromal cells (pseudoemperipolesis) or cobblestone structure-like formation26 that could interfere with microscopy readouts. Median cell viability on MSCs was 69% of seeded cells for BCP-ALL and 94% for T-ALL compared with 1.2% and 45.5% in monoculture after 96 hours. A high rate of survival on this platform (viability of >70% at day 3 compared with day 0) correlated with a higher ratio (r >1) of cells in S phase vs apoptotic cells (Figure 3A). To determine whether these differences are the result of stromal coculture effects or intrinsic features of ALL cells, we compared leukemia proliferation and drug sensitivity patterns ex vivo and in vivo in leukemia xenografts. Marked differences in sensitivity were detected in both BCP-ALL and T-ALL for drugs whose mechanisms of action require active cycling (Figure 2, cluster E; supplemental Figure 3), including mitotic spindle formation inhibitors, DNA synthesis, cell cycle, and mitosis regulatory kinases. We used a mixture model fit to distinguish high-proliferating (>40% of cells in S phase) from low-proliferating (<40% of cells in S phase) ALL patients. ALL samples with high proliferative activity in vitro engrafted significantly faster (P < .001) than samples with low proliferative activity (Figure 3B). As expected, drugs with the highest differential activity in high- and low-proliferating samples inhibit targets involved in cell cycle control (supplemental Figure 4). Most importantly, samples with rapid engraftment kinetics (Figure 3B) were more sensitive to cytarabine and docetaxel ex vivo (Figure 3C), which correlates with stronger antileukemic effects in vivo (Figure 3D). Thus, the ex vivo coculture model captures leukemia-specific characteristics with respect to cell cycle activity, which are preserved in vivo in the leukemia xenograft model and are not caused by the coculture conditions.
Drug profiling reveals leukemia-intrinsic features. (A) Coculturing on MSCs supports survival of T-ALL (n = 22) and BCP-ALL (n = 25). Data at day 4 are given, normalized to seeded viable cell numbers at day 0 in both monoculture and coculture (left panel). The symbols identify the patients in the 2 culture conditions. Cell cycle and apoptosis rates of primary T-ALL (n = 18) and BCP-ALL (n = 14) cells in coculture are provided on the right. Samples are ranked from highest (top) to lowest (bottom) survival. Ratio of cells in S phase and apoptosis is given on the far right. (B) Engraftment kinetics for ALL cases with >40% and <40% of cells in S phase are given (i). Time to engraftment with 25% ALL blasts in the 2 groups is indicated in the lower panel (ii). (C) In vitro ALL proliferation correlates with drug response to cytarabine (antimetabolite), docetaxel (antimitotic), and other drugs that target the cell cycle (supplemental Figure 4). ALL cells with >40% of cells in S phase respond to cytarabine and docetaxel with lower IC50 compared with samples with <40% of cells in S phase. (D) Cytarabine and docetaxel response profiles predict in vivo ALL response (n = 8). ****P < .001 (paired Student t test); ***P < .001 (2-sided Student t test); **P = .0087. i.p., intraperitoneal; i.v., intraveous; PB, peripheral blood.
Drug profiling reveals leukemia-intrinsic features. (A) Coculturing on MSCs supports survival of T-ALL (n = 22) and BCP-ALL (n = 25). Data at day 4 are given, normalized to seeded viable cell numbers at day 0 in both monoculture and coculture (left panel). The symbols identify the patients in the 2 culture conditions. Cell cycle and apoptosis rates of primary T-ALL (n = 18) and BCP-ALL (n = 14) cells in coculture are provided on the right. Samples are ranked from highest (top) to lowest (bottom) survival. Ratio of cells in S phase and apoptosis is given on the far right. (B) Engraftment kinetics for ALL cases with >40% and <40% of cells in S phase are given (i). Time to engraftment with 25% ALL blasts in the 2 groups is indicated in the lower panel (ii). (C) In vitro ALL proliferation correlates with drug response to cytarabine (antimetabolite), docetaxel (antimitotic), and other drugs that target the cell cycle (supplemental Figure 4). ALL cells with >40% of cells in S phase respond to cytarabine and docetaxel with lower IC50 compared with samples with <40% of cells in S phase. (D) Cytarabine and docetaxel response profiles predict in vivo ALL response (n = 8). ****P < .001 (paired Student t test); ***P < .001 (2-sided Student t test); **P = .0087. i.p., intraperitoneal; i.v., intraveous; PB, peripheral blood.
Because other groups opted for systems based on monocultures, we compared our coculture data to a readout in serum-supplemented liquid leukemia cultures.18 By selecting 8 BCP-ALL and 19 T-ALL samples, we confirmed improved survival of most of these samples on MSCs compared with cell suspension cultures (supplemental Figure 5A-B). Although IC50 values from coculture and monoculture were significantly correlated for 22 drugs tested under both conditions (Spearman ρ = 0.64; P < .001), there were several discrepancies in drugs of interest. We observed a significant correlation of drug activity in cocultures with in vivo drug responses (supplemental Figure 5D-F), most evident for venetoclax (supplemental Figure 5D), docetaxel and dasatinib, and to a lesser extent for cytarabine (supplemental Figure 5E). In contrast, drug activity data obtained from liquid monocultures showed a predictive trend only for cytarabine but not for the other agents tested (supplemental Figure 5F). These observations support the potential of our system for identifying relevant vulnerabilities in a clinical setting.
Drug profiling reveals individual patterns of drug sensitivity and resistance in relapsed and refractory ALL
Drug profiling may convey relevant information for selecting new agents for salvage therapy in patients with highly drug-resistant disease. To compare the activity of different substances in different patients, it is important to evaluate drug activity in a single patient against the full response range obtained on the same platform for other leukemia cases, including clinically relevant subsets. We profiled PDX samples from 12 patients with relapsed ALL refractory to salvage therapy (RR) who did not achieve the second or third remission required for inclusion in early clinical trials (Figure 4A) and primary leukemia cells from 5 patients with refractory disease in real time prospectively (Figure 4B; Table 1). Figure 4 shows IC50 values for a selection of therapeutic agents in samples of interest against those obtained for all other samples on our platform (gray). RR ALL samples were generally more resistant to agents used for induction in ALL such as dexamethasone (10 of 12 patients), cytarabine (9 of 12 patients), and doxorubicin (9 of 12 patients) compared with other diagnostic and relapse ALL patients (Figure 4A). In contrast, individual samples were highly sensitive to dexamethasone, idarubicin, and mitoxantrone, which are included in the standard of care for relapsed ALL,38 and to new agents from different classes, such as venetoclax, dasatinib, bortezomib, nutlin, JQ1, and panobinostat. Again, we noticed unexpected responses to venetoclax and dasatinib in a few patients, which we discuss later in this article. In addition, sensitivity patterns could be associated with cytogenetic groups. For example, patients with MLL-AF4 ALL were sensitive to PI3K/mTOR/AKT or HSP90 inhibitors, consistent with previous reports39 (supplemental Figure 6).
Distinct drug activity patterns can be detected for individual samples and patient groups of interest. (A) RR PDX (n = 12) samples exhibit general resistance to conventional clinical compounds but remain sensitive to some experimental drugs. (B) Primary RR patients (n = 5) tested before last salvage therapy demonstrate persistent resistance to standard chemotherapy and individual sensitivity to experimental molecules. All responses are represented as IC50 (log[nM]) and are compared with other diagnostic and relapse ALL patients depicted in the background. **P < .005 (2-sided Student t test); *P < .05.
Distinct drug activity patterns can be detected for individual samples and patient groups of interest. (A) RR PDX (n = 12) samples exhibit general resistance to conventional clinical compounds but remain sensitive to some experimental drugs. (B) Primary RR patients (n = 5) tested before last salvage therapy demonstrate persistent resistance to standard chemotherapy and individual sensitivity to experimental molecules. All responses are represented as IC50 (log[nM]) and are compared with other diagnostic and relapse ALL patients depicted in the background. **P < .005 (2-sided Student t test); *P < .05.
To assess the feasibility of our approach in the clinical setting, we tested 5 patients with highly refractory ALL at the time of relapse (Figure 4B). Results could be obtained within 5 days. These patients did not respond to standard-of-care drugs on the platform (dexamethasone, vincristine, doxorubicin, or mitoxantrone) but were individually sensitive to venetoclax (patients 1, 2, and 3) and panobinostat (patient 5). Thus, drug profiling may provide important information when exploring options for patients with drug-resistant disease.
Response to venetoclax ex vivo correlates with strong in vivo antileukemic activity as a single agent and in combination
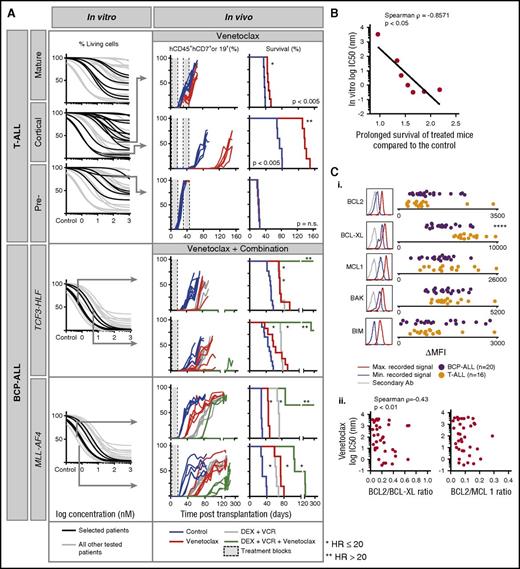
Given the strong in vitro activity of venetoclax across various ALL subtypes, including a subset of T-ALL, BCP-ALL, TCF3-HLF ALL, and all MLL-AF4 ALL patients, we tested venetoclax (n = 7) in vivo in the xenograft model (Figure 5A). Several patients with T-ALL responded to venetoclax in vitro with IC50 values in the nanomolar range (Figure 5A), consistent with reports describing activity in early thymic precursor ALL and T-ALL.40-42 These results were verified by flow cytometry using 7-aminoactinomycin D staining to quantify cell death (supplemental Figure 7). As expected, the response to oral administration of venetoclax in vivo correlated with in vitro activity for 3 T-ALL patients with strong, intermediate, and low venetoclax sensitivity. Single-agent venetoclax treatment delayed leukemia progression significantly in the patient with strong in vitro sensitivity (hazard ratio, 20; IC50 <1 nM; low Emax; treated vs untreated P < .005) compared with patients with low IC50 (<100 nM) but higher Emax (HR, 0.07; treated vs untreated P < .005) or high IC50 (>1 μM). In addition, complete response was detected when treating the T-VHR-03 case in mice with high leukemia burden (75% engraftment; supplemental Figure 8). We recently reported similar venetoclax efficacy in 3 TCF3-HLF ALL patients in vivo35 ; comparable correlations were obtained in 2 patients with TCF3-HLF– or MLL-rearranged ALL (Figure 5A). For all tested cases, venetoclax-induced delays in in vivo leukemia progression correlated with in vitro responses (Spearman ρ = –0.86; P < .05; Figure 5B).
In vitro sensitivity to the BCL-2 antagonist venetoclax correlates with the response in leukemia xenografts. (A) In vitro response to venetoclax for indicated ALL subtypes (black) compared with other ALL (gray). From top to bottom: mature T-ALL (n = 6), cortical T-ALL (n = 13), pre-T-ALL (n = 6), TCF3-HLF ALL (n = 4), and MLL-AF4 ALL (n = 3). Cell viability (7-aminoactinomycin D) was measured by flow cytometry after 72 hours of treatment and was normalized against controls treated with dimethyl sulfoxide. Arrows indicate samples whose response had been validated in vivo for venetoclax (top to bottom: T-VHR-03, T-HR-11, and T-HR-10) or venetoclax in combination with vincristine and dexamethasone (top to bottom: B-HR-24, B-HR-20, B-HR-26, and B-VHR-07). The left panel shows the number of leukemia cells compared with mouse lymphocytes over time. The right panel shows corresponding Kaplan-Meier survival curves (event defined as 25% of mCD45–hCD45+hCD19+ or hCD7+ leukemia cells detected by flow cytometry). (B) In vitro response to venetoclax correlates with fold increase of survival comparing treatment with venetoclax with treatment with vehicle (n = 7). (C) BCL2 protein family expression (i) analyzed by flow cytometer in T-ALL (n = 16) and BCP-ALL (n = 20). Correlation of BCL2:BCL-XL and BCL2:MCL1 ratio (ii) with in vitro venetoclax response. ****P < .001 (2-tailed Student t test). Ab, antibody; HR, hazard ratio; Max., maximum; MFI, mean fluorescent intensity; Min., minimum.
In vitro sensitivity to the BCL-2 antagonist venetoclax correlates with the response in leukemia xenografts. (A) In vitro response to venetoclax for indicated ALL subtypes (black) compared with other ALL (gray). From top to bottom: mature T-ALL (n = 6), cortical T-ALL (n = 13), pre-T-ALL (n = 6), TCF3-HLF ALL (n = 4), and MLL-AF4 ALL (n = 3). Cell viability (7-aminoactinomycin D) was measured by flow cytometry after 72 hours of treatment and was normalized against controls treated with dimethyl sulfoxide. Arrows indicate samples whose response had been validated in vivo for venetoclax (top to bottom: T-VHR-03, T-HR-11, and T-HR-10) or venetoclax in combination with vincristine and dexamethasone (top to bottom: B-HR-24, B-HR-20, B-HR-26, and B-VHR-07). The left panel shows the number of leukemia cells compared with mouse lymphocytes over time. The right panel shows corresponding Kaplan-Meier survival curves (event defined as 25% of mCD45–hCD45+hCD19+ or hCD7+ leukemia cells detected by flow cytometry). (B) In vitro response to venetoclax correlates with fold increase of survival comparing treatment with venetoclax with treatment with vehicle (n = 7). (C) BCL2 protein family expression (i) analyzed by flow cytometer in T-ALL (n = 16) and BCP-ALL (n = 20). Correlation of BCL2:BCL-XL and BCL2:MCL1 ratio (ii) with in vitro venetoclax response. ****P < .001 (2-tailed Student t test). Ab, antibody; HR, hazard ratio; Max., maximum; MFI, mean fluorescent intensity; Min., minimum.
As with most chemotherapeutic agents, single-agent venetoclax therapy is unlikely to be effective. Currently, most investigational agents will be tested in combination with a standard-of-care antileukemic regimen that includes 2 to 4 drugs such as vincristine, dexamethasone, asparaginase, and an anthracycline typically used for reinduction chemotherapy at relapse.38 We detected synergy in vitro by using co-titration experiments, but this assay is challenging when assessing a drug with such strong in vitro activity as venetoclax43 (supplemental Figure 9; supplemental Table 4). Because it is impossible to provide supportive care to mice after myelotoxic chemotherapy in vivo, we next tested the combination of venetoclax, dexamethasone, and vincristine without anthracyclines (Figure 5A). Venetoclax or chemotherapy alone delayed leukemia progression for TCF3-HLF– and MLL-AF4–rearranged patients (P < .005). The 3-drug combination prevented leukemia progression for more than 300 days in 2 TCF3-HLF samples and in 3 of 5 MLL-AF4 ALL samples. Leukemia progression was significantly delayed in remaining samples.
The identification of response-predictive biomarkers, in addition to drug profiling, is important for the clinical development of BH3 mimetics. The BCL2:BCL-XL and BCL2:MCL1 ratios were suggested as biomarkers for venetoclax sensitivity in ALL44 and in multiple myeloma,45 respectively. We determined levels of BCL2 family members by intracellular flow cytometry and western blotting (Figure 5C; supplemental Figure 10). In vitro response to venetoclax neither correlated with BCL2 family protein expression levels nor BCL2:MCL1 or BCL2:BCL-XL ratios in 36 BCP-ALL and T-ALL samples tested by flow cytometry (Figure 5C; supplemental Figure 11). It will be important to perform further BH3 profiling in parallel with drug response profiling in clinical trials to establish predictive biomarkers.
Drug profiling identifies a subset within T-ALL highly responsive to dasatinib
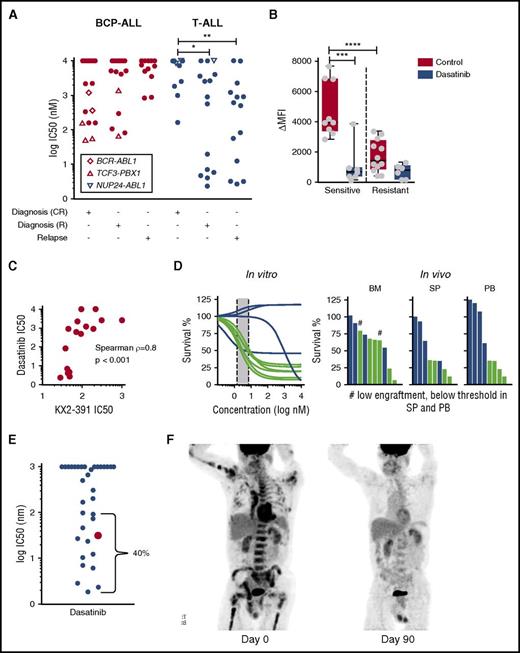
We detected unexpected responses to the ABL1/SRC inhibitor dasatinib (IC50 <100 nM) in 12 T-ALL patients (30%) without the typical ABL1 kinase translocation (Figure 6A). Importantly, these responses were detected in both diagnostic and relapse samples from high-risk patients by MRD. Moreover, the IC50 for dasatinib in these patients was at least tenfold lower than in any of the best BCP-ALL responders tested, including 5 ALL patients with rearranged TCF3-PBX1 (recently linked to active BCR signaling19,46 ) who were sensitive to dasatinib but not imatinib (Figure 6A). No known recurrent genetic abnormality could be linked to this phenotype (supplemental Table 3). We found neither recurrent mutations nor gene fusions by exome and transcriptome sequencing that associated with dasatinib sensitivity in T-ALL (supplemental Table 6). RNA sequencing also indicated that 1 dasatinib-sensitive patient had low FYN, but high SRC expression (supplemental Figure 12). Given that the dasatinib response did not correlate with the response to other BCR-ABL inhibitors, we hypothesized that dasatinib acts via SRC inhibition. By phospho-flow cytometry, we detected higher levels of activated, phosphorylated SRC in dasatinib-sensitive samples (Figure 6B); SRC phosphorylation was abrogated after exposure to dasatinib. The SRC inhibitor KX2-391, which inhibits SRC at nanomolar concentrations,47 induced cell death in dasatinib-sensitive T-ALL patients at concentrations below 100 nM (Figure 6C), supporting the relevance of the SRC pathway in this T-ALL subset. Apart from KX2-391, dasatinib response also correlated with responses to other receptor tyrosine kinase inhibitors (eg, midostaurin, crenolinib; adjusted P < .005; supplemental Figure 13), consistent with the central role of SRC in receptor tyrosine kinase signaling.48 Importantly, in vitro response to dasatinib correlated with antileukemic activity in vivo in T-ALL xenografts (Figure 6D).
In vitro sensitivity of T-ALL to dasatinib correlates with antileukemic efficacy in the patient. (A) Subset of T-ALL patients at diagnosis that relapsed (R) and at relapse are highly sensitive to dasatinib in vitro. (B) Dasatinib-sensitive T-ALL cells have higher levels of phosphorylated SRC that decreases after treatment with 1 µM dasatinib for 2 hours as measured by flow cytometry. (C) Dasatinib response correlates with sensitivity to the SRC inhibitor KX2-391 (n = 16). (D) In vitro captured response correlates with in vivo response to dasatinib (n = 10). Indicated is the percent of T-ALL blasts compared with mouse lymphocytes normalized to vehicle-treated controls. (E) Sensitivity of adult and pediatric T-ALL patients to dasatinib reveals 40% of patients with IC50 below 100 nM. (F) Left: positron emission tomography/computed tomography (PET/CT) scan demonstrates significant disease burden throughout the marrow in bilateral upper and lower extremities, the pelvis, vertebrae, and contiguous nodes within the mediastinum. Right: PET/CT ∼15 months after the original presentation, shortly after initiation of dasatinib monotherapy. This image demonstrates complete response (CR) with no signs of marrow or nodal involvement. *P = .0272; **P = .0072; ***P = .0004; ****P = .0001. BM, bone marrow; PB, peripheral blood; SP, spleen.
In vitro sensitivity of T-ALL to dasatinib correlates with antileukemic efficacy in the patient. (A) Subset of T-ALL patients at diagnosis that relapsed (R) and at relapse are highly sensitive to dasatinib in vitro. (B) Dasatinib-sensitive T-ALL cells have higher levels of phosphorylated SRC that decreases after treatment with 1 µM dasatinib for 2 hours as measured by flow cytometry. (C) Dasatinib response correlates with sensitivity to the SRC inhibitor KX2-391 (n = 16). (D) In vitro captured response correlates with in vivo response to dasatinib (n = 10). Indicated is the percent of T-ALL blasts compared with mouse lymphocytes normalized to vehicle-treated controls. (E) Sensitivity of adult and pediatric T-ALL patients to dasatinib reveals 40% of patients with IC50 below 100 nM. (F) Left: positron emission tomography/computed tomography (PET/CT) scan demonstrates significant disease burden throughout the marrow in bilateral upper and lower extremities, the pelvis, vertebrae, and contiguous nodes within the mediastinum. Right: PET/CT ∼15 months after the original presentation, shortly after initiation of dasatinib monotherapy. This image demonstrates complete response (CR) with no signs of marrow or nodal involvement. *P = .0272; **P = .0072; ***P = .0004; ****P = .0001. BM, bone marrow; PB, peripheral blood; SP, spleen.
To validate our observations, we checked the drug sensitivity of 33 adult and pediatric T-ALL patients obtained on a liquid monoculture platform.18 Remarkably, 4 of 33 responded to dasatinib with IC50 <10 nM (Figure 6E), and 9 of 33 with IC50 between 10 nM and <100 nM. One of these samples was from an adult male patient with refractory T-ALL with mediastinal and abdominal lymph node involvement after 8 cycles of hyper-CVAD (cyclophosphamide, vincristine, doxorubicin, dexamethasone, methotrexate, and cytarabine) chemotherapy, allogeneic stem cell transplantation, and relapse treatment with nelarabine, mitoxantrone, and cytarabine. On the basis of these results, dasatinib (140 mg/day) was initiated, first in combination with pegylated asparaginase, which was interrupted after 1 dose because of intolerance. Dasatinib monotherapy was continued, and interval resolution of all lesions was evidenced on a repeat positron emission tomography/computed tomography scan 2 months after initiation of dasatinib (Figure 6F). In total, the disease could be controlled over 5 months. Although the short exposure to asparaginase may have contributed to this response, disease control with dasatinib monotherapy over several months is indicative of clinical activity. These results confirm that a subset of drug-resistant and relapsed T-ALL can be identified by drug profiling to be particularly sensitive to dasatinib; thus, further exploration of underlying molecular mechanisms is warranted. Given the experience with established combinations of dasatinib with chemotherapy for the treatment of BCR-ABL–positive ALL,49 our data provide a strong rationale for drug repurposing on the basis of drug profiling results for selected patients with drug-resistant T-ALL in pediatric and adult patient populations.
Discussion
Here we provide compelling evidence that informative and reproducible differences in drug response profiles can be detected in patient groups of interest while simultaneously revealing patient-to-patient response variations. Heterogeneous and strong activity was found for different classes of agents in patients with refractory ALL. We did not observe any correlations between drug response phenotypes and somatic mutations, which may be partly a result of the limited size of our cohort; multivariable analyses based on whole genome or exome sequencing results on a larger cohort would be of interest in the future to definitively establish correlations. However, our results, which indicate that it will be challenging to infer drug activity based solely on genomic data, are consistent with reports in adult hematologic malignancies.16,18
As a basis for standardization, we opted for coculture on human MSCs,27,28 which efficiently supported most of the primary ALL samples that we tested in serum-free conditions. Our assay provides better ALL cell survival and stronger correlation with in vivo drug activity in PDX models compared with monocultures (supplemental Figure 5). This model also increases the possibilities for multidimensional expansion. Effects can be analyzed not only on the target leukemia cells but also on nonhematopoietic microenvironmental cells, and use of additional markers that indicate metabolic states, distinct differentiation processes, or signaling activity could be envisaged. The fact that we detected subsets with more proliferative activity both in vitro and in vivo, based on drug profiling, indicates that important leukemia-intrinsic features are maintained and captured under the in vitro cell culture conditions. We and others16,18 have demonstrated the use of drug profiling for identifying phenotypes that are responsive to new therapeutic agents. We have identified recurrent ALL patients who are highly sensitive to triggering RIP1-dependent cell death with SMAC mimetics37 or to BCL2 inhibition in BCP-ALL subsets, including TCF3-HLF ALL35 and T-ALL subsets. Moreover, we discovered a subgroup in T-ALL that is highly sensitive to dasatinib, which should be further evaluated first in patients with highly drug-resistant and refractory disease.
Drug response profiling may contribute to defining cohorts that could benefit from new agents. The profiles that we detected for venetoclax,30 which was recently approved for treatment of chronic lymphocytic leukemia, illustrate the type of information that could be used to improve patient selection in early clinical trials. We show that a relatively large proportion of BCP-ALL patients may respond to venetoclax, including very high-risk subtypes such as TCF3-HLF–35 and MLL-AF4–positive ALL. Our findings are confirmed independently by others who showed strong venetoclax activity in MLL-rearranged ALL.50 Supportive information using other biomarkers would be desirable for monitoring response in clinical trials. The BCL2:BCL-XL expression ratio was proposed as a predictive biomarker for venetoclax response,44 but our results and other data50 suggest that this approach may not detect all cases. BH3 profiling using synthetic peptides instead of targeted small-molecule drugs44,51,52 may provide complementary information, as evaluated in a phase 2 study that assessed venetoclax monotherapy in patients with refractory and relapsed AML.53 We propose that in vitro drug profiling be incorporated into upcoming clinical trials for venetoclax to determine its predictive potential.
New treatment options are urgently needed for relapsed T-ALL.1 We discovered a T-ALL subset highly sensitive to dasatinib. We also show good response to this TKI in a patient with previously refractory T-ALL whose treatment was designed on the basis of drug profiling data. A patient with NUP1-ABL1–positive T-ALL was also reported to respond to dasatinib-based therapy,54 but none of the patients with high sensitivity to dasatinib were NUP1-ABL1–positive in our series. We did not identify activating mutations that may directly explain dasatinib sensitivity, indicating that the underlying mechanisms may occur at a different level, which will motivate follow-up studies. Given that dasatinib combinations with ALL standard-of-care chemotherapy49 and a pediatric dose55 are established, its inclusion in chemotherapy for patients with drug-resistant T-ALL that displays a dasatinib-responsive phenotype should be evaluated. Here, we demonstrated that in vitro drug profiling captures functional information of clinical importance and reveals new biological entities in ALL. Given the growing interest of clinicians in this approach, prospective evaluation is warranted to establish its value for more precise selection of therapeutic agents for patients with drug-resistant disease.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
M.P.D. thanks Edoardo Missiaglia for helpful discussions.
This work was supported by the Cancer League of the Canton of Zurich, the Empiris Foundation, the “Kinderkrebsforschung Schweiz” Foundation, the Sassella Foundation, the “Stiftung für Krebsbekämpfung,” the Swiss National Science Foundation (310030-133108, 310030-156407), the Fondation Panacée, the Human Hemato-Lymphatic Diseases clinical research program of the University of Zurich, the German Childhood Cancer Foundation, the German Cancer Consortium, and the European Union’s Seventh Framework Programme for research, technological development, and demonstration (Grant agreement No. 278514 IntReALL). P.H. was supported by the Hungarian National Brain Research Program (MTA-SE-NAP B-BIOMAG) and Finnish TEKES FiDiPro Fellow Grant 40294/13. J.W.T. was supported by The Leukemia & Lymphoma Society, the V Foundation for Cancer Research, Gabrielle’s Angel Foundation for Cancer Research, and the National Institutes of Health, National Cancer Institute (5R00CA151457-04 and 1R01CA183947-01). V.S. is a Wellcome-Department of Biotechnology (India) Margdarshi Fellow. M.P.D. is a SystemsX fellow.
Authorship
Contribution: J.-P.B., B.C.B., V.F., M.P.D., and A.R. jointly designed the project; J.-P.B., B.C.B., J.W.T., V.F., A.R., and S.H.D. designed the experiments; P.H. supervised the microscopy platform setup and provided components of the image analysis pipeline; V.F., A.R., J.T., J.K., P.R.-P., P.V., O.P., S.J., B.M., S.H., J.W.T., S.E., and S.H.D. performed experiments; G.C., M. Schrappe, M. Stanulla, M.P.D., T.R., P.V., M.D., A.E.K., M.U.M., A.V.S., C.E., B.H.C., T.J.B., R.H.C., S.U., G.P.B., O.R.B., E.D.-F., M.L.L., J.T., R.M., V.S., J.A.I., and S.H.D. provided reagents, analysis tools, samples and clinical data; M.P.D., V.F., A.R., P.R.-P., J.K., J.W.T., O.P., S.J., and S.H.D. analyzed data and prepared tables and figures; J.-P.B. and B.C.B. supervised research; and J.-P.B., B.C.B., M.P.D., and V.F. wrote the manuscript. All authors critically reviewed the manuscript for its content.
Conflict-of-interest disclosure: T.R. is a full-time employee of Novartis Pharma AG. The remaining authors declare no competing financial interests.
Correspondence: Jean-Pierre Bourquin, University Children’s Hospital Zurich, Steinwiesstr 75, CH-8032 Zurich, Switzerland; e-mail: jean-pierre.bourquin@kispi.uzh.ch.
References
Author notes
V.F., M.P.D., A.R., B.C.B., and J.-P.B. contributed equally to this work.


![Figure 4. Distinct drug activity patterns can be detected for individual samples and patient groups of interest. (A) RR PDX (n = 12) samples exhibit general resistance to conventional clinical compounds but remain sensitive to some experimental drugs. (B) Primary RR patients (n = 5) tested before last salvage therapy demonstrate persistent resistance to standard chemotherapy and individual sensitivity to experimental molecules. All responses are represented as IC50 (log[nM]) and are compared with other diagnostic and relapse ALL patients depicted in the background. **P < .005 (2-sided Student t test); *P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/11/10.1182_blood-2016-09-738070/4/m_blood738070f4.jpeg?Expires=1768053951&Signature=3aarTi2GRI2Oxdd68TsXLLpS8bSKBrgF2EfocWoMCm~oZVqB~JL-mUX66vFQz~A49oWyjacUTDlSDEwootmu7364LldlPkg23-2~iaFkuvuJJlppwR233kWAwtmYZiFXkkuLj8yLOnOxd12uB7AG~Hlzsk6zJssPdLIThT0gqhnei6h3Bdj1TjI5n3L5oUH~PE220~Kt36~7wnSVLUXNhTAsaU4qENuxBX23UhmJn31pbQmcTLsKQtGgn8plkYjsmXbCRzXx~p9EEiliQBbZVbEmCwB-rwCoD~-QARBl5~FlLfv8PU-yA1kmMi5CloyYC4~HrT~m3Sr5hd6JQaxlaQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)