Key Points
Patient-derived leukemia xenografts are highly polyclonal and show stochastic and clone-size–driven selection.
Leukemia clones are asymmetrically distributed in xenografts and preferentially localize to the bone marrow or to extramedullary sites.
Abstract
Genetic and phenotypic heterogeneity of human leukemia is thought to drive leukemia progression through a Darwinian process of selection and evolution of increasingly malignant clones. However, the lack of markers that uniquely identify individual leukemia clones precludes high-resolution tracing of their clonal dynamics. Here, we use cellular barcoding to analyze the clonal behavior of patient-derived leukemia-propagating cells (LPCs) in murine xenografts. Using a leukemic cell line and diagnostic bone marrow cells from 6 patients with B-progenitor cell acute lymphoblastic leukemia, we demonstrate that patient-derived xenografts were highly polyclonal, consisting of tens to hundreds of LPC clones. The number of clones was stable within xenografts but strongly reduced upon serial transplantation. In contrast to primary recipients, in which clonal composition was highly diverse, clonal composition in serial xenografts was highly similar between recipients of the same donor and reflected donor clonality, supporting a deterministic, clone-size–based model for clonal selection. Quantitative analysis of clonal abundance in several anatomic sites identified 2 types of anatomic asymmetry. First, clones were asymmetrically distributed between different bones. Second, clonal composition in the skeleton significantly differed from extramedullary sites, showing similar numbers but different clone sizes. Altogether, this study shows that cellular barcoding and xenotransplantation providea useful model to study the behavior of patient-derived LPC clones, which provides insights relevant for experimental studies on cancer stem cells and for clinical protocols for the diagnosis and treatment of leukemia.
Introduction
Leukemia is thought to arise through a Darwinian type of clonal evolution, in which acquisition of genetic mutations drives the selection and progression of preleukemic clones toward overt leukemia.1-5 Acquisition of mutations has long been considered a sequential, linear process.6 However, with the progress of sequencing technology and collection of massive amounts of data, a more complex picture of cancer clonal composition has emerged.7,8 Insight into clonal heterogeneity of leukemia and its evolution is of utmost importance because it may fuel emergence of novel, clinically relevant biological features, such as more aggressive growth or drug resistance.9 However, especially in acute lymphoblastic leukemia (ALL), many aspects of clonal evolution are subject to ongoing debate. For instance, the existence of leukemia stem cells (LSCs), their frequency within the total leukemic population, and the dynamics of their clonal offspring remain unknown.10,11
A crucial aspect of studying clonal evolution of leukemia is how to define, identify, and trace individual LSC clones. Currently, identification and enumeration of LSCs is hampered by the lack of phenotypic markers that unambiguously discriminate LSCs from the bulk leukemic cell population.10,12,13 Therefore, LSCs are commonly defined by their capacity to clonally expand and propagate the disease in immune deficient xenografts. Although the term clone is commonly used, its definition is instrumental for the interpretation of clonal tracking studies. Conceptually, a clone represents a group of cells that descends from a common ancestor and therefore shares certain heritable and unique genetic features. Experimentally, clones are identified by detecting those features (markers). Clonal markers can be cell-intrinsic (eg, genetic mutations, immunoglobulin heavy chain rearrangements), induced by cell manipulation (eg, vector integration, barcodes) or by the experimental design (eg, limiting dilution or single cell-sorting strategies), and should be defined based on the particular experimental question.14,15 In the present study, we use cellular barcodes to mark and trace patient-derived leukemia cells. We use the term clone to refer to a group of cells that carry the same barcode and therefore derive from the same ancestor, which we call the “leukemia-propagating cell” (LPC).
Previous studies investigating clonal evolution of leukemia have relied on (deep) sequencing of somatic mutations, either in bulk cell populations4,16 or in single leukemia cells.17 These studies have been instrumental to demonstrating the existence of clonal heterogeneity,4,10 identifying certain driver mutations,18 and reconstructing leukemia clonal architecture.4,8 However, these studies generally provide “snapshots” of mutational landscapes at certain time points (eg, diagnosis and relapse), and complex mathematical models are used to reconstruct clonal ancestry.19,20 To validate current views on the clonal pathogenesis of leukemia, therefore, there is a need for complementary strategies to ultimately derive a quantitative description of individual leukemia clones and the competitive behavior of their clonal offspring.
Lineage tracking using cellular barcodes provides such a strategy, allowing for simple, high-resolution tracking of individual clones over time.21-23 Cellular barcoding relies on the viral integration of random DNA sequences of fixed length (“barcodes”) into the genome of target cells.22,24,25 After transplantation of barcoded cells, their offspring can be traced by quantifying the barcode abundance using deep sequencing.25 So far, few studies have used cellular barcodes to study the clonal dynamics of human leukemia, and they differ in their conclusions. In vitro, culture of barcoded patient-derived leukemia cells on human mesenchymal stromal cells resulted in polyclonal cultures that retained their clonal complexity for 4 to 6 weeks.26 In addition, barcoding of a KCL-22 chronic leukemia cell line allowed for the detection of small therapy-resistant subclones that would have gone undetected with standard next-generation sequencing strategies.27 In contrast, in vivo, leukemia induced by artificial oncogenic overexpression rapidly evolved toward mono- or oligoclonality.28,29 It is unclear to what extent the murine environment contributes to these differences30 and whether these models reflect the competitive biological behavior of patient-derived leukemia clones.
In this study, we applied cellular barcoding and xenotransplantation to comprehensively characterize the dynamics of LPC clones derived from pediatric patients with B-progenitor cell ALL (B-ALL). We identified hundreds of LPC that steadily contributed to the leukemic population in murine xenografts. Many clones were asymmetrically distributed, preferentially localized in either bone marrow or extramedullary sites. Overall, our results demonstrate that cellular barcoding is a reliable method to count and trace patient-derived LPC clones, which can be used to obtain important insights on leukemia clonal evolution.
Methods
Cells
The pediatric BCR-ABL+ SupB15 cell line was maintained in Dulbecco’s modified Eagle medium supplemented with 10% fetal calf serum (Stemcell Technologies, Grenoble, France) and 1% penicillin and streptomycin (Gibco, ThermoScientific, Waltham, MA). Cryopreserved diagnostic bone marrow cells from pediatric patients with B-ALL had been collected as part of routine diagnostics for suspected leukemia. Residual mononuclear cells left after clinical testing were cryopreserved for research purposes (Department of Pediatric Hematology/Oncology, University Medical Center Groningen). Written informed consent was provided by all patients and/or their caregivers. All procedures were approved by the Medical Ethical Committee of the University Medical Center Groningen.
Mice
Immune deficient Nod/SCID/IL2Rγ−/− mice were bred and housed at the Central Animal Facility of the Groningen University. All animal experiments were approved by the Groningen University Animal Care Committee and by the Dutch National Central Animal Experiments Committee.
Barcoded vector library
A barcode library was prepared as previously described, with the modification that each barcoded vector in the current library was isolated and stored separately as DNA prep and Escherichia coli stock.25,31 Details on barcode library production appear in supplemental File 1, available on the Blood Web site.
Barcode library validation
The SupB15 pediatric B-ALL cell line was transduced with a lentiviral barcoded pEGZ2 vector library containing ∼800 barcodes. Forty-eight hours after the initial transduction, cells were sorted based on green fluorescent protein (GFP) expression (43 × 106 cells, 13% GFP+), expanded in vitro to 18 × 106 cells, and then transplanted into sublethally irradiated NSG mice (2.5 × 106 cells/mouse; n = 5) or split into 5 parallel cultures (106 cells/parallel culture). In vitro cultures were maintained for 20 weeks. Twice weekly, 1 × 106cells of each passage were seeded into the next well (supplemental File 2). Remaining cells were stored as pellets at −20°C for barcode analysis.
Cellular barcoding of patient-derived leukemia cells
Diagnostic B-ALL cells were thawed rapidly. From each patient sample, part of the cells was transplanted directly into immune deficient NSG mice, and part was used for lentiviral barcoding. The direct transplant served as a control to assess loss of engraftment potential during the barcoding procedure and to allow for a potential second barcoding attempt upon euthanization (Figure 2A; supplemental Files 3 and 4). Cells for barcoding were resuspended into Stem Span medium (Stemcell Technologies) containing 10% fetal calf serum, recombinant human interleukin-7 (10 ng/mL, Preprotech, London, United Kingdom), TPO (100 ng/mL, Preprotech), FLT3-L (20 ng/mL, R&D Systems, Minneapolis, MN), and recombinant human SCF (50 ng/mL, R&D Systems, Oxon, UK). Twelve hours after thawing, Polybrene was added (2 µg/mL, Sigma Aldrich, Zwijndrecht, The Netherlands) and cells were transduced in 2 rounds of 12 hours with a lentiviral barcoded pEGZ2 vector library containing ∼800 barcodes (ALL-58) or ∼500 barcodes (ALL-70). Two days after the initial transduction, GFP+ cells were sorted using a MoFlo flow cytometer (Beckman Coulter, Woerden, The Netherlands) and transplanted.
Validation of the barcoding method in vitro and in vivo. (A) Experimental design. (B) Barcode composition in 1 representative cell culture; each color represents 1 barcode. (C) Quantification of barcode complexity in vitro. (D) Determinism in vitro barcode composition over time, calculated as Spearman rank coefficient between sequential passages of the same parallel culture. (E) In vivo human chimerism in xenografts of barcoded SupB15 leukemia cells, measured by flow cytometry for human CD19+ cells. One mouse did not engraft. (F-G) Barcode composition in blood at 5, 6, and 7 weeks after transplant and bone marrow (BM) of SupB15 xenografts. Each color represents 1 barcode; the height of the bars corresponds to the level of human chimerism. Depicted are 2 representative xenografts. (H) Comparison of barcodes in murine blood and in the in vitro culture over time. (I) Comparison of barcode clones in BM at the time of euthanization with in vitro clonality at the corresponding time points.
Validation of the barcoding method in vitro and in vivo. (A) Experimental design. (B) Barcode composition in 1 representative cell culture; each color represents 1 barcode. (C) Quantification of barcode complexity in vitro. (D) Determinism in vitro barcode composition over time, calculated as Spearman rank coefficient between sequential passages of the same parallel culture. (E) In vivo human chimerism in xenografts of barcoded SupB15 leukemia cells, measured by flow cytometry for human CD19+ cells. One mouse did not engraft. (F-G) Barcode composition in blood at 5, 6, and 7 weeks after transplant and bone marrow (BM) of SupB15 xenografts. Each color represents 1 barcode; the height of the bars corresponds to the level of human chimerism. Depicted are 2 representative xenografts. (H) Comparison of barcodes in murine blood and in the in vitro culture over time. (I) Comparison of barcode clones in BM at the time of euthanization with in vitro clonality at the corresponding time points.
Xenotransplantation of barcoded leukemia cells
Barcoded leukemia cells were transplanted into sublethally irradiated (1 to 2 Gy) NSG mice by IV injection into the retro-orbital plexus (ALL-58, primary transplant) or tail vein (all other transplants, supplemental Files 3 and 4). Patient sample ALL-58 required in vivo expansion to allow successful barcoding. Bone marrow from 2 xenografts of unbarcoded leukemia cells was barcoded and transplanted in 2 separate experiments. Transduction efficiencies were 0.4% (ALL-58.1) and 2.5% (ALL-58.2), resulting in cell doses of 35 000 (ALL-58.1) and 400 000 (ALL-58.2) per mouse. Patient sample ALL-70 was barcoded successfully immediately upon thawing at a transduction efficiency of 1% and transplanted at 14 000 cells/mouse (2 mice) or 140 000 cells/mouse (2 mice).
Leukemia development
Leukemia development was monitored based on clinical symptoms and monthly blood analysis. Differential blood counts were performed on a Medonic CA620 Hematology Analyzer (Boule Medical AB, Spanga, Sweden). Leukemic and GFP chimerism were analyzed by flow cytometry31,32 (supplemental Files 5 and 6). When mice developed symptomatic leukemia, they were euthanized by cervical dislocation under isoflurane anesthesia, and blood, bones, spleen, and liver were collected. Bones were isolated according to anatomic location: (1) sternum; (2) spine; (3) left femur and tibia (left hind); and (4) right femur and tibia (right hind). Single cell suspensions were prepared by crushing in lysis solution (NH4Cl) and/or filtering through a 100-M filter, as previously described.29,32 Cells were analyzed by flow cytometry (Figure 3B; supplemental File 5) and stored in pellets of 1 to 5 × 106 for barcode analysis.
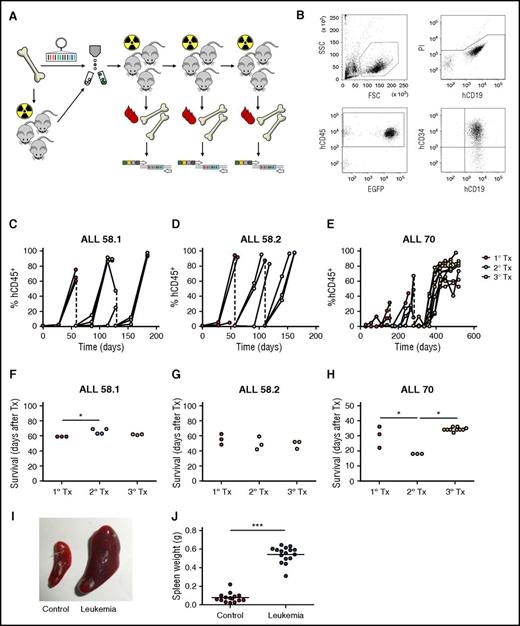
Lentiviral barcoding of primary pediatric leukemia. (A) Experimental design. (B) Gating strategy for flow cytometric detection of human leukemia cells in mouse blood. FSC, forward scatter; SSC, side scatter. (C-E) Percentage of human chimerism (%hCD45+ of live cells) in blood of serial xenografts of patient-derived leukemia. Connected dots represent samples from a single mouse. (F-H) Survival of serial xenografts of patient-derived leukemia. Each dot represents a single mouse. (I-J) Spleen size in control and leukemic mice. Each dot is 1 mouse. ***P < .001.
Lentiviral barcoding of primary pediatric leukemia. (A) Experimental design. (B) Gating strategy for flow cytometric detection of human leukemia cells in mouse blood. FSC, forward scatter; SSC, side scatter. (C-E) Percentage of human chimerism (%hCD45+ of live cells) in blood of serial xenografts of patient-derived leukemia. Connected dots represent samples from a single mouse. (F-H) Survival of serial xenografts of patient-derived leukemia. Each dot represents a single mouse. (I-J) Spleen size in control and leukemic mice. Each dot is 1 mouse. ***P < .001.
Barcode library production, deep sequencing, barcode data processing, and analysis and clonal analysis of nonleukemic hematopoiesis
For methods used for barcode library production, deep sequencing, barcode analysis, the random sampling model, estimation of stochasticity and determinism, clonal analysis of nonleukemic hematopoiesis, and general statistical analysis, see supplemental Files 1 and 7.
Results
Construction and validation of the barcode library
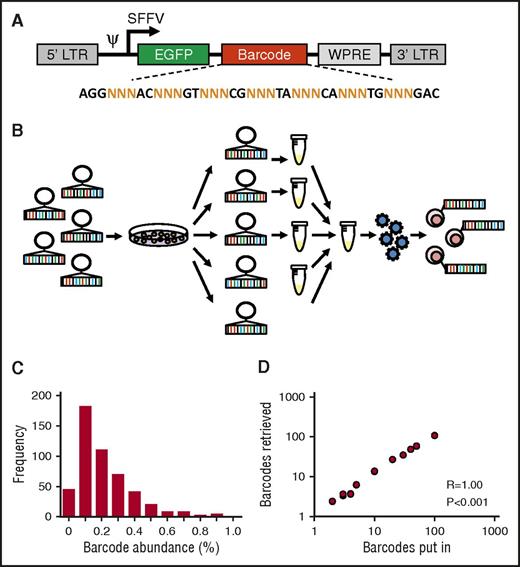
To allow for robust and reproducible clonal labeling, we generated ∼800 individual barcode preps that were pooled into libraries of varying complexity (Figure 1A-B). Individual preparation of barcodes is useful, because it allows for construction of libraries of known, equimolar barcode content and complexity. In addition, each barcode can be regenerated endlessly from E. coli stock, thus enhancing reproducibility of the library and of experimental data.22 Sequencing analysis of a 500-barcode library confirmed that most barcodes were evenly distributed across the library and present with frequencies of 0.1% to 0.2% (Figure 1C). To further validate our library and methods for barcode analysis, we mixed individual barcode DNA samples at equimolar ratios into libraries of different sizes and sequenced each mixture in duplicate. We found a nearly perfect 1:1 correlation between the number of barcodes put in each sample and the number of barcodes retrieved from deep sequencing results (Spearman r = 1.00, P < .001; Figure 1D).
Construction and validation of an equimolar barcode library. (A) Structure of the barcode linker. The linker contains a variable sequence part that consists of triplets of random (N) nucleotides that generate barcode diversity, flanked by fixed nucleotides that facilitate barcode retrieval in deep sequencing results. EGFP, enhanced green fluorescent protein. (B) Schematic representation of barcode library production and barcoding protocol. (C) Frequency distribution of barcodes in a 500-barcode library, determined by polymerase chain reaction and deep sequencing. (D) Correlation between the number of barcodes in samples submitted for sequencing and the number of barcodes retrieved in deep sequencing results.
Construction and validation of an equimolar barcode library. (A) Structure of the barcode linker. The linker contains a variable sequence part that consists of triplets of random (N) nucleotides that generate barcode diversity, flanked by fixed nucleotides that facilitate barcode retrieval in deep sequencing results. EGFP, enhanced green fluorescent protein. (B) Schematic representation of barcode library production and barcoding protocol. (C) Frequency distribution of barcodes in a 500-barcode library, determined by polymerase chain reaction and deep sequencing. (D) Correlation between the number of barcodes in samples submitted for sequencing and the number of barcodes retrieved in deep sequencing results.
Validation of the barcoding method for highly polyclonal samples
We subsequently aimed to validate our methods for clone identification and tracking in which we characterized the clonal behavior of a barcoded pediatric leukemia cell line (SupB15) in vitro and in vivo. We presumed that all cells from a cell line are equal, resulting in the highest possible barcode complexity, which would put the maximum challenge on our methods for clone retrieval and data analysis.
In vitro, we initiated 5 parallel cultures of 106 barcoded SupB15 cells each (details in “Methods”; growth curve in supplemental File 2). Barcode analysis revealed high clonal complexity in the first passages (711 barcodes at week 0, 696 at week 2), closely approaching the size of the used library (Figure 2B-C). Afterward, complexity slowly declined, yet remained polyclonal (Figure 2B-C).
To validate our methods in vivo, we transplanted 2.5 × 106 cells/mouse of the same barcoded cell line into sublethally irradiated NSG mice (n = 5). All xenografts developed a polyclonal leukemia, consisting of 7 to 71 clones in blood (Figure 2F-H) and 208 to 255 clones in bone marrow (Figure 2I). Notably, the number of clones retrieved from blood gradually increased over time. This may be due to presence of clones with more dormant behavior, which only reach the threshold for detection at later time points. In addition, the number of clones in bone marrow was markedly higher than in blood.
Comparison of in vivo and in vitro clonal complexity at corresponding time points demonstrated lower frequencies of LPC clones in vivo, both in blood (Figure 2H) and bone marrow (average 390 clones in vitro vs 231 clones in vivo; P < .0001, Figure 2I). To calculate the survival rate of the transplanted cells needed to achieve such a reduction in clone frequency, we used an in silico simulation script (supplemental File 7). We found that a survival rate of 1:3000 resembled our observations closely, resulting in approximately 200 surviving clones.
Altogether, these findings validate the usefulness of cellular barcoding for detection and high-resolution tracking of individual leukemia clones in vitro and in vivo.
Engraftment of patient-derived leukemia
We subsequently aimed to use cellular barcodes to characterize the behavior of patient-derived leukemia clones. Diagnostic bone marrow cells of 6 patients with B-ALL were subjected to barcoding and transplanted into immune deficient NSG mice. Two samples engrafted and caused GFP+ leukemia (Figure 3A, Table 1; details on experimental procedures in supplemental Files 2 and 3). The remaining 4 samples did not produce any human chimerism up to 28 weeks after transplantation.
Leukemia development was characterized by high chimerism for human CD19+CD34+ cells in peripheral blood and bone marrow (Figure 3B-E), of which the majority was GFP+ (supplemental File 6). In addition, leukemic mice displayed clinical symptoms necessitating euthanization (Figure 3F-H), and significantly enlarged spleen size (0.07 vs 0.54 g, P < .001, Figure 3I-J) with >90% CD19+CD34+ B cells (not shown). Interestingly, compared with nonengrafting samples, the engrafting samples were obtained from patients with more aggressive types of leukemia (BCR-ABL+ of delIKZF1; Table 1) and the dynamics of engraftment differed between these samples. Whereas xenografts of the BCR-ABL+ leukemia (ALL-58) all had >90% leukemic chimerism in peripheral blood within 6 weeks after transplant (Figure 2C-D), xenografts of the IKZF1-deleted leukemia (ALL-70) demonstrated slower leukemogenesis and lower levels of blood chimerism at euthanization, especially in primary xenografts (Figure 3E). No engraftment with human T cells (CD45+CD3+), granulocytes (CD45+CD16+), or mature B cells (CD45+CD19+CD34−) was observed in any mouse.
Because transduction efficiency of patient samples was rather low, we tested whether the transduction procedure selected for a specific population of more aggressive and/or quiescent clones. We found similar levels of chimerism and rates of survival between recipients of barcoded leukemia cells and nonbarcoded cells (supplemental File 8). Analysis of the sorted cells before transplantation showed that the patient samples were highly polyclonal, containing 402 (ALL-58.2) and 403 (ALL-70) barcode clones (supplemental File 9). This suggests that the barcoded LPC provide a biologically representative sample of the total leukemic population, although some selection may have occurred.
Patient-derived leukemia xenografts are highly polyclonal
Barcode analysis in blood throughout the experiment (Figure 4A-C) and bone marrow at euthanization (Figure 4D-F) demonstrated that patient-derived xenografts were highly polyclonal, consisting of tens to hundreds of LPC clones. Within individual xenografts, the numbers and types of clones were stable over time (Figure 4A-C), indicating their steady contribution to the leukemic population. Each primary xenograft was repopulated with different types of clones (Figure 4A-C). The number of retrieved clones was dependent on the patient sample (Figure 4A-F) and on the transplanted cell dose (Figure 4H-I). When more cells were transplanted, we retrieved a higher number of barcodes (Figure 4H), corresponding to calculated LPC frequencies ranging from 10−3 to 10−4 (Figure 4I). None of the patient-derived leukemia samples proliferated in vitro. The observed patterns of engraftment and clonal dynamics were different from nonleukemic cord blood CD34+ hematopoietic progenitors, which produced slower, multilineage engraftment, with an average of 8 clones (range, 3-15) in bone marrow of primary xenografts (supplemental File 10). Notably, despite the fact that considerably lower cell doses were used for patient samples than for transplantation of the SupB15 cells (35 000 to 400 000 cells for patient samples vs 2.5 × 106 cells for the cell line), the number of retrieved clones was significantly higher in patient-derived xenografts compared with cell line xenografts, which may reflect increased capacity of patient-derived clones to survive the xenograft barrier.
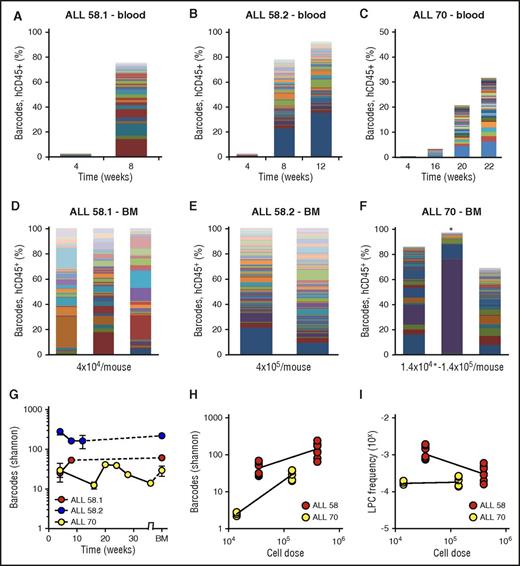
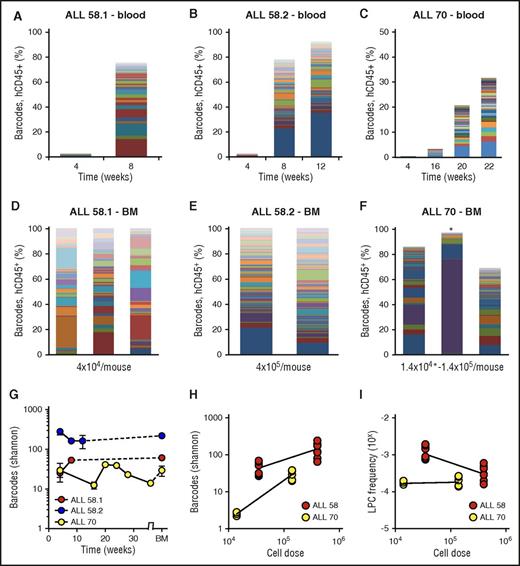
Clonal complexity of patient-derived leukemia subclones. (A-C) Clonal composition in blood of primary xenografts of patient-derived leukemia. Each bar represents 1 time point; each colored rectangle 1 barcode. The height of the bars corresponds to the %hCD45+ cells, determined by flow cytometry. (D-F) Clonal composition in BM of primary xenografts at euthanization. Each bar represents spine BM of 1 mouse. For ALL-70 (F), the mouse transplanted with 1.4 × 104 cells is marked with an asterisk. (G) Barcode complexity in blood and BM of primary patient-derived xenografts over time. (H) Plot of retrieved barcode complexity in primary xenografts vs transplanted cell dose. Each dot represents 1 sample (ie, 1 anatomic location). (I) Plot of LPC frequency in primary xenografts vs transplanted cell dose. LPC frequency was calculated by dividing the number of retrieved clones by the administered cell dose.
Clonal complexity of patient-derived leukemia subclones. (A-C) Clonal composition in blood of primary xenografts of patient-derived leukemia. Each bar represents 1 time point; each colored rectangle 1 barcode. The height of the bars corresponds to the %hCD45+ cells, determined by flow cytometry. (D-F) Clonal composition in BM of primary xenografts at euthanization. Each bar represents spine BM of 1 mouse. For ALL-70 (F), the mouse transplanted with 1.4 × 104 cells is marked with an asterisk. (G) Barcode complexity in blood and BM of primary patient-derived xenografts over time. (H) Plot of retrieved barcode complexity in primary xenografts vs transplanted cell dose. Each dot represents 1 sample (ie, 1 anatomic location). (I) Plot of LPC frequency in primary xenografts vs transplanted cell dose. LPC frequency was calculated by dividing the number of retrieved clones by the administered cell dose.
Heritability of clonal dominance after serial transplantation
We noticed striking differences in the clonal patterns in primary xenografts transplanted with the same patient sample (Figure 4). Theoretically, outgrowth of an LPC clone can be either stochastic (based on chance, thus poorly predictable) or deterministic (reflecting certain clone- of cell-dependent properties that confer a proliferative advantage). To investigate these possibilities, we assessed heritability and predictability of barcode patterns over time in vitro and in vivo in a serial transplantation model.
First, we revisited our in vitro cell culture data (Figure 2). As expected, in the first passages, clonal selection was largely stochastic, reflected by a low Spearman rank coefficient (Figure 2D). In later passages, clonal patterns became increasingly deterministic, meaning that clone order (by size) did not change much.
We subsequently assessed clonal selection patterns of patient-derived leukemia samples in serial transplantation experiments (Figure 5; supplemental File 11). We observed a strong reduction of clonal complexity in secondary and tertiary recipients compared with their donors (Figure 5A-B). With each serial transplantation, 30% to 90% of clones were lost (Figure 5A-C). Using the random sampling simulation model used previously for the cell line (supplemental File 7), we estimated survival rates of 1:20 to 1:100 from primary to secondary xenografts, or from secondary to tertiary xenografts.
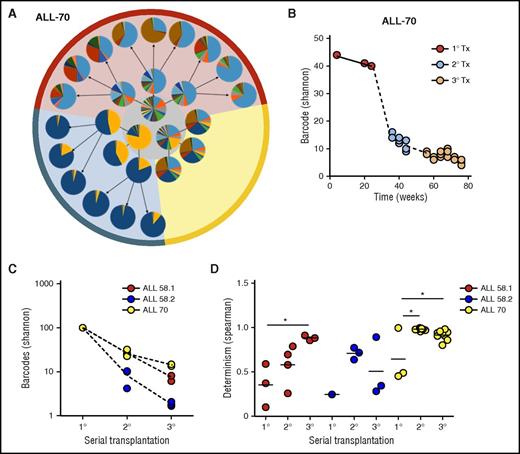
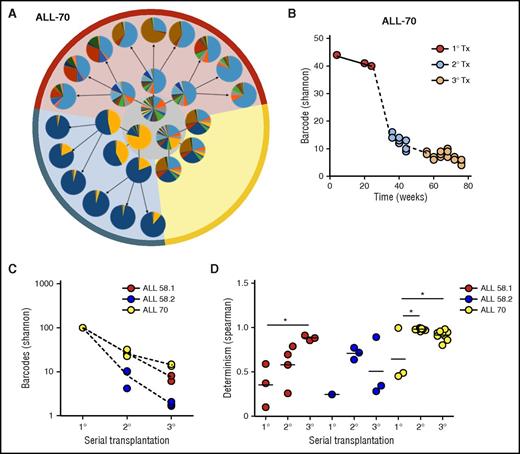
Clonal selection upon serial transplantation (Tx). (A) Barcode composition in serial xenografts of donor ALL-70. Each circle represents 1 mouse. Primary xenografts are depicted in the center gray circle; arrows connect each donor to its serial recipients. The background colors mark descendants of the same ancestor. (B) Quantification of barcode complexity in blood of serial xenografts of donor ALL-70. Each dot represents 1 mouse at a certain time point. Dashed lines connect the donor to its serial recipients. (C) Barcode complexity in spine bone marrow of serial recipients. Each dot represents 1 mouse. Detailed information on bone marrow composition of all recipients of ALL-58 can be found in supplemental File 11. (D) Degree of determinism in barcode composition between recipients of the same donor is expressed by Spearman rank coefficient. For example, for donor ALL-70, we calculated Spearman rank coefficients for barcode composition in spine bone marrow among the 3 primary recipients (A, gray circle), between the secondary recipients, and between their tertiary recipients. Only mice receiving the same cell dose (red and yellow area) are plotted.
Clonal selection upon serial transplantation (Tx). (A) Barcode composition in serial xenografts of donor ALL-70. Each circle represents 1 mouse. Primary xenografts are depicted in the center gray circle; arrows connect each donor to its serial recipients. The background colors mark descendants of the same ancestor. (B) Quantification of barcode complexity in blood of serial xenografts of donor ALL-70. Each dot represents 1 mouse at a certain time point. Dashed lines connect the donor to its serial recipients. (C) Barcode complexity in spine bone marrow of serial recipients. Each dot represents 1 mouse. Detailed information on bone marrow composition of all recipients of ALL-58 can be found in supplemental File 11. (D) Degree of determinism in barcode composition between recipients of the same donor is expressed by Spearman rank coefficient. For example, for donor ALL-70, we calculated Spearman rank coefficients for barcode composition in spine bone marrow among the 3 primary recipients (A, gray circle), between the secondary recipients, and between their tertiary recipients. Only mice receiving the same cell dose (red and yellow area) are plotted.
In contrast to the primary recipients, which were highly different in their clonal composition (Figure 5A, middle circle), we noted that clonal compositions were more similar in secondary and tertiary recipients (Figure 5A, outer circles). This was confirmed using unsupervised clustering analysis, in which recipients from the same donor clustered together (supplemental File 12), and by Spearman rank analysis, showing increasing similarity over serial transplantation (Figure 5D).
Spatial asymmetry of patient-derived leukemia clones
Despite increasing insight into the importance of the niche microenvironment for leukemia evolution,33-35 the anatomical distribution of individual leukemia clones is unknown.36 In our study, we compared barcode composition at euthanization of the mice in 4 skeletal sites (sternum, left hind leg, right hind leg, spine) and 3 extramedullary sites (liver, spleen, and blood). We identified 2 types of clonal asymmetry: (1) within the skeleton between different bone locations and (2) between the skeleton and extramedullary sites (Figure 6; supplemental Files 13 and 14).
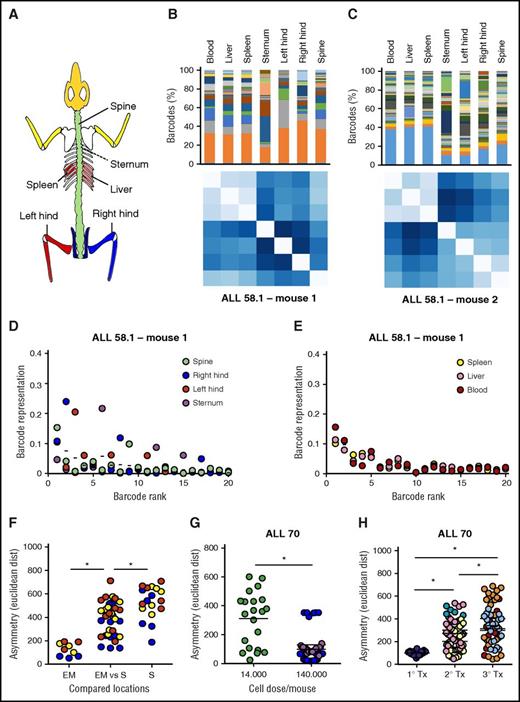
Asymmetric distribution of patient-derived LSC clones in murine xenografts. At euthanization of the mice, leukemia cells from patient-derived xenografts were isolated from 7 anatomic locations; barcode composition was determined by polymerase chain reaction and deep sequencing. (A) Schematic representation of the skeletal locations analyzed. (B,C) Anatomic distribution of barcoded clones in 2 representative xenografts of ALL-58.1. (Top) Relative barcode abundance per location. Each color is 1 barcode clone. (Bottom) Similarity in clonal distribution, assessed by Euclidean distance (for detailed information on methods, see supplemental File 8). (D-E) Relative clonal abundance per anatomic location. Clones are ranked based on their overall abundance (x-axis). The y-axis shows the relative abundance of each clone to the total pool in 1 location. Dots on the same vertical line reflect the size of a particular clone in different anatomic locations. The mean contribution of each clone is shown with a horizontal line. (F-H) Clonal diversity related to anatomical location (F); transplanted cell dose (G) or serial transplantation (H). Depicted are Euclidean distances over all combinations of anatomic locations within individual mice. Different colors are used to depict individual mice.
Asymmetric distribution of patient-derived LSC clones in murine xenografts. At euthanization of the mice, leukemia cells from patient-derived xenografts were isolated from 7 anatomic locations; barcode composition was determined by polymerase chain reaction and deep sequencing. (A) Schematic representation of the skeletal locations analyzed. (B,C) Anatomic distribution of barcoded clones in 2 representative xenografts of ALL-58.1. (Top) Relative barcode abundance per location. Each color is 1 barcode clone. (Bottom) Similarity in clonal distribution, assessed by Euclidean distance (for detailed information on methods, see supplemental File 8). (D-E) Relative clonal abundance per anatomic location. Clones are ranked based on their overall abundance (x-axis). The y-axis shows the relative abundance of each clone to the total pool in 1 location. Dots on the same vertical line reflect the size of a particular clone in different anatomic locations. The mean contribution of each clone is shown with a horizontal line. (F-H) Clonal diversity related to anatomical location (F); transplanted cell dose (G) or serial transplantation (H). Depicted are Euclidean distances over all combinations of anatomic locations within individual mice. Different colors are used to depict individual mice.
Within the skeleton, we noted that the relative abundance of certain clones differed between bones. For example, in Figure 6B (top), the dark gray clone dominated in the left hind leg, but not in any of the other locations. We visualized and quantified this in different ways. First, we compared the variation in clonal abundance per location around their average (overall) abundance (Figure 6D-E). We noted that the abundance of some clones was variable, whereas others were symmetrically distributed (Figure 6D). Interestingly, extramedullary sites did not show such asymmetry, demonstrated by minimal variation around average (Figure 6E). Second, we compared the cumulative variation in clone sizes using Euclidean distances (Figure 6B-C,Figure 6F-H), which also showed that extramedullary sites are highly similar to each other, whereas bones are more diverse (Figure 6F). Of note, the Spearman rank order of clones did not vary much by location (supplemental File 13), indicating that clonal asymmetry is primarily the result of variation in clone sizes, rather than their absolute presence/absence.
Notably, when all anatomic locations were crosscompared, we noted a higher degree of asymmetry in xenografts transplanted with a relatively low cell dose (Figure 6G). In addition, clonal anatomic asymmetry increased over serial transplantations, with the highest degree of asymmetry observed in the tertiary recipients (Figure 6H).
Together, these data demonstrate that LSC are asymmetrically distributed in xenografts, especially in situations or sites where the number of contributing clones is low.
Discussion
Patient-derived leukemia xenografts are highly polyclonal
Insight into the frequency of LPC and the dynamics of their clonal offspring is important for our understanding of leukemia pathogenesis and progression. Here, we describe the tracking of clonal dynamics of xenografted patient-derived ALL using cellular barcodes. The barcoding model has several advantages over previously used methods of clonal analysis: It is relatively simple, quantitative, and allows for prospective, high-throughput analysis of clonal complexity.20,37 In addition, it allows to address questions on leukemia clonal evolution that cannot be addressed in patients.
We demonstrate that patient-derived leukemia xenografts are highly polyclonal, consisting of tens to hundreds LPC clones, and provide insights into the mechanisms of their selection. We identify significant skewing in LPC clonal distribution, both within the skeleton and between the skeleton and extramedullary sites. Altogether, these findings are relevant for experimental studies on ALL biology and for clinical protocols for diagnosis and follow-up of patients with leukemia.
Cellular barcoding to characterize clonal evolution of leukemia
In contrast to acute myeloid leukemia, in which a stepwise model of tumor progression is well established,38,39 published studies regarding the cell of origin and its trajectory toward overt leukemia in ALL have been contradictory. Genome-wide analyses of DNA copy number alterations and loss-off-heterozygosity have identified various genetic aberrations associated with ALL progression (eg, PAX5, IKZF1, RAG1/2, CREBBP),40 which are present in different combinations and frequencies, indicative of the polyclonal nature of the disease. Importantly, in >95% of patients, at least part of the genetic aberrations present at diagnosis are retrieved at relapse, suggesting a common clonal ancestor.4 With the advent of modern sequencing technology, more genetic aberrations and an even higher degree of diversity will likely be uncovered. This will increase the challenge of discriminating driver mutations from innocent bystander effects, identifying mutations causing chemotherapeutic resistance, and reconstructing clonal hierarchies. Our results provide a novel perspective on ALL clonal evolution, which identifies and counts clones based on their proliferative capacity, rather than their mutational load (which may change over time). In the future, correlated clonal dynamics, simultaneously assessed by genomic aberrations and cellular barcodes, may provide a more thorough picture of leukemia clonal dynamics.
Patterns of clonal selection
The observation that leukemia clonal complexity is greatly reduced upon serial transplantation raises the question to which extent functional differences between individual leukemia clones dictate their survival and/or drive their selection. Here, we provide evidence for both stochastic (random) and deterministic (predictable) patterns of clonal behavior. We show that clonal selection is largely stochastic in initial in vitro passages and in primary xenografts and becomes increasingly deterministic over time. Deterministic behavior has also been shown to apply to other tumor types41 and is commonly explained by the existence of certain clonal inequalities driving their selection. However, the observed patterns of selection here may also be explained by the very properties of the barcoding method, which marks any cell and counts those that proliferate. Although initial marking will be random, upon proliferation, each clone will be represented by a large number of cells. Accordingly, serial transplantation/passaging will likely result in reproducible clonal hierarchies if the number of passaged cells representing each clone is sufficiently high. In the future, ascertainment of potential functional differences between cancer clones will need to take into account the stochastic patterns intrinsic to these types of experiments.
Asymmetric distribution of patient-derived leukemia clones
The observation that leukemia clones are asymmetrically distributed within the skeleton and between the skeleton and extramedullary sites is in line with earlier observations from our group, demonstrating skewed skeletal localization of murine normal (nonmalignant) hematopoietic clones.32 Based on this, one may speculate that interactions between LPCs and niche cells may define clone homing preferences32,42-44 ; however, our results support an alternative explanation, indicating that stochastic effects play substantial role in clone distribution. Accordingly, it may not be surprising that asymmetry increases when fewer LPCs are present. As an extreme example, a single LPC can only home to and engraft in 1 site. Nonetheless, it is striking that despite weeks of incubation and massive proliferation, the offspring of certain LPC still fail to mobilize and fully equilibrate over the body. Notably, once they do occupy extramedullary sites, they efficiently and equally distribute over different anatomic locations (resembling our previously reported granulocyte colony-stimulating factor mobilized hematopoietic stem cell).32 We did not find any evidence for selective clone preference to certain sites, suggesting that clonal distribution is stochastic42 and that, once engrafted, differences in proliferation and/or mobilization, perhaps dictated by the bone marrow niche, account for the observed asymmetry.
Asymmetry in spatial distribution of leukemia clones has important experimental and translational implications, suggesting that sampling of a single anatomic site may underestimate the clonal repertoire and may miss certain (potentially relevant) clones located elsewhere. Further identification of potential skeletal asymmetry in patients and of the clone- and/or niche-specific differences dictating this asymmetry will be relevant for the diagnosis, treatment, and follow-up of leukemia.
In conclusion, this study demonstrates that cellular barcoding and xenotransplantation is a simple and useful strategy to model the clonal evolution of human leukemia cells. In the future, combining cellular barcoding with in vivo chemotherapy and/or genomic mutation analysis will provide a powerful tool to study the clonal evolution of chemotherapeutic resistance and to optimize patient treatment.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank H. Moes, G. Mesander, and R.J. van de Lei for expert cell sorting assistance; H. Schepers for assistance in setting up the transduction methods; and R. van Os for valuable discussions.
This study was supported by research funding from the University Medical Center Groningen Mandema stipend (M.E.B.), the European Research Institute for the Biology of Ageing startup grant (M.E.B.), and the Dutch Cancer Society (grants RUG 2014-6957 and RUG 2015-7964) (M.E.B.).
Authorship
Contribution: M.E.B., G.d.H., and L.V.B. designed the research. M.E.B., T.K., S.J., B.A., S.S., and L.V.B. performed the research. E.d.B. contributed vital reagents and patient samples. M.E.B., E.Z., and L.V.B. analyzed the data. M.E.B., G.d.H., and L.V.B. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mirjam E. Belderbos, Princess Máxima Center for Pediatric Oncology, Room KE.01.129.2, PO Box 85090, 3508 AB Utrecht, The Netherlands, e-mail: m.e.belderbos@prinsesmaximacentrum.nl; and Gerald de Haan, Department of Stem Cell Biology and Ageing, European Research Institute for the Biology of Ageing, University Medical Center Groningen, Building 3226, Room 03.34, PO Box 197, Internal Zip Code FA50, 9700 AD Groningen, The Netherlands; e-mail: g.de.haan@umcg.nl.