Abstract
Hematopoietic cancers are often initiated by deregulation of the transcriptional machinery. Prominent among such regulators are the sequence-specific DNA-binding transcription factors (TFs), which bind to enhancer and promoter elements in the genome to control gene expression through the recruitment of cofactors. Remarkably, perturbing the function of even a single TF or cofactor can modulate the active enhancer landscape of a cell; conversely, knowledge of the enhancer configuration can be used to discover functionally important TFs in a given cellular process. Our expanding insight into enhancer function can be attributed to the emergence of genome-scale measurements of enhancer activity, which can be applied to virtually any cell type to expose regulatory mechanisms. Such approaches are beginning to reveal the abnormal enhancer configurations present in cancer cells, thereby providing a framework for understanding how transcriptional dysregulation can lead to malignancy. Here, we review the evidence for alterations in enhancer landscapes contributing to the pathogenesis of leukemia, a malignancy in which enhancer-binding proteins and enhancer DNA itself are altered via genetic mutation. We will also highlight examples of small molecules that reprogram the enhancer landscape of leukemia cells in association with therapeutic benefit.
Introduction
Leukemias are cancers marked by aberrant transcription. Sequencing of acute myeloid leukemia (AML) genomes revealed a preponderance of DNA mutations occurring in genes related to transcription, chromatin regulation, and DNA methylation.1,2 Transcriptional deregulation is also central to lymphoid malignancies, as leukemias in this lineage are frequently marked by B- or T-cell–specific transcription factor (TF) mutations.3-6
However, mutations in protein-coding genes may not completely capture the means by which transcription is dysregulated in leukemias. Broader DNA sequencing efforts have revealed that only 2% of the human genome codes for proteins, and the majority of disease-associated DNA sequence variants identified in genome-wide association studies (GWASs) map to this noncoding space.7-11 An estimated 88% of disease-associated single-nucleotide polymorphisms (SNPs) in the National Human Genome Research Institute catalog of GWASs are found in noncoding regions of the genome.10,12 Noncoding SNPs have been implicated in numerous disease processes, including variation of fetal hemoglobin levels in sickle cell anemia13-15 and the risk of developing both childhood and adult leukemias.16-18 Understanding these regions of DNA is therefore critical to understanding the pathogenesis of many diseases, including hematopoietic cancers. While noncoding DNA sequences can be devoted to myriad functions, many of these elements function as cis-regulatory elements that influence transcription of vicinal genes. An important class of cis elements is enhancers, which are clusters of TF binding sites uniquely capable of influencing gene transcription over large genomic distances.
Enhancer elements are especially important to control transcription in a time-, stimulus-, cell type–, or developmental stage–specific manner, and the genes regulated by enhancers are often required in specific developmental or other carefully controlled contexts.19 DNA sequences within the enhancer are recognized by sequence-specific DNA-binding TFs, which recruit a number of proteins that enable transcription of target genes.20 These coactivators include histone-modifying enzymes such as p300/CBP, elongation-promoting proteins such as Brd4 and PTEF-b, and a large number of proteins that compose the preinitiation complex and ultimately promote RNA polymerase II activity.20 The presence of these proteins and their activities enables identification of enhancers via chromatin immunoprecipitation followed by deep sequencing (chromatin immunoprecipitation sequencing [ChIP-seq]) using a number of markers, including acetylation of histone 3 lysine 27 (H3K27Ac), monomethlyation of histone H3 at lysine 4 (H3K4me1), TFs, or coactivators such as BRD4, Mediator, and p300, or by DNA accessibility measurements.21-23
As mentioned above, enhancers can regulate gene transcription from a distance. The intervening sequences can be looped out to allow juxtaposition of enhancer and promoter DNA, which is thought to be essential for transcriptional activation.24-27 The development of chromatin conformation capture assays determined that this phenomenon occurs in cells with DNA loop stabilization by the cohesin complex and may occur prior to productive transcriptional activation.28-32 Enhancer function is typically confined within larger topological domains (TADs) of chromosomes, which have borders defined in part by binding sites for the architectural zinc-finger protein CTCF.33,34
The application of assays to comprehensively map enhancer activity in cancer cells has unveiled global reprogramming of enhancer activity associated with malignant transformation. Enhancer activity can vary between normal and malignant tissues and even within a disease. The repertoires of active enhancers in a cell type have been dissected to reveal important insights about the hematologic malignancies and define novel subsets of the disease that exhibit different behaviors and treatment responses (Table 1).
For example, ChIP-seq analysis of H3K27ac was used to profile the enhancer landscape of AML patient samples and cell lines and nontransformed hematopoietic cell lines.35 Several enhancer features were able to define subsets of AML, such as large enhancer clusters (aka superenhancers) at hematopoiesis-related genes or monocyte-lineage genes, depending on the subtype of AML.35 Profiling the enhancer landscapes identified an active superenhancer at the retinoic acid receptor α (RARA) locus in 25% of samples. The presence of this superenhancer was capable of predicting differentiation response to RARA-directed therapy despite a lack of genetic alterations of RARA, demonstrating a utility of enhancer mapping experiments in exposing vulnerabilities in cancer.35 A similar approach carried out in T-cell leukemias and lymphomas identified similarities between cancerous and activated T-cells, suggesting a link between immune cell activation and malignant transformation.36
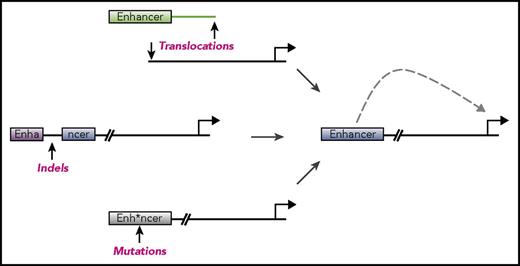
The enhancer landscape in hematopoietic malignancies can reveal important information about the origins and vulnerabilities of these diseases, but enhancers can also be altered to drive cancers. We discuss below specific mechanisms by which enhancer deregulation promotes the pathogenesis of hematologic malignancies, with an emphasis on illustrative examples rather than a comprehensive list of each alteration (Figure 1).
Mechanisms of enhancer dysregulation in hematopoietic malignancies. Enhancers are cis-regulatory elements that provide control over gene expression in specialized cellular contexts. A number of mechanisms have been discovered whereby enhancers are deregulated in order to drive cancers. These include chromosomal structural abnormalities such as translocations (top), insertions or deletions (middle), or point mutations (bottom). Any of these abnormalities can lead to enhancers aberrantly driving expression of genes important for cancer pathogenesis. These abnormalities can also lead to disruption of enhancers, shutting off genes and also contributing to disease.
Mechanisms of enhancer dysregulation in hematopoietic malignancies. Enhancers are cis-regulatory elements that provide control over gene expression in specialized cellular contexts. A number of mechanisms have been discovered whereby enhancers are deregulated in order to drive cancers. These include chromosomal structural abnormalities such as translocations (top), insertions or deletions (middle), or point mutations (bottom). Any of these abnormalities can lead to enhancers aberrantly driving expression of genes important for cancer pathogenesis. These abnormalities can also lead to disruption of enhancers, shutting off genes and also contributing to disease.
Translocations involving enhancers
One way for a cancer cell to deregulate oncogene expression is through chromosomal translocations that rearrange the position of enhancer elements in the genome. This mechanism of chromosomal reconfiguration can be frequently appreciated in hematopoietic malignancies, in part due to the normal utilization of DNA breakage during the development of these cell types, particularly lymphoid lineages.37
One of the first translocations identified that deregulates enhancers was found in human B-cell lymphomas harboring t(8;14) translocations involving the proto-oncogene MYC and the immunoglobulin heavy-chain (IgH) locus.38,39 Many of the breakpoints involved in the heavy chain-MYC translocations occur at sites of V(D)J recombination, suggesting that they occur in immature B cells still utilizing this type of recombination before maturing and generating lymphomas.40 V(D)J-like events also occur in lymphoma between the IgH locus and other oncogenes such as BCL2.40 Peripheral B-cell lymphomas instead appear to use a distinct regulatory region at the IgH locus that bears hallmarks of class-switching recombination, a process active in mature B cells typically found in peripheral tissues.40,41
Translocations involving MYC also occur in 42% of cases of the plasma cell malignancy multiple myeloma.42 The breakpoints in these cells bear no hallmarks of V(D)J or class-switching recombination, processes thought to be inactive in the terminally differentiated plasma cell.42 This suggests that the translocation event takes place after maturation of the B cell rather than as an early event that is carried through to the plasma cell stage. Taken together, these studies highlight how translocations of regulatory regions can be mined for important insights into the events that lead to the initiation of the tumor as well as into the biology of the cell of origin.
T-cell acute lymphoblastic leukemias (T-ALLs) can be driven by aberrant expression of the TF TAL1, and several enhancer rearrangements that aberrantly activate TAL1 expression have been discovered.43 Approximately 3% of T-ALLs harbor t(1;14) translocations that link TAL1 to the T-cell receptor (TCR) locus, thereby driving its expression through TCR enhancers.44 Deletions resembling V(D)J recombination events can also occur in T-ALL, and bring TAL1 to alternative regulatory elements that drive its expression.45,46
Translocations that place oncogenes under the control of novel enhancers can also be found in myeloid malignancies (Figure 2). Analysis of AMLs marked by inv(3) and t(3;3) karyotypes, known to involve the hematopoietic TF EVI1, revealed that the breakpoints contained a putative enhancer region, marked by H3K27ac, H3K4me1, and p300, that was essential for EVI1 activation in inv(3) AML cell lines.47-49 The region was confirmed to have enhancer activity in a luciferase-based assay and, by 4C analysis, loops to contact the EVI1 locus.48 On unaltered chromosomes, the enhancer regulates expression of GATA2, a TF critical for the maintenance of hematopoietic stem and progenitor cells.50 These chromosomal rearrangements therefore offer a means through which EVI1 overexpression and reduction of GATA2 expression can occur in progenitor cells that normally harbor active GATA2 expression, driving AML in a highly selective context.48
Inversion or translocation of chromosome 3 drives AML through enhancer dysregulation. An enhancer on chromosome 3 normally drives expression of the TF GATA2 in hematopoietic progenitor cells. Elsewhere on chromosome 3 is the TF EVI1. Structural abnormalities of chromosome 3 found in one type of AML, such as inv(3) or t(3;3), lead to the movement of this enhancer away from GATA2, resulting in loss of expression, and nearer to EVI1, driving its expression. A series of studies of these chromosome 3 abnormalities have determined that ectopic transcription of EVI1 is sufficient to drive AML, whereas concomitant loss of GATA2 expression accelerates this process.
Inversion or translocation of chromosome 3 drives AML through enhancer dysregulation. An enhancer on chromosome 3 normally drives expression of the TF GATA2 in hematopoietic progenitor cells. Elsewhere on chromosome 3 is the TF EVI1. Structural abnormalities of chromosome 3 found in one type of AML, such as inv(3) or t(3;3), lead to the movement of this enhancer away from GATA2, resulting in loss of expression, and nearer to EVI1, driving its expression. A series of studies of these chromosome 3 abnormalities have determined that ectopic transcription of EVI1 is sufficient to drive AML, whereas concomitant loss of GATA2 expression accelerates this process.
An interesting question that arose from these studies was whether the more important aspect of the enhancer translocation event is the activation of EVI1 expression or the loss of GATA2 expression. To address this question, one group generated a bacterial artificial chromosome with the GATA2 enhancer element in cis with an EVI1 transgene. Their results indicated that this mechanism of EVI1 activation was sufficient to drive leukemia.49 Follow-up work demonstrated that hemizygous GATA2 expression was insufficient to initiate leukemia, but, in tandem with the enhancer-driven EVI1 bacterial artificial chromosome, significantly accelerated leukemia initiation and progression.51 Thus, a single chromosomal event creates 2 distinct but synergistic transcriptional anomalies that underlie this type of AML.
Amplifications of enhancers to drive oncogenesis
Another mechanism of enhancer alteration to deregulate oncogene expression is via copy-number variation (CNV). An analysis of 160 T-ALL cases identified a region on chromosome 8q24 that appeared to harbor no protein-coding genes and yet was the site of recurrent focal duplications spanning a minimum conserved region of 40 kb. This region, ∼1.47 Mb downstream of the MYC gene, is bound in T-ALL by Notch1 and was found to loop to MYC to regulate its expression.52 This enhancer is required for normal T-cell development, as its knockout results in a hypoplastic and hypocellular thymus gland while preserving other hematopoietic lineages and without affecting MYC levels in other tissues.52 Deletion of this element in mice prevented initiation of T-ALL driven by a constitutively active NOTCH1, underscoring its importance to the disease.52 In a similar mechanism, a cluster of enhancers ∼1.7 Mb downstream of the MYC gene have been shown to loop to MYC and to regulate its expression in myeloid cells, and this region is contained in a number of rare copy-number amplifications in AML.53-55
Although CNVs commonly occur in protein-coding genes and increase expression of known oncoproteins, they have also been found in gene-poor regions with unclear tumorigenic value.56,57 When these gene-poor regions contain enhancers, however, a possible role of the amplification is to create a concentrated environment with multiple copies of the enhancers and their enhancer-bound activator and coactivator proteins, thus leading to similar effects as CNVs containing the target oncogenes themselves.52,58,59
Mutations and indels disrupting or creating enhancers
Although the previous examples highlight large chromosomal alterations that deregulate enhancer function, the DNA sequences comprising the enhancer itself can also be mutated to alter function in cancer cells. As mentioned above, T-ALLs can be driven by TAL1 expression mediated by enhancer translocations. However, T-ALLs lacking chromosomal rearrangements can also have enhancer-mediated deregulation of TAL1 expression. H3K27ac ChIP-seq uncovered a putative enhancer element that was only present in a subset of T-ALL samples, and analysis of the DNA sequence underlying this element identified small insertions and deletions that resulted in creation of a de novo MYB binding site.60 MYB binding to this altered DNA sequence led to the establishment of a novel super-enhancer that drives TAL1 expression and T-cell leukemia.
ChIP-seq enhancer profiling was also recently used to identify novel enhancers in T-ALL that drive aberrant expression of the oncogenic gene LMO1.61 Jurkat cells, which express high levels of this gene without an LMO1 gene rearrangement, were found to contain an enhancer element 4 kb upstream of the LMO1 gene.61 This element was bound by MYB at a site that contained a SNP that created a de novo MYB motif. Validation of this polymorphism in 187 pediatric T-ALLs identified the same mutation in 2% of patient samples.61 In addition to the MYB TF, this MYB motif also attracts other members of a core circuit of TFs defined in T-ALL, including TAL1, RUNX1, and GATA3 (all of which are absent from the wild-type version of this sequence), underscoring the centrality of MYB in establishing this oncogenic enhancer.61,62
Another example of somatic enhancer element mutations was observed in a noncoding region of chromosome 9p13 of the chronic lymphocytic leukemia (CLL) genome. Whole-genome sequencing analysis of 150 paired normal and CLL patient samples identified this region, which was then nominated as a potential enhancer region by DNase 1 sensitivity and H3K27ac/H3K4me1 ChIP-seq analyses.63 Chromosome conformation capture analysis showed that this region makes contacts with the PAX5 gene, and mutant samples expressed less PAX5 than CLLs with a wild-type 9p13 region.63 Clustered regularly interspaced short palindromic repeats disruption of this enhancer to model the effect of the mutation resulted in reduced PAX5 expression, supporting that the element regulates expression of this gene.63 The PAX5 gene itself is recurrently mutated in CLLs that have undergone transformation into diffuse large B-cell lymphoma, and these results highlight the ability of whole-genome sequencing to reveal new enhancers and novel mechanisms of leukemia pathogenesis.64,65
In addition to driving expression of oncogenes, germline variations in enhancer sequences can also be associated with risk of developing cancer. CLL is a leukemia with a strong inherited risk, with an approximately eightfold increase in the likelihood of eventual diagnosis if a person’s first-degree relative has CLL.66 GWAS has identified polymorphisms associated with increased risk for CLL at chromosome 15q15.1, ∼5.4 kb upstream of BMF, a negative regulator of BCL2.16 Loss of BMF leads to B-cell expansion and lymphomas in mice.67,68 4C analysis found that the GWAS polymorphisms occur in a region that contacts the BMF locus as part of a superenhancer and disrupt a RELA motif.69 This reduces enhancer activity and correlates with reduced expression of BMF in CLL expression data, providing one possible explanation for the increased risk of CLL in individuals with these SNPs.69
Enhancer deregulation by alteration of enhancer-binding factors
The core of an enhancer element is composed of clustered TF binding sites, which function cooperatively to recruit multiple cofactors to promote transcriptional activation. Although enhancers themselves can be altered to drive cancer, mutations in any of the proteins that bind to enhancer DNA can lead to powerful alterations of the enhancer landscape. Numerous components involved in enhancer-mediated gene regulation, including TFs and chromatin regulators, are recurrently mutated in leukemia to reprogram enhancer activity and promote cancer initiation and progression. For example, aberrantly active NOTCH1 was recently demonstrated to bind to an enhancer to drive expression of a long noncoding RNA in T-ALL, but not untransformed T cells.70 This T-ALL–specific long noncoding RNA, LUNAR1, subsequently co-occupies an intronic IGFR1 enhancer with NOTCH1 to promote IGFR1 expression, and LUNAR1 depletion leads T-ALL cell-cycle arrest via reduced expression of IGFR1.70
One dynamically regulated feature of enhancers, DNA methylation, is mediated by DNA methyltransferases. Mutations of DNA methyltransferase-encoding gene DNMT3A occur in ∼20% of AMLs and appear to be an initiating mutation in this disease.71 Analysis of the R882H variant of DNMT3A revealed that this mutant protein localizes to broad enhancer elements in the AML genome.72 At some of these sites, this causes a loss of 5-methylcytosine that leads to recruitment of histone acetyltransferases (HATs) and a switch to active chromatin.72 Affected sites appear to regulate genes related to hematopoietic stemness; thus, DNMT3A mutations alter enhancers to activate self-renewal and contribute to the development of AML.72-74
In contrast, more than one-quarter of AMLs harbor loss-of-function mutations in TET2, a gene encoding an enzyme that promotes DNA demethylation by hydroxylating 5-methylcytosine.75,76 Studies in TET2-mutant preleukemia cell models and AML patient samples showed that these mutations result in hypermethylated DNA preferentially at enhancers and in consequent suppression of expression of genes regulated by these enhancers, underscoring how hyper- or hypomethylation of distinct sets of enhancers can lead to AML.77
To regulate target genes, enhancers typically form DNA loops that can be stabilized by the cohesin complex. Loss-of-function mutations in several genes encoding cohesin proteins are frequently found in AML.2,78 These mutations appear to enforce a leukemia stem cell gene expression signature that results in impaired differentiation of all hematopoietic lineages and increased self-renewal ability.78-81 Detailed analysis revealed that the blockade of differentiation occurs preferentially in highly undifferentiated cells, including hematopoietic stem cells (HSCs) and multipotent progenitors, which exhibit an overall reduction of chromatin accessibility at transcription start sites and fewer nucleosome-free regions in their chromatin.79,80 However, there is also an increase of accessibility at genomic sites that harbor motifs bound by HSC TFs, including ERG, GATA2, and RUNX1, paralleled by increased occupancy of these TFs.79 It is unclear how loss of function of the cohesin complex, which likely regulates enhancer properties in all hematopoietic stages, affects only early cells such as HSCs. One possible explanation is that cohesin mutations preferentially impair the function of differentiation-promoting TFs over stemness-related TFs.
As mentioned above, enhancer activity is typically confined within TADs that can be defined by CTCF boundaries. CTCF mutations were recently identified in a subset of T-ALLs, suggesting that alteration of CTCF function may be important in this cancer.82 In T-ALL, deletions of the DNA encompassing CTCF binding sites have been identified that disrupt insulation of the TADs surrounding the TFs TAL1 and LMO2. These genes are then activated by enhancers in neighboring TADs, contributing to the transformation of T cells.83 Studies in gliomas revealed that a distinct mechanism, methylation of the underlying DNA, restricts CTCF binding and promotes enhancer deregulation in cancer.84 Early work demonstrated that imprinting at the H19/IGFR2 gene cluster could be carried out by methylation at a CTCF site in the cluster, and methylation of CTCF binding sites has been found to perturb CTCF occupancy across 19 different human cell lines, suggesting this mechanism is not unique to gliomas.85-88 Because DNA methylation is commonly deregulated in leukemia, it seems likely that methylation of CTCF sites could be a driver mechanism that occurs in this hematopoietic context as well.89
Therapeutic reprogramming of enhancers with small molecules
Enhancer elements enable spatial-, temporal-, and lineage-specific control of gene expression to a cell. By creating, disrupting, or moving enhancer elements, cancer cells are able to take the existing transcriptional apparatus and redirect it for the purpose of promoting cancer. Interfering with this aberrant transcription is therefore an attractive approach for therapy. If enhancer alterations enforce an oncogenic transcription program, then therapies directed at these aberrations may be able to reprogram these affected cells. In recent years, several novel therapies have aimed to exploit enhancer-associated dependencies, which are currently under investigation in preclinical models and in the clinic.
One of the most prominent examples are BET bromodomain inhibitors, which target a family of coactivator proteins, including BRD4, that have been found to be crucial in a number of malignancies. The first molecules to target BET proteins, such as JQ1 and I-BET, are acetyl-lysine mimetics that compete with acetylated proteins such as histone tails and TFs for binding with BRD4.90,91 BRD4 occupancy at regions of active transcription is thereby suppressed, leading to loss of transcriptional activity. In AML, for example, genetic or chemical inhibition of BRD4 led to rapid extinguishing of MYC expression, corresponding with loss of BRD4 from a large cluster of enhancers located downstream of the MYC gene.53,92
Some chromatin regulators disproportionately occupy a small number of enhancer elements, such as TRIM33 in B-ALL, and their inhibition therefore selectively perturbs a small number of transcripts in the genome.93 In contrast, BRD4 is globally present at regions of active transcription, yet only a subset of transcripts appears sensitive to BET inhibition.92,94 Based on observations like those at the large AML MYC enhancer, it was hypothesized that superenhancers, marked by high levels of H3K27ac, BRD4, and Mediator, define sensitivity of gene expression to BET inhibitors.94,95 However, enhancer size alone is a limited predictor sensitivity to BET inhibition, and a better correlate of transcriptional sensitivity to BET inhibition instead appears to be the degree of loss of Mediator from an element after treatment with a BET inhibitor.96,97 In AML, Mediator loss from chromatin following BET inhibition occurred at elements that harbored disproportionately more MYB occupancy, suggesting a role for this TF in influencing BET-inhibitor sensitivity.96 Modified BET inhibitors that degrade rather than simply block BRD4 show global effects on transcription that stand in contrast to the relative selectivity of conventional BET inhibitors.98 This result suggests that the BET protein itself may function similarly at all sites in the genome and raises the possibility that properties intrinsic to BET inhibitor–sensitive enhancers, such as the on/off kinetics of such sites in the genome, may be unique. Thus, the selectivity underlying a BET inhibitor’s therapeutic window might only be achieved by competitive blockade of BRD4 activity in the cell.
Leukemias resistant to BET inhibitors have emerged in the laboratory setting through mechanisms that appear to restore MYC expression via Wnt machinery either establishing alternative enhancer elements or replacing BRD4 at key cis elements altogether.99,100 A deeper understanding of BET inhibitor resistance may yield useful insights into selective properties of these molecules.
Studies in erythroblast cell lines have revealed that inhibition of BET protein BRD2 disrupts CTCF insulation of enhancer activity, an additional property that may also contribute to the selective response of some enhancers to BET inhibitors.101 In the same cell line, BRD4 occupancy of an enhancer was not a good predictor of its JQ1 sensitivity, but the presence of a cooperating TF, GATA1, was a strong correlate of transcriptional response to BET inhibition.102
The pattern of BRD4 occupancy at enhancer elements in the T-ALL genome can also provide insight into acquired resistance to other targeted therapies. In an effort to understand why some T-ALLs failed to respond to NOTCH1-directed inhibitors, Knoechel et al were able to isolate cells that persisted in spite of exposure to a γ secretase inhibitor.103 In these resistant cells, they observed that there was a marked increase in chromatin compaction and repressive chromatin marks and a concomitant overall decrease in the enhancer marker H3K27ac. Profiling of regulatory elements in T-ALL thus revealed heterogeneity in the transcriptional makeup of individual pretreatment cells. These cells were noted to be sensitive to BRD4 knockdown in a short hairpin RNA screen and to BET inhibitors, and it was postulated that these cells were particularly dependent on BRD4 to maintain open, active chromatin at crucial cancer-promoting genes.103,104
The Mediator coactivator complex contains a single kinase subunit, CDK8, or its paralog, CDK19.105 These subunits were historically described as inhibitory, acting to restrain transcriptional activity when associated with the Mediator complex.105-107 Cortistatin A is a natural product that inhibits CDK8/19, which provides a pharmacological means by which transcriptional activity can be released from regulation by the Mediator kinases.108,109 This was used to demonstrate that hyperactivation of superenhancers could lead to growth suppression of AML cells, providing an alternative means of enhancer-directed therapy in this disease.110
Another strategy for enhancer modulation is inhibition of CDK7, a kinase subunit of the general TF TFIIH, with a small-molecule covalent inhibitor THZ1.111 T-ALL cell lines and primary patient xenografts are highly sensitive to THZ1 exposure.112 Unlike some types of cancer cells that suffered global reduction in transcriptional activity upon THZ1 treatment, more selective transcriptional effects occurred in T-ALL cells.112 Known master regulator TFs in T-ALL RUNX1, TAL1, and GATA3 were disproportionately affected by THZ1 treatment in T-ALL and are adjacent to superenhancer elements in this context, implying that transcription regulated by this type of element is vulnerable to perturbation of CDK7.112 Peripheral T-cell lymphomas were also found to be sensitive to THZ1 treatment, although in this cancer sensitivity appeared to be achieved via reduction in phospho-STAT3 protein, but not STAT3 transcript levels. This raises the possibility that additional mechanisms of efficacy may have a role in the sensitivity to THZ1.113
Maintenance of the H3K27ac mark, and of enhancer activity, is regulated by the balance between HATs and histone deacetylases (HDACs), and targeting these classes of proteins may enable reprogramming of enhancers. Recent development of potent small-molecule inhibitors of the p300 and CBP may allow targeting of these HATs, which localize to enhancers and acetylate histones and other enhancer-bound proteins, including TFs, and are required for the survival of AML.114-118 HDAC inhibitors (HDACi’s) have already been approved by the US Food and Drug Administration for use in patients with CTCL, yet fewer than half of all patients with the disease respond to this therapy.119 To understand this response heterogeneity, Qu et al used chromatin accessibility data obtained in their study of CTCL versus nonmalignant T cells and found that in patient responders to HDACi’s, the inhibitors accentuated transcription from already open chromatin rather than causing an opening of closed chromatin.119 The combination of diminished accessibility of a JUN-AP1 TF network and elevated accessibility at CTCF sites was predictive in patient samples of response to an HDACi.119 Thus, profiling of regulatory elements in a given cancer can offer insights that enable investigators to understand not only the disease itself but also how it will respond to therapies, especially those directed at enhancer elements.
Concluding remarks
Genomics studies are providing volumes of data that link regulatory DNA sequence alterations with cancer. Focusing on protein-coding regions of DNA has led to many advances in our understanding of cancers and exposing novel treatment strategies. However, most of the variants unearthed by GWAS lie in the noncoding genome, and this space will undoubtedly continue to provide powerful insights. Here, we have highlighted the many ways in which noncoding regulatory elements in the genome can be mutated, translocated, amplified, or deleted to promote leukemia and other hematopoietic cancers. However, even in the absence of DNA sequence alterations, understanding the enhancer configuration in a tumor sample can provide crucial information about the cell state and active signaling pathways, explaining how individual tumors will respond to therapy. With an expanded effort to advance enhancer-directed therapies, mapping the enhancer configuration of a cancer may one day be as important as sequencing the exomes.
During the course of treatments, cancer cells accumulate new somatic mutations and chromosomal abnormalities that can contribute to development of resistance and result in relapse.120-126 How the enhancer landscape changes in similarly treated cells is unknown. If changes in enhancers contribute to resistance, then perhaps enhancer-directed therapy would reverse or prevent this evolution of resistance to other targeted therapies.
As therapeutics that reprogram enhancer activity progress in the clinic, we require a deeper understanding of molecular mechanisms of these agents. A fundamental unanswered question is the molecular basis for specificity. In other words, why are chromatin regulators that bind pervasively across the genome disproportionately required for a subset of cis elements? Understanding the molecular principles of specificity might provide much needed biomarkers that predict patients that will have exceptional responses in the clinic.
Acknowledgments
The authors thank members of the C.R.V. laboratory for advice and critical discussions. They also thank all of the investigators who have contributed to the field and the many ideas presented herein and apologize to those whose work could not be cited owing to space constraints.
C.R.V. is supported by the Leukemia and Lymphoma Society, the Pershing Square Sohn Cancer Research Alliance, the Starr Cancer Consortium, and the National Institutes of Health, National Cancer Institute grants RO1 CA174793 and 5P01CA013106.
Authorship
Contribution: A.S.B., B.L., and C.R.V. wrote the manuscript.
Conflict-of-interest disclosure: C.R.V. is an advisor to KSQ Therapeutics and receives research funding from Boehringer-Ingelheim. The remaining authors declare no competing financial interests.
Correspondence: Christopher R. Vakoc, Cold Spring Harbor Laboratory, 1 Bungtown Rd, Cold Spring Harbor, NY 11724; e-mail: vakoc@cshl.edu.