Abstract
The genome is constantly attacked by genotoxic insults. DNA damage has long been established as a cause of cancer development through its mutagenic consequences. Conversely, radiation therapy and chemotherapy induce DNA damage to drive cells into apoptosis or senescence as outcomes of the DNA damage response (DDR). More recently, DNA damage has been recognized as a causal factor for the aging process. The role of DNA damage in aging and age-related diseases is illustrated by numerous congenital progeroid syndromes that are caused by mutations in genome maintenance pathways. During the past 2 decades, understanding how DDR drives cancer development and contributes to the aging process has progressed rapidly. It turns out that the DDR factor p53 takes center stage during tumor development and also plays an important role in the aging process. Studies in metazoan models ranging from Caenorhabditis elegans to mammals have revealed cell-autonomous and systemic DDR mechanisms that orchestrate adaptive responses that augment maintenance of the aging organism amid gradually accumulating DNA damage.
DNA damage drives the aging process
Aging is a nearly universal property of life forms ranging from single-cell bacteria to humans. Fundamentally, aging is the default fate of life forms whose building blocks (DNA, RNA, and proteins) are constantly subjected to chemical alterations that impair their function. Although RNA is chemically more vulnerable than DNA, RNAs are usually rather rapidly turned over, and the consequences of damaged RNA molecules are temporarily restricted. Damaged proteins are either kept in shape by chaperones or are degraded, and their amino acids are recycled through the ubiquitin proteasome system or autophagy. But damaged proteins aggregate during aging and result in functional deterioration, especially in neurons where they give rise to dementia, including the most prevalent type, Alzheimer’s disease. The consequences of DNA damage, however, are much more widespread because DNA contains the information for all of the RNA and proteins a cell produces.
It has been estimated that tens of thousands of damaging events occur each day in every single one of our cells.1 Genotoxic attacks can originate from extrinsically inflicted radiation damage or chemicals as well as from endogenous sources such as metabolic byproducts or reactive oxygen species (ROS). Oxidative modifications and single-strand breaks (SSBs) are the most frequently encountered lesions and are rapidly repaired by base excision repair (BER) and DNA ligases, respectively.2,3 Both systems rely on the redundancy of several glycosylases that initiate BER and numerous ligases, because complete failure to repair those lesions is incompatible with life. Helix-distorting lesions are removed by nucleotide excision repair (NER), a highly complex repair pathway with intriguing connections to cancer and the aging process, which we discuss later for their conceptually instructive character.4 Even highly cytotoxic double-strand breaks (DSBs) and interstrand crosslinks occur on a daily basis. Those lesions are particularly dangerous because they impair the cell’s ability to replicate DNA and segregate the chromosomes, potentially giving rise to aneuploidy, which results in functional distortion of the genetic programs of the cell.5 The 2 major routes for DSB repair are nonhomologous end joining (NHEJ) and homologous recombination (HR), but even in their complete absence, there are other options such as backup end joining or microhomology-directed repair.6,7
The distinction between the modes and results of NHEJ and HR make an interesting point about constraints on genome maintenance in the context of other cellular functions. The rapid NHEJ process is used mostly when cells are not replicating their DNA and when no undamaged repair template is available. Before the end joining, the broken chromosome ends are resected, potentially because a high-energy ionization event produces ROS that inflict base damage adjacent to the break site and aid re-ligation. Given that only a fraction of the human genome encodes for genes, the local loss of a DNA stretch might be tolerable in most instances.8,9 Speed at the expense of accuracy is particularly important when cells divide rapidly. In contrast, HR is highly accurate because it uses the homologous sequence as a repair template. However, during replication, HR might also boost genome instability when replication forks collapse amid the engagement of chromosomes in the HR process.10,11 HR is mostly used during late S and within the G2 phases of the cell cycle.12 Some of the most prominent tumor suppressor genes are operating in the HR process. Mutations in the BRCA1 and BRCA2 genes predispose to breast and ovarian cancer and are both involved in HR. Interestingly, despite the backup DSB repair pathways, a combination of HR and NHEJ deficiency renders cells highly susceptible even to endogenous levels of DNA damage. The synthetic lethality of defects in the HR and NHEJ pathways led the way for tumor therapies that are customized for HR defects that result from a BRCA1 mutation and use inhibition of the NHEJ process, for example.13 A clinical strategy for using the sensitivity of BRCA1-deficient cells is the application of catalytic PARP1 inhibitors, which results in replication-associated DSBs that cannot be resolved in the absence of functional HR.14
There are numerous instances in which speed matters over accuracy. The speed argument particularly applies to cells engaged in replication. Even though the replicative DNA polymerases have a low error rate because of their proofreading activity (which is accomplished through their exonuclease domain), occasional errors might slip through. Mismatch repair scans the genome after replication for such errors by detecting structural aberrations of mispaired nucleotides.15 Some DNA lesions (eg, helix-distorting lesions) cannot be circumvented by replicative DNA polymerases. In those cases, translesion synthesis polymerases take over.16 They handle the base pairing with less stringency but keep the replication forks going even though they tend to make mistakes.
Cancer and aging: the flip side of DNA damage
There are 2 principally distinct outcomes of DNA damage: erroneous repair and persistent DNA damage. Erroneous repair can lead to mutations and chromosomal aberrations, both of which are causal events in cancer development. In contrast, persistent DNA damage can block transcription and replication, thus hampering cellular functionality and promoting cellular senescence and apoptosis. Consequently stem cell compartments become depleted, tissues degenerate, and homeostasis declines; ultimately, persistent DNA damage drives the aging process.
Erroneous repair can result from mistakes in placing the correct nucleotides, aberrant recombination, or lesion bypass and could alter the genetic information in numerous ways. When a mutation inactivates a tumor suppressor gene such as p53, the checkpoints that control the proliferation rate of a cell lose their function.17 When distinct sections of a chromosome are erroneously joined, the promoters might aberrantly drive the expression of an oncogene. One prominent example for this is the MYC gene, which is normally expressed in a highly controlled fashion to drive cell cycle entry. However, when the V(D)J recombination during B-cell maturation fails and the Eμ enhancer that normally boosts the expression of the immunoglobulin heavy chain gene is fused to the MYC gene, the B cells are driven into uncontrolled proliferation.18 A recent study by the Nussenzweig laboratory revealed that even during interphase, DSBs that result from topoisomerase 2B activity at topologic domain boundaries of chromatin result in the chromosomal rearrangements observed in cancers.19
Through its mutagenic effect, DNA damage thus triggers tumor development. Importantly, the mutations need to affect genes that control the cell’s DNA damage response (DDR) because a functional DDR prevents the cells from uncontrolled proliferation. The DDR controls (through DNA damage checkpoints) circumstances under which cells may enter and proceed through the cell cycle.20 The tumor suppressor p53 plays an important role in controlling cellular proliferation in the context of DNA damage (Figure 1). Normally, p53 is a short-lived protein whose activity is tightly regulated by various processes, including transcriptional and translational control and posttranslational modifications (PTMs).21 Moreover, p53 is rapidly ubiquitylated by MDM2 for example and consequently targeted for degradation by the ubiquitin-dependent proteosomal system.22,23 However, in the presence of DNA damage, p53 is stabilized by DDR signaling. For instance, p53 is activated by several posttranslational modifications, including phosphorylation on serine-15 by the phosphatidylinositol kinase-related kinases ataxia-telangiectasia mutated (ATM) and ATM and Rad3-related (ATR) or on serine-20 by the checkpoint kinase (CHK2) at the N-terminal transactivation domain, which interfere with the inhibitory interaction between MDM2 and p53, rendering p53 stabilization and accumulation. The specific DNA binding activity of p53 can be further enhanced or fine-tuned by phosphorylation and acetylation at the DNA-binding domain and C-terminal regulatory domain, modulating the distinct cell fate decisions.24-27 Many regulatory events determine the quality of the p53 DDR response and there are probably many more waiting to be discovered.28
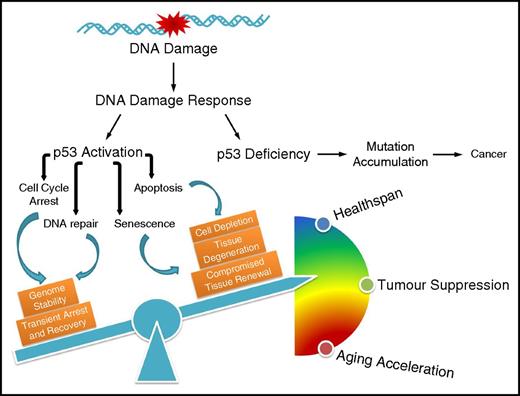
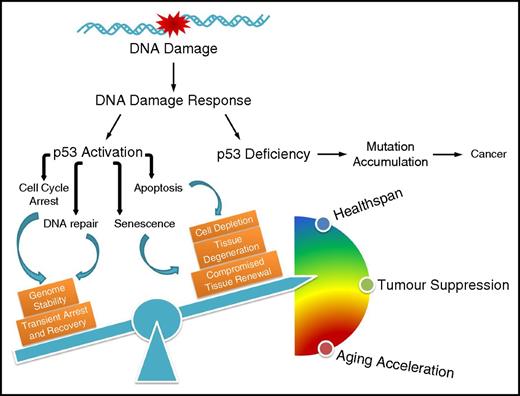
The influence of p53-mediated cell fate decision on cancer development and the aging process. Defective p53 leads to accumulation of mutations that drive carcinogenesis; on the contrary, p53 regulates diverse outcomes of the DDR, the fine-tuning of which balances healthspan, tumor suppression, and accelerated aging.
The influence of p53-mediated cell fate decision on cancer development and the aging process. Defective p53 leads to accumulation of mutations that drive carcinogenesis; on the contrary, p53 regulates diverse outcomes of the DDR, the fine-tuning of which balances healthspan, tumor suppression, and accelerated aging.
To exemplify the outcomes of the p53 response in a simplistic way, we can turn to its most ancestral form, which is the Caenorhabditis elegans p53-like CEP-1 protein.29,30 The homology to human p53 is mainly restricted to the DNA binding domain, with most of the hot-spot mutations associated with human cancers highly conserved. In addition, the residues that form the zinc finger are present in the nematode form. The most well characterized function of CEP-1 is the regulation of DNA damage–induced apoptosis in meiotic pachytene cells. The pachytene is the critical meiotic phase during which recombination takes place. During meiotic recombination, the SPO-11 endonuclease induces DSBs that are used for exchange with the homologous chromosome. When the cells reach the late pachytene stage, all DSBs should have been properly processed, and the Holliday junctions should have been resolved. However, when DSBs are still present, the meiotic recombination has failed and those cells cannot proceed to form genomic integer gametes. At this stage, the CEP-1 messenger RNA is translated because, before this point in meiosis, its translation has been repressed by the KH-motif RNA-binding protein and quaking homolog GLD-1.31 In addition to translational control, the stability of CEP-1 is also regulated through ubiquitylation.32 Once CEP-1 becomes available for activation through DDR triggered by the persistent DSBs, it transcriptionally induces the 2 proapoptotic BH3-only domain genes egl-1 and ced-13.33-35 As in mammals, the BH3-only domain proteins inhibit Bcl2, which in worms is encoded by the ced-9 gene. CED-9 then alleviates its sequestration of the Apaf1-like CED-4, which in turn sets off the CED-3 caspase to seal the fate of the cells carrying unprocessed meiotic DSBs or those induced by ionizing radiation.36
The apoptotic DDR regulation of p53 is highly conserved in humans in which the BH3-only domain genes p53-upregulated modulator of apoptosis (PUMA) and NOXA (named for “damage”) are induced in a similar fashion.37,38 In addition to triggering the apoptotic demise of genomically compromised cells, p53 also induces cell cycle arrest, best characterized by the transcriptional activation of the cyclin-dependent kinase inhibitor 1A (CDKN1A) gene p21 that inhibits mitotic cyclin-dependent kinases.39 Cell cycle arrest is critical for allowing cells time to repair the damage, during which p53 enhances several DNA repair pathways to facilitate the clearance of DNA lesions.40 p53 is involved in NER by transcriptionally upregulating the xeroderma pigmentosum complementation group C (XPC) and damage-specific DNA binding protein 2 (DDB2), both of which are important damage-recognizing factors required for initiating global-genome NER.41,42 p53 has also been implicated in the transcriptional control of the mismatch respair component human MutS homolog 2 (hMSH2) upon DNA damage.43,44 The expression of the Fanconi anemia complementation group C (FANCC) gene, the lack of which causes an inherited DDR deficiency syndrome, is also closely related to the promoter abundance of p53.45 Beyond transcriptional regulation, p53 also modulates BER in a transcriptionally independent manner. The activities of the pivotal BER enzymes 8-oxoguanine glycosylase and apurinic/apyrimidinic endonuclease are augmented by direct interaction with p53, thus enhancing the efficiency of excising oxidative DNA lesions.46 Moreover, a subpool of ATM-phosphorylated p53 upon ionizing radiation was found directly associated with the lesions, thus promoting DNA repair.47 In parallel to the repair pathways, p53 upregulates the p53-controlled ribonucleotide reductase (p53R2) for supplementing sufficient nucleotides during DNA re-synthesis, thereby further facilitating the DNA repair process.48
Amid unrepairable damage, however, p53 may also induce cellular senescence, thus permanently withdrawing cells from cycling yet keeping them metabolically active.49 The individual contributions of transient cell cycle arrest, cellular senescence, and apoptosis to the tumor suppressor function of p53 might vary depending on the cell type and other tumor suppressor gene and oncogene mutations that may be present. However, we can extract some conceptually instructive insights into the outcome of DDR. A failure to arrest the cell cycle either transiently or permanently amid DNA damage might fuel mutation rates and thus boost tumorigenesis. Even heavily genomically compromised cells might survive when p53 is dysfunctional.
Importantly, however, there is the other extreme of DDR. When p53 puts on the breaks too much, cells might not proliferate well enough to ensure a physiological level of tissue regeneration (Figure 1). This can be detrimental, especially for tissues that require proliferative activity of their stem and progenitor cell compartments such as those in the hematopoietic system.50 In addition, extraneous apoptosis might result in loss of tissue integrity regardless of which cell type is affected. Cellular senescence impairs proliferation and thus tissue regeneration and homeostasis; beyond that, it impacts neighboring cells and potentially even the organism through the senescence-associated secretory phenotype (SASP).51 SASP is a collection of heterogeneous cytokines that promote inflammation, tissue remodeling, and proliferation of recipient cells. In recent years, the contribution of senescent cells to aging has revealed the widespread physiological and pathological consequences of this cell fate. Highly pathological levels of cellular senescence have been observed in mice that carry mutations in the mitotic spindle checkpoint gene Bubr1.52 However, when the senescence pathway was abrogated by a mutation in the Ink4a-encoded p16 gene or the cells were eliminated when the p16 promoter was fused to a caspase that consequently eliminated senescent cells, the premature aging phenotype of the Bubr1 mutants was alleviated.53 Moreover, even the functional decline during normal aging could be slowed down by elimination of senescent cells.54 Currently, senolytic drugs are being tested (eg, Bcl-XL inhibitors) that could selectively eliminate senescent cells and supposedly slow the aging process of the organism.55 However, it is important to note that cellular senescence might also have positive functions; for example, the SASP component PDGF-A was demonstrated to support wound repair in murine skin.56
The outcome of enhanced p53 function was first demonstrated in 2 different mutant mice that expressed hyperactive forms of p53.57,58 Tyner and colleagues57 generated p53+/m mice, in which the m allele comprises a truncated form of p53 containing only exons 7-11 of the p53 coding sequence. With enhanced stability and transactivation activity of the m allele product, p53+/m mice were highly protected from tumors but at the expense of accelerated aging. Further analysis of the hematopoietic stem cells (HSCs) revealed a reduced number of proliferating HSCs with age in p53+/m mice, confirming that increased p53 activity may lead to decreased production of progenitor and mature differentiated cells from the stem cells thereby contributing to the aging phenotype.59 In mice that overexpress another truncated isoform of p53, p44, signs of premature aging were observed as early as 4 months of age and the mice consistently displayed a low incidence of cancer.58 The fine balance between keeping the brakes on proliferation and controlling cellular survival has been preserved in mice that carry an extra copy of the p53 gene (super-p53 mice).60 These mice are protected from cancer development, likely because they are equipped with 3 p53 alleles that are much less likely to lose heterozygosity. For extended lifespan, however, this was not sufficient and instead required an additional copy of the ARF tumor suppressor.61 The mice with increased basal level of p53 as a result of low expression levels of MDM2 (mdm2puro/Δ7-12 mice) were also cancer resistant but they had a normal lifespan,62 further echoing the notion that a balanced p53 level is of great significance for tumor suppression without accelerated aging.
When it comes to humans, previous studies have revealed a Pro/Arg polymorphism at amino acid residue 72 of p53 that alters the potential of inducing apoptosis.63-65 A meta-analysis of the published literature has indicated that carriers of the p53 Pro/Pro form, which is less potent in inducing apoptosis than the p53 Arg/Arg form, had increased survival albeit higher mortality from cancer.65 Another independent study recruiting more than 9000 participants demonstrated a higher overall survival for p53 Pro/Pro carriers and, most importantly, an increased survival after cancer or other life-threatening disease.66 However, contrasting results were obtained in a more recent study with an even larger population,67 leaving open the question of whether hyperactive p53 plays an unfavorable role in human lifespan.
Persistent DNA damage: a driving force of aging
While DDR prevents tumorigenesis, its constitutive activation, such as that in hyperactive p53 mutants, accelerates the aging process. The continuous activation of DDR arises by genetic mutations that augment DDR and can also result from DNA lesions that are not repaired and thus persist. For instance, critically shortened and thus unprotected telomeres are recognized as DNA DSBs.68 Indeed, the inability of telomerase to maintain the telomere length cause premature aging through activating the DDR and p53.69,70 Mice that are lacking the catalytic subunit or the RNA component of telomerase have shorter lifespans with early onset of aging phenotypes, even though in mice, these phenotypes require several generations of defective telomere extension to arrive at critical telomere shortening, which precipitates the progeroid pathologies.71,72 Premature replicative senescence also shortens lifespan in Ku80−/−-mutant mice that lack functional NHEJ.73 Intriguingly, the accelerated aging phenotypes of both animal models can be alleviated by the loss of p53, again underlining the pivotal role of p53 in DDR-mediated premature aging.74,75
Another prominent example that demonstrates the link between persistent DNA damage and premature aging comes from the autosomal genetic disorder Fanconi anemia (FA), which is characterized by progressive bone marrow failure resulting from the functional decline of hematopoietic stem and progenitor cells (HSPCs). In addition to their sensitivity to interstrand crosslinks,76,77 FA cells are hypersensitive to oxidative stress and have excessive levels of oxidative DNA damage.78,79 Ceccaldi et al80 further demonstrated that the unresolved DNA damage leads to constitutive activation of p53, which results in a p21-dependent G0/G1 cell-cycle arrest in HSPCs from FA patients. Remarkably, the depletion of p53 or p21 could alleviate the defects in HSPCs. Another example of the important role of p53 in controlling the DDR in HSPCs is provided by a mouse mutant that expresses the hypomorph p53515C allele in an Mdm2-deficient background. Here, p53 activation amid high ROS levels leads to exacerbated cell cycle arrest, senescence, and apoptosis in HSPCs.81
In genetic diseases caused by mutations in genome stability factors and also during normal aging, the ability of maintaining genome integrity declines, especially in highly regenerative tissues such as the hematopoietic system. By comparing transcription profiles of purified HSCs from mice age 2 to 21 months, Chambers et al82 have reported that with age genes associated with stress response were upregulated, whereas genes involved in maintaining genome integrity, including DNA repair genes, were downregulated. Consistent with this observation, the efficiency of DNA repair in aged HSCs is limited, leading to gradual accumulation of DNA damage and thereby attenuating the self-renewal and tissue regenerative capacity of HSCs.83-86
For understanding the role of persistent DNA damage in the aging process, congenital NER deficiencies have been particularly illuminating. NER recognizes helix-distorting lesions that are most prominently formed when short-wavelength UV light links adjacent bases to form cyclobutane pyrimidine dimers (CPDs) and pyrimidine-pyrimidone 6-4 photoproducts (6-4PPs).87 Indeed, CPDs are the primary instigator of UV-induced skin carcinogenesis.88 The lesions are recognized either by scanning through global genome NER (GG-NER) or when RNA polymerase II (RNAPII) stalls at a lesion and transcription-coupled NER (TC-NER) is activated. Both pathways then trigger a common NER pathway that verifies the damage, unwinds the double helix, incises on either side of the lesion, excises the stretch containing the damage, resynthesizes the gap, and finally ligates the remaining gap.
Congenital defects affecting one or the other recognition pathway could not possibly be more distinct.89 Xeroderma pigmentosum (XP) patients display pigmentation abnormalities, atrophic skin, and a several-thousand-fold elevated skin cancer susceptibility.90 In stark contrast, Cockayne syndrome (CS) patients suffer from postnatal growth retardation and neurologic defects, and they display numerous manifestations of premature aging but remain cancer free.91,92 Several NER genes are associated with XP but those involved in GG-NER most clearly show the most distinctive skin phenotypes and cancer susceptibility, whereas XP mutations associated with factors operating in the common NER pathway typically display neurodegenerative symptoms in addition.93 CS patients, in contrast, carry mutations in the CSA or CSB genes that operate in the initial steps of TC-NER. NER deficiencies are highly complex on both the molecular and the pathologic levels and are not restricted to XP and CS alone; however, they are highly instructive regarding the distinct outcomes of DDR.4 Whereas GG-NER defects are mutagenic when unrepaired lesions pass through replication where they might result in replicative errors or genome instability when replication forks collapse, TC-NER defects result in stalling RNAPII. At high levels of RNAPII stalling, transcription is blocked and cells might no longer fulfill their function or might even die. Whereas mutagenic DNA lesions fuel cancer development, persistent DNA lesions drive the aging process by hampering the DNA metabolism because replication and transcription are blocked.
Intriguingly, cells respond to transcription-blocking lesions even when they are detected at very low levels. Regardless of whether cells proliferate or are terminally differentiated, in response to those lesions, they reduce the expression levels of genes that are involved in the somatic growth axis, particularly the insulin-like growth factor-1 receptor (IGF-1R) and growth hormone receptor (GHR).94 Both receptors are central elements of the somatic growth axis, and their reduction has been established as a bona fide longevity assurance mechanism that extends lifespan in mice that lack the pituitary or the GHR gene.95-97 Progeroid (premature aging-like) mice that carry defects in NER also show reduced somatic growth gene expression and dampened IGF-1 levels.98-100 It is likely that cells respond by attenuating the somatic growth axis to ensure that the organism survives when DNA damage accumulates.101 Indeed, stress resistance (a feature strongly correlated with longevity) was observed in cells that acquired transcription-blocking lesions as they became exquisitely resistance to oxidative stress,94 similar to mice with reduced IGF-1R gene dosage.102
The consequences of this type of DDR have recently been studied in the nematode C elegans in which the mechanisms of longevity have been most extensively investigated. The FOXO transcription factor DAF-16 is normally kept inactive by insulin-like signaling but, consistent with IGF-1R dampening in mammals, it is activated upon transcription-blocking DNA lesions. Here, DAF-16 promotes developmental growth and maintains the integrity and functionality of tissues in adult animals even when the DNA damage persists.103 DAF-16 is a bona fide longevity assurance factor, and its involvement in DDR suggests a distinct mechanism of adaptations to genome instability. DNA damage tolerance by augmented maintenance of tissues is particularly relevant for cell types that are not proliferating, such as the adult nematode’s cells outside of the germline that are entirely postmitotic. Therefore, it seems that there are 2 types of longevity assurance mechanisms: (1) DNA repair systems that prevent and delay the accumulation of DNA damage and (2) lifespan regulators that determine the threshold to which DNA lesions are tolerated before they become detrimental. The latter are regulated by transcription factors such as DAF-16, which mediates gene expression programs that comprise stress resistance genes as well as developmental growth programs.103
Intriguingly, the consequences of DDR are not confined to the genomically compromised cell alone. Somatic growth signaling, for instance, is mediated through the secretion of IGF-1 and GH, which have endocrine activities, and SASP factors that exert non-cell-autonomous effects.50 In addition, p53 has non-cell-autonomous consequences when it regulates the cytokine secretion of tumor cells that determine whether they are cleared by macrophages.104 In C elegans, innate immune factors that are induced in response to DNA damage in germ cells trigger a systemic stress resistance program that elevates somatic endurance thus extending reproductive lifespan to allow the generation of offspring, once genome stability in the germ cells is reconstituted.105 It is important to gain more insights into the non-cell-autonomous regulation of DDR and which role p53 might play in this process.
Alleviating DNA damage responses
Experiments discussed above, especially those on additional p53 gene dosage and p53 ablation in several DNA repair deficiencies associated with exacerbated replicative senescence, have indicated that a balance in DDR is important for cancer suppression and longevity and this balance could potentially be targeted for promoting health in old age (Figure 1). In addition, modulations of the consequences of DNA repair defects on cellular metabolism have been suggested to alleviate pathologies resulting from genome instability. Xrcc1-mutant mice that are defective in SSB repair show highly elevated protein poly-ADP-ribosylation (PARylation) levels and develop neurologic disorders characterized by ataxia. Interestingly, genetic ablation of Parp1 could restore ADP-ribose levels, reduce neuronal loss, and alleviate the ataxia.106 One consequence of PARP1 activity is that excessive PARylation could dampen the NAD+ levels that result in defective mitophagy, which could be alleviated by replenishing NAD+, thus improving the health of progeroid NER and ATM mouse models.107,108 Elevated NAD+ levels have numerous consequences on cellular metabolism and could also promote longevity through sirtuin-mediated activation of the mitochondrial unfolded protein response and FOXO signaling.109 NAD+ could also alleviate PARP1 inhibition mediated by the NAD+ binding protein DBC1.110 It is thus conceivable that intervention strategies could promote longevity and healthspan by affecting the activity of genome maintenance systems.
Concluding remarks
DNA damage invariably occurs as a result of a plethora of endogenous and exogenous genotoxic insults. A complex arsenal of DNA repair mechanisms efficiently removes the lesions and ensures the maintenance of the soma during lifespans that are relevant for supporting the generation of offspring during the evolutionary history of a species. The capacity to keep genomes of somatic cells intact vanishes thereafter, which leads to the functional decline of cells and tissues. How germ cells retain the levels of DNA repair accuracy that has allowed maintaining the gene pool of a species for hundreds of millions of years is an intriguing and yet incompletely understood question. The aging organism responds to DNA damage by cell-autonomous and systemic DDR. The p53 gene is an important tumor suppressor and also an orchestrator of an extensive network of DDR factors that are relevant for adaptation to DNA damage in the aging organism. Studies in metazoan model systems have begun to shed light on how signaling pathways and endocrine axes respond to the buildup of DNA damage with aging. Approaches such as combining proteome assessments with studies of posttranslational modifications and metabolic alterations have begun to shed new light on the complexity of DDR and the consequences of persistent DNA damage in the aging organism.111 Results of further explorations will be critical to understanding how longevity assurance mechanisms respond to DNA damage and how they influence the aging phenotype and the etiology of aging-associated diseases.
Acknowledgments
This work was supported by a fellowship from the Cologne Graduate School on Aging (CECAD-CGA) (H.-L.O.), by the Deutsche Forschungsgemeinschaft (CECAD, SFB 829, SFB 670, and KFO 286) (B.S.), a starting grant from the European Research Council (ERC 260383), Marie Curie Initial Training Network (FP7 ITN CodeAge 316354, aDDRess 316390, MARRIAGE 316964), FLAG-ERA JTC 2015 (G-Immunomics, SCHU 2494/3-1), the German-Israeli Foundation (GIF 1104-68.11/2010), the Deutsche Krebshilfe (109453), the Bundesministerium für Bildung und Forschung (Sybacol FKZ0315893A-B), and the European Cooperation in Science and Technology (COST) action (BM1408, GENiE).
Authorship
Contribution: H.-L.O. and B.S. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Björn Schumacher, University of Cologne, Joseph-Stelzmann Str 26, D-50931 Cologne, Germany; e-mail: bjoern.schumacher@uni-koeln.de.