Key Points
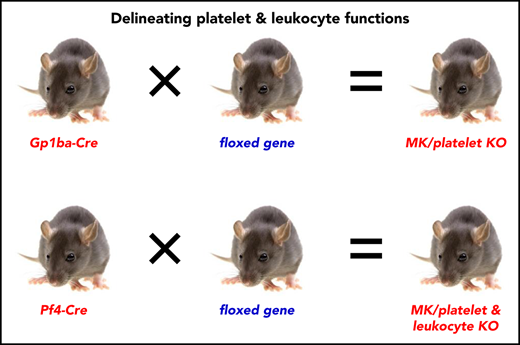
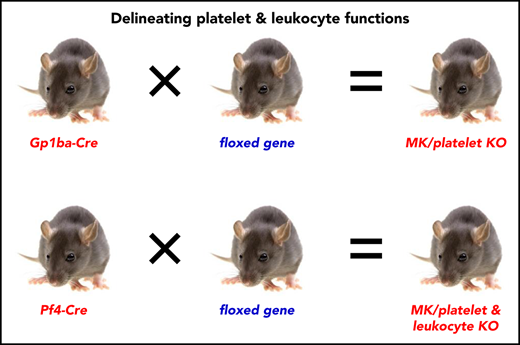
The Gp1ba-Cre mouse allows highly specific deletion of floxed genes in the megakaryocyte lineage.
The Gp1ba-Cre mouse enables differentiation of platelet and leukocyte functions.
Abstract
Conditional knockout (KO) mouse models are invaluable for elucidating the physiological roles of platelets. The Platelet factor 4-Cre recombinase (Pf4-Cre) transgenic mouse is the current model of choice for generating megakaryocyte/platelet-specific KO mice. Platelets and leukocytes work closely together in a wide range of disease settings, yet the specific contribution of platelets to these processes remains unclear. This is partially a result of the Pf4-Cre transgene being expressed in a variety of leukocyte populations. To overcome this issue, we developed a Gp1ba-Cre transgenic mouse strain in which Cre expression is driven by the endogenous Gp1ba locus. By crossing Gp1ba-Cre and Pf4-Cre mice to the mT/mG dual-fluorescence reporter mouse and performing a head-to-head comparison, we demonstrate more stringent megakaryocyte lineage-specific expression of the Gp1ba-Cre transgene. Broader tissue expression was observed with the Pf4-Cre transgene, leading to recombination in many hematopoietic lineages, including monocytes, macrophages, granulocytes, and dendritic and B and T cells. Direct comparison of phenotypes of Csk, Shp1, or CD148 conditional KO mice generated using either the Gp1ba-Cre or Pf4-Cre strains revealed similar platelet phenotypes. However, additional inflammatory and immunological anomalies were observed in Pf4-Cre-generated KO mice as a result of nonspecific deletion in other hematopoietic lineages. By excluding leukocyte contributions to phenotypes, the Gp1ba-Cre mouse will advance our understanding of the role of platelets in inflammation and other pathophysiological processes in which platelet-leukocyte interactions are involved.
Introduction
Platelets and leukocytes have closely interlinked functions. Platelets are increasingly recognized to contribute to inflammation,1 immunity,2,3 atherogenesis,4 cancer metastasis,5 and the separation of blood and lymphatic vasculature.6 Concomitantly, leukocytes play a critical role in thromboinflammatory diseases, including ischemic stroke, myocardial infarction, and deep vein thrombosis.7 Megakaryocyte (MK)/platelet-specific conditional knockout (KO) mouse models have become invaluable means of determining the molecular mechanisms regulating platelet production and pathophysiological functions in which they are involved. The current model of choice for generating MK/platelet-specific conditional KO mice is the Platelet factor 4-Cre recombinase (Pf4-Cre) transgenic mouse, developed by Skoda and coworkers.8 The Pf4-Cre mouse has been used in more than 160 studies to date, to generate a variety of MK/platelet-specific conditional KO mice (supplemental Table 1, available on the Blood Web site), providing numerous novel insights into platelet production and function.
However, the utility of the Pf4-Cre strain to delineate the role of platelets from leukocytes is limited. Recent studies demonstrate transgene expression outside the MK lineage, including hematopoietic stem cells, a subpopulation of circulating leukocytes and macrophages.9-12 Recombination in these cells can result in phenotypes unrelated to deletion of proteins in platelets, a case in point being reduced plasma levels of factor (F)XIII-A in F13a1fl/fl;Pf4-Cre mice being a result of FXIII-A ablation in Pf4-expressing macrophages.13 A particularly confounding issue with Pf4-Cre transgene is that a considerable proportion of leukocytes undergoes recombination on inflammatory stimulation,9,12 complicating in vivo experiments.
Importantly, findings on broader Pf4-Cre expression are in agreement with reports on endogenous Pf4 (also referred to as C-X-C motif chemokine ligand 4) expression pattern. Although this chemokine was for many years thought to be expressed exclusively in MKs and platelets,14,15 there is now unequivocal evidence that endogenous Pf4 is also expressed in a variety of immune cells, including monocytes,16-20 macrophages,13,16,18-24 microglia,25,26 dendritic cells,20,24,27-30 granulocytes,31 mast cells,32 T cells,12,17 and B cells,20,24 either constitutively or on stimulation. Moreover, broader endogenous Pf4 expression is also supported by gene expression databases.33-35 Unexpectedly, endogenous Pf4 expression has also been described outside the hematopoietic lineage in a population of intestinal epithelial cells.36 Furthermore, Pf4-Cre-mediated recombination was also observed in a subset of epithelial cells in the colon, resulting in the development of colon cancer in Adenomatous polyposis colifl/fl;Pf4-Cre mice.9 Thus, caution must be taken when interpreting phenotypes arising from Pf4-Cre-generated conditional KO mouse models. Notably, there are currently no alternative Cre deleter strains available that are specific for the MK lineage.
To overcome these limitations of the Pf4-Cre deleter strain, we developed a Gp1ba-Cre transgenic mouse model, using the MK/platelet-specific endogenous Gp1ba locus to drive Cre expression. Gp1ba encodes the glycoprotein (GP)Ibα subunit of the GPIb-IX-V complex, the receptor for von Willebrand factor.37 Our results demonstrate that the Gp1ba-Cre deleter mouse enables highly efficient and specific ablation of floxed genes in the MK lineage. We provide a new tool for generating MK/platelet-specific KO mice that allows researchers to differentiate the functional roles of platelets and leukocytes in any pathophysiological condition.
Materials and methods
Mouse models
All mice used were on a C57BL/6 background. The Gp1ba-Cre transgenic mouse was generated as described here. Membrane-targeted tandem dimer tomato (mT; tdTomato)/membrane-targeted enhanced green fluorescent protein (mG; EGFP; mT/mG) double-fluorescent Cre reporter mice were obtained from Jackson Laboratory.38 Pf4-Cre+/KI, CD148fl/fl, Cskfl/fl, and Shp1fl/fl mice were generated as previously described.8,39-41 mT/mG+/fl;Pf4-Cre+/KI, CD148fl/fl;Pf4-Cre+/KI, Cskfl/fl;Pf4-Cre+/KI, and Shp1fl/fl;Pf4-Cre+/KI mice were generated by crossing Pf4-Cre+/KI male mice with mT/mG+/fl, CD148fl/fl, Cskfl/fl, or Shp1fl/fl female mice, respectively.42,43 Similarly, mT/mG+/fl;Gp1ba-Cre+/KI, CD148fl/fl;Gp1ba-Cre+/KI, Cskfl/fl;Gp1ba-Cre+/KI, and Shp1fl/fl;Gp1ba-Cre+/KI mice were generated by crossing Gp1ba-Cre+/KI male mice with mT/mG+/fl, CD148fl/fl, Cskfl/fl, or Shp1fl/fl female mice. All procedures were undertaken with UK Home Office approval in accordance with the Animals (Scientific Procedures) Act of 1986.
Generation of Gp1ba-Cre transgenic mouse
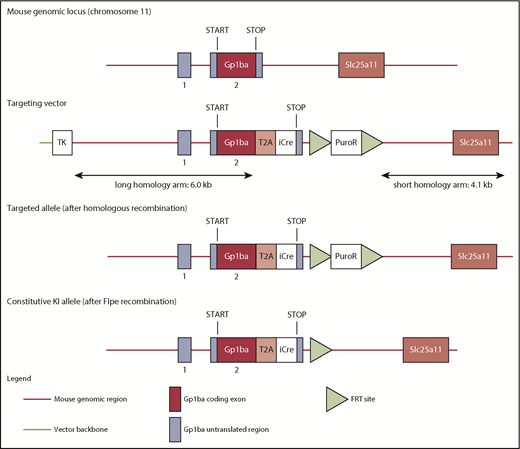
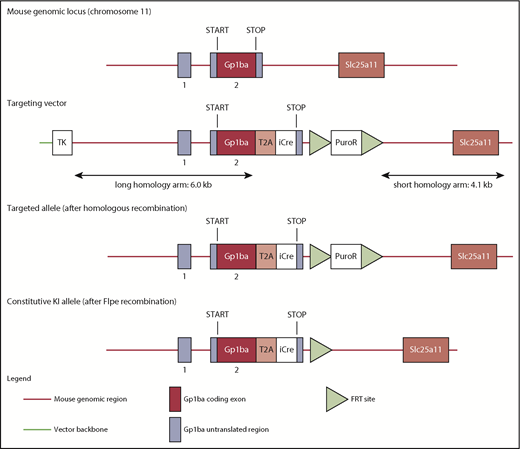
The targeting strategy enabled the generation of a constitutive knock-in (KI) of a T2A-improved-Cre (iCre) in the endogenous Gp1ba gene (Taconic Biosciences, Hudson, NY; Figure 1). Exon 2 of the Gp1ba gene contains the complete open reading frame. The sequences for the T2A and the open reading frame of iCre have been inserted between the last amino acid and the translation termination codon in exon 2 of Gp1ba (National Center for Biotechnology Information Reference Sequence: NM_010326_2). The positive selection marker (Puromycin resistance) was flanked by FRT sites and inserted downstream of the mouse 3′ untranslated region. The targeting vector was transfected into the Taconic Biosciences C57BL/6N Tac ES cell line, and homologous recombinant clones were isolated using positive (puromycin resistance) and negative (thymidine kinase) selections. The constitutive KI allele expresses a chimeric transcript harboring the Gp1ba gene fused to the T2A and the iCre sequences and was obtained after in vivo Flp-mediated removal of the selection marker, which was confirmed by polymerase chain reaction on genomic DNA. To detect the constitutive KI allele (372-bp fragment), as well as the wild-type (WT) allele (297-bp fragment), by polymerase chain reaction, the following primers were used: 5′-GAACACAACTCTCCTTGCTGG-3′, forward; 5′-GAAGAGTTAATGGCAGGAAAGAG-3′, reverse.
Targeting strategy to generate the Gp1ba-Cre mouse. The mouse Gp1ba gene consists of a 5′ untranslated exon (1), followed by a short intron and an exon (2) containing the open reading frame encoding GPIbα protein.58 The targeting strategy enables the generation of a constitutive KI of a T2A-iCre in the endogenous Gp1ba locus. The sequences for the T2A and the open reading frame of iCre have been inserted between the last amino acid and the translation termination codon in exon 2 of Gp1ba. Puromycin resistance (positive selection marker) is flanked by FRT sites and inserted downstream of the mouse 3′ untranslated region. The constitutive KI allele is obtained after in vivo Flp-mediated removal of the selection marker and expresses a chimeric transcript harboring the iCre gene fused to the Gp1ba gene via the T2A sequence. TK, thymidine kinase.
Targeting strategy to generate the Gp1ba-Cre mouse. The mouse Gp1ba gene consists of a 5′ untranslated exon (1), followed by a short intron and an exon (2) containing the open reading frame encoding GPIbα protein.58 The targeting strategy enables the generation of a constitutive KI of a T2A-iCre in the endogenous Gp1ba locus. The sequences for the T2A and the open reading frame of iCre have been inserted between the last amino acid and the translation termination codon in exon 2 of Gp1ba. Puromycin resistance (positive selection marker) is flanked by FRT sites and inserted downstream of the mouse 3′ untranslated region. The constitutive KI allele is obtained after in vivo Flp-mediated removal of the selection marker and expresses a chimeric transcript harboring the iCre gene fused to the Gp1ba gene via the T2A sequence. TK, thymidine kinase.
All other materials and methods, along with the statistical analysis, are described in supplemental Materials and methods.
Results
Generation of the Gp1ba-Cre transgenic mouse strain
The Gp1ba gene was selected to drive Cre expression, because it is well established that GPIbα is highly and specifically expressed in MKs and platelets.44 The T2A sequence was employed in the targeted insertion strategy, which allows multiple proteins to be expressed from a single multicistronic transcript.45 To enable the expression of the recombinase under control of the endogenous Gp1ba locus, T2A was inserted in-frame with iCre between the last amino acid and the translation termination codon in Gp1ba (Figure 1). On translation of the chimeric Gp1ba-T2A-iCre mRNA, the T2A sequence will lead to ribosome skipping, resulting in the coexpression of GPIbα and Cre as discrete proteins in cells in which GPIbα is endogenously expressed. As a result of the T2A technology, the GPIbα protein is expected to possess an additional 17 amino acids (EGRGSLLTCGDVEENPG) at its C terminus, and a proline on the N terminus of iCre. Gp1ba-Cre mice are viable and fertile, displayed no overt developmental or behavioral defects, and are born at expected Mendelian frequencies (supplemental Table 2).
Minor alterations in platelet volume and receptor expression in Gp1ba-Cre+/KI mice
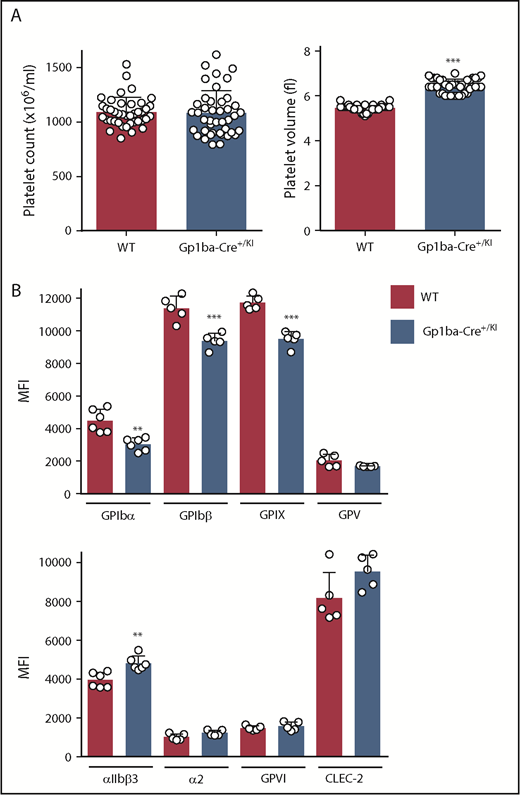
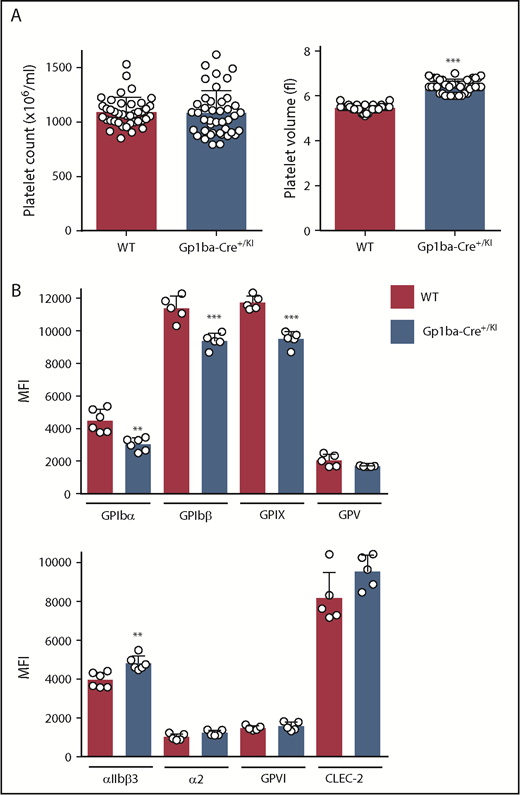
The Gp1ba-Cre mice used in the present study were heterozygous for the KI allele (Gp1ba-Cre+/KI). Platelet count and other hematological parameters were normal in these mice, except platelet volume, which was increased by 18% (Figure 2A and supplemental Table 3). To test whether expression of the GPIb-IX-V complex was affected, we measured surface receptor expression levels of each subunit of the complex by flow cytometry. GPIbα expression was reduced by 33%, likely accounting for the increase in platelet volume (Figure 2B).46,47 GPIbβ and GPIX were concomitantly reduced by 18% and 19%, respectively, whereas GPV expression was normal. In contrast, surface expression of the integrin αIIbβ3 was increased by 18%, likely reflecting the increase in platelet volume. No significant differences were detected in surface expression of integrin α2, GPVI, and CLEC-2 (Figure 2B).
Platelet parameters of the Gp1ba-Cre mouse. (A) Platelet counts and platelet volumes, n = 40 to 42 mice per genotype. (B) Platelet surface receptor expression of GPIbα, GPIbβ, GPIX, GPV, integrin αIIbβ3, integrin α2, GPVI, and CLEC-2 were measured by flow cytometry and shown as median fluorescence intensity (MFI), n = 5 to 6 mice per genotype. **P < .01; ***P < .001, unpaired, 2-tailed t-test, mean ± standard deviation (SD).
Platelet parameters of the Gp1ba-Cre mouse. (A) Platelet counts and platelet volumes, n = 40 to 42 mice per genotype. (B) Platelet surface receptor expression of GPIbα, GPIbβ, GPIX, GPV, integrin αIIbβ3, integrin α2, GPVI, and CLEC-2 were measured by flow cytometry and shown as median fluorescence intensity (MFI), n = 5 to 6 mice per genotype. **P < .01; ***P < .001, unpaired, 2-tailed t-test, mean ± standard deviation (SD).
To better understand the consequences of the Gp1ba-Cre targeted insertion strategy on GPIbα expression and other platelet parameters, we analyzed mice homozygous for the KI allele (Gp1ba-CreKI/KI). These mice had a 49% reduction in platelet count, a 71% increase in platelet volume, and reduced surface expression of GPIbα (by 72%), GPIbβ (by 51%), GPIX (by 51%), and GPV (by 50%) compared with WT mice (supplemental Figure 1). The relationship between GPIbα levels and platelet volume and count is well established both in humans and mice.46,47 Our results suggest that the targeted insertion might interfere with mRNA stability, translation, or GPIbα protein stability or trafficking.
Normal platelet function and hemostasis in Gp1ba-Cre+/KI mice
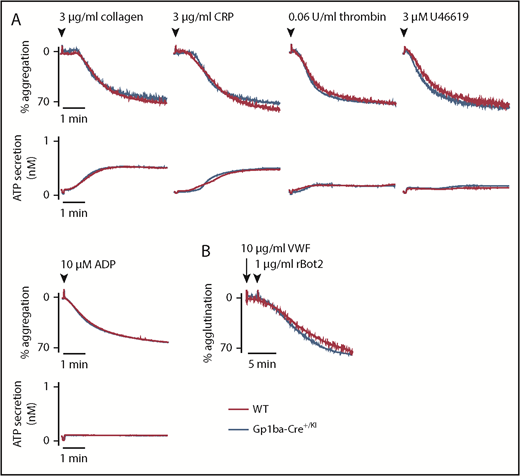
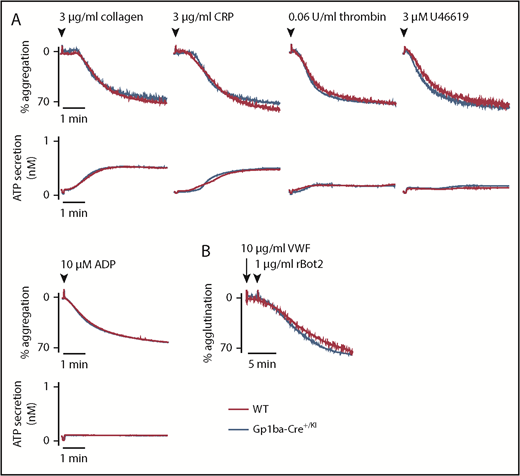
To study whether the reduction in GPIb-IX complex levels translates to aberrant platelet function in the Gp1ba-Cre+/KI mice, platelet aggregation and adenosine triphosphate secretion were measured in response to a range of platelet agonists. No differences were found in aggregation or secretion in response to collagen (3 and 10 μg/mL), the synthetic GPVI-specific agonist collagen-related peptide (3 and 10 μg/mL), thrombin (0.06 and 0.1 U/mL), the thromboxane A2 analog U46619 (3 and 10 μM), or adenosine diphosphate (10 and 30 μM; Figure 3A and supplemental Figure 2). Recombinant botrocetin-2 (1 μg/mL) and mouse von Willebrand factor (10 μg/mL) were used in combination to specifically test GPIb-IX-V-mediated platelet agglutination.48 Despite the reduced expression of GPIb-IX complex on the platelet surface, no differences were observed in the function of GPIbα in platelets from Gp1ba-Cre+/KI mice (Figure 3B).
Aggregation and agglutination of Gp1ba-Cre platelets. (A) Mean platelet aggregation and secretion traces in response to the indicated agonists, n = 4 to 8 mice per condition per genotype. (B) Mean platelet agglutination traces in response to 10 µg/mL von Willebrand factor (for 2 minutes) followed by 1 µg/mL recombinant botrocetin-2 stimulation,48 n = 3 mice per condition per genotype. ATP, adenosine triphosphate; rBot2, recombinant botrocetin-2; VWF, von Willebrand factor.
Aggregation and agglutination of Gp1ba-Cre platelets. (A) Mean platelet aggregation and secretion traces in response to the indicated agonists, n = 4 to 8 mice per condition per genotype. (B) Mean platelet agglutination traces in response to 10 µg/mL von Willebrand factor (for 2 minutes) followed by 1 µg/mL recombinant botrocetin-2 stimulation,48 n = 3 mice per condition per genotype. ATP, adenosine triphosphate; rBot2, recombinant botrocetin-2; VWF, von Willebrand factor.
The ability of Gp1ba-Cre+/KI platelets to adhere and spread on a fibrinogen-coated surface under static conditions was subsequently assessed. Nonstimulated and thrombin preactivated Gp1ba-Cre platelets spread to the same extent as WT platelets on fibrinogen (supplemental Figure 3), demonstrating normal integrin αIIbβ3-mediated functional responses.
To investigate whether minor defects described here translated into hemostatic complications in Gp1ba-Cre+/KI mice, we assessed tail bleeding after excision of a 5-mm portion of the tail tip. No significant differences in blood loss were observed in Gp1ba-Cre+/KI mice compared with WT mice (supplemental Figure 4). Collectively, these findings demonstrate that platelet function is maintained in Gp1ba-Cre+/KI mice. Gp1ba-Cre+/KI mice were used to generate conditional KO mice and as controls in all subsequent experiments.
Highly specific recombination in the MK lineage with the Gp1ba-Cre transgene
The expression pattern of Cre is expected to mirror that of endogenous GPIbα in the Gp1ba-Cre+/KI mouse. To test whether Cre is specifically expressed in the MK lineage, we crossed the Gp1ba-Cre+/KI mouse with the mT/mG double-fluorescent Cre reporter mouse.38 Cells in the mT/mG+/fl;Gp1ba-Cre+/KI mouse express mT; tdTomato before Cre-mediated recombination and mG; EGFP after Cre-mediated excision. Although the EGFP signal appears rapidly, within 24 hours after Cre expression, the tdTomato fluorescence disappears slowly because of gradually declining tdTomato mRNA and protein levels.38 Monitoring both EGFP and tdTomato fluorescence can provide unique insights on the onset of Cre expression, which is especially useful for studying megakaryopoiesis. For direct comparison, we also crossed the mT/mG reporter strain with the Pf4-Cre mouse to generate mT/mG+/fl;Pf4-Cre+/KI mice, and analyzed them in parallel with the mT/mG+/fl;Gp1ba-Cre+/KI mice.
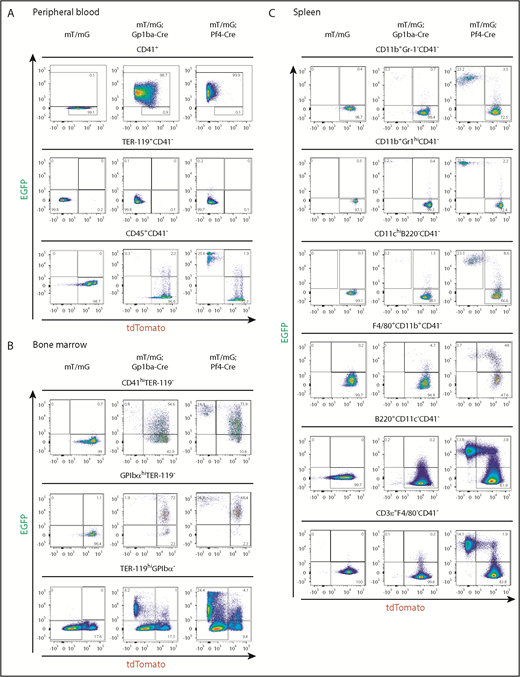
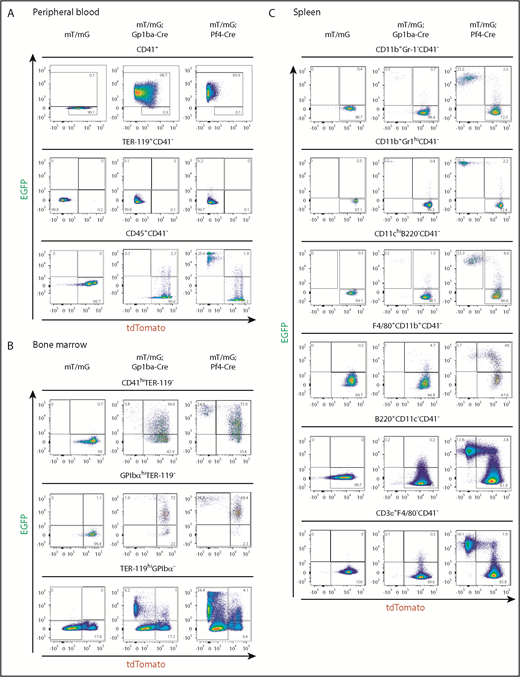
We measured recombination in platelets from peripheral blood using flow cytometry by counting EGFP+ events in the CD41+ (integrin αIIb+) population. We found that 98.7% and 99.9% of platelets were EGFP+, thus undergoing recombination in Gp1ba-Cre-generated and Pf4-Cre-generated mice, respectively (Figure 4A). Interestingly, 64.1% and 2.4% of mT/mG+/fl;Gp1ba-Cre+/KI and mT/mG+/fl;Pf4-Cre+/KI platelets have retained tdTomato expression, respectively, suggesting recombination takes place later in Gp1ba-Cre-generated mice (supplemental Figure 5). We next determined EGFP+ fractions of red blood cells and leukocytes by gating on CD41− cells to exclude platelet/erythrocyte and platelet/leukocyte aggregates (supplemental Figure 6). EGFP expression was virtually absent in red blood cells (TER-119+CD41−) in mT/mG+/fl;Gp1ba-Cre+/KI (0.1%) and mT/mG+/fl;Pf4-Cre+/KI (0.2%) mice (Figure 4A). For CD45+(pan-leukocyte marker)CD41− blood cells, we found 0.3% EGFP+tdTomato− and 2.3% EGFP+tdTomato+ cells in mT/mG+/fl;Gp1ba-Cre+/KI mice. Whereas the former population likely represents bona fide recombined cells, we cannot exclude the possibility that the latter population includes EGFP+ platelet-leukocyte aggregates. Notably, we found that 25.6% of CD45+ cells were EGFP+tdTomato− in mT/mG+/fl;Pf4-Cre+/KI mice (Figure 4 and supplemental Table 4), suggesting recombination in a large population of circulating leukocytes.
Gp1ba-Cre-mediated and Pf4-Cre-mediated recombination of blood, bone marrow, and spleen cells. (A) Flow cytometry analysis of peripheral blood cells showing EGFP and tdTomato fluorescence of platelets (CD41+), n = 6 mice per genotype; erythrocytes (TER-119+CD41−), n = 6 mice per genotype; and leukocytes (CD45+CD41−), n = 5 to 7 mice per genotype from mT/mG+/fl, mT/mG+/fl;Gp1ba-Cre+/KI, and mT/mG+/fl;Pf4-Cre+/KI mice. (B) Flow cytometry analysis of bone marrow cells showing EGFP and tdTomato fluorescence of CD41hiTER-119− bone marrow cells, n = 4 to 5 mice per genotype; GPIbαhiTER-119− bone marrow cells, n = 4 to 5 mice per genotype; and TER-119+GPIbα- bone marrow cells, n = 3 mice per genotype from mT/mG+/fl, mT/mG+/fl;Gp1ba-Cre+/KI, and mT/mG+/fl;Pf4-Cre+/KI mice. (C) Flow cytometry analysis showing EGFP and tdTomato fluorescence of the indicated spleen cell populations, n = 5 mice per genotype from mT/mG+/fl, mT/mG+/fl;Gp1ba-Cre+/KI, and mT/mG+/fl;Pf4-Cre+/KI mice. (A-C) Numbers in gates represent mean percentages of respective cell populations. Mean ± SD are shown in supplemental Table 4. Flow cytometry gating strategies for peripheral blood and bone marrow cells are shown in supplemental Figure 6. Gating strategies for spleen cell populations are shown in supplemental Figure 7.
Gp1ba-Cre-mediated and Pf4-Cre-mediated recombination of blood, bone marrow, and spleen cells. (A) Flow cytometry analysis of peripheral blood cells showing EGFP and tdTomato fluorescence of platelets (CD41+), n = 6 mice per genotype; erythrocytes (TER-119+CD41−), n = 6 mice per genotype; and leukocytes (CD45+CD41−), n = 5 to 7 mice per genotype from mT/mG+/fl, mT/mG+/fl;Gp1ba-Cre+/KI, and mT/mG+/fl;Pf4-Cre+/KI mice. (B) Flow cytometry analysis of bone marrow cells showing EGFP and tdTomato fluorescence of CD41hiTER-119− bone marrow cells, n = 4 to 5 mice per genotype; GPIbαhiTER-119− bone marrow cells, n = 4 to 5 mice per genotype; and TER-119+GPIbα- bone marrow cells, n = 3 mice per genotype from mT/mG+/fl, mT/mG+/fl;Gp1ba-Cre+/KI, and mT/mG+/fl;Pf4-Cre+/KI mice. (C) Flow cytometry analysis showing EGFP and tdTomato fluorescence of the indicated spleen cell populations, n = 5 mice per genotype from mT/mG+/fl, mT/mG+/fl;Gp1ba-Cre+/KI, and mT/mG+/fl;Pf4-Cre+/KI mice. (A-C) Numbers in gates represent mean percentages of respective cell populations. Mean ± SD are shown in supplemental Table 4. Flow cytometry gating strategies for peripheral blood and bone marrow cells are shown in supplemental Figure 6. Gating strategies for spleen cell populations are shown in supplemental Figure 7.
We next quantified EGFP-expressing bone marrow (BM) cells. We observed fewer EGFP+tdTomato− and EGFP+tdTomato+ BM cells within the CD41 high (CD41hi) population of mT/mG+/fl;Gp1ba-Cre+/KI mice compared with mT/mG+/fl;Pf4-Cre+/KI mice (0.8% vs 14.9% and 54.6% vs 73.9%, respectively; Figure 4B, supplemental Figure 6, and supplemental Table 4). Similarly, when gated on mature GPIbαhi MKs, we found that in mT/mG+/fl;Gp1ba-Cre+/KI mice, 73.9% of GPIbαhi MKs were EGFP+, whereas in the mT/mG+/fl;Pf4-Cre+/KI mice, it was 95.2% (Figure 4B). These results indicate that Pf4-Cre is expressed earlier in development than Gp1ba-Cre, and that not all cells have yet produced sufficient EGFP to fall into the EGFP+ gate. This is most likely a result of the 24-hour delay in generating detectable levels of EGFP after recombination.38 Next, we focused on the TER-119+GPIbα− BM cell population representing erythroid lineage cells. We observed 6.2% EGFP+tdTomato− cells in the mT/mG+/fl;Gp1ba-Cre+/KI mice, likely representing a common MK/erythroid progenitor population (Figure 4B). In contrast, 24.4% of TER-119+ BM cells were EGFP+tdTomato− in the mT/mG+/fl;Pf4-Cre+/KI mice, suggesting recombination in maturing erythroid lineage cells in these mice. The lack of EGFP+ or tdTomato+ peripheral blood erythrocytes in either mouse model is most likely a result of protein degradation during the long lifespan of these anucleated cells (Figure 4A).
We then investigated EGFP expression in different subsets of spleen cells. We found less than 0.3% EGFP+tdTomato− myeloid cells (CD11b+Gr-1−), granulocytes (CD11b+Gr-1hi), conventional dendritic cells (CD11chiB220−), B cells (B220+CD11c−), and T cells (CD3ε+F4/80−) in mT/mG+/fl;Gp1ba-Cre+/KI mice (Figure 4C, supplemental Figure 7, and supplemental Table 4). In addition, we observed 4.7% EGFP+tdTomato+ macrophages (F4/80+CD11b+) in these mice, which represent either bona fide recombined macrophages or cells that have phagocytosed EGFP+ platelets.
Strikingly, we detected high proportions of EGFP+tdTomato− myeloid and lymphoid cell subsets in mT/mG+/fl;Pf4-Cre+/KI spleens, including 23.2% myeloid cells (CD11b+Gr-1−), 25.2% granulocytes (CD11b+Gr-1hi), 23.3% conventional dendritic cells (CD11chiB220−), 13.8% B cells (B220+CD11c−), and 14.1% T cells (CD3ε+F4/80−; Figure 4C). Furthermore, we identified 48% EGFP+tdTomato+ macrophages (F4/80+CD11b+) in these mice, suggesting recombination in immature macrophages.
Collectively, these findings demonstrate Cre-mediated recombination in a large proportion of platelets and MKs in both Gp1ba-Cre+/KI and Pf4-Cre+/KI mice. However, the earlier onset of Pf4-Cre also results in recombination in ∼15% to 50% of erythroid, myeloid, and lymphoid cells, which is not observed in Gp1ba-Cre mice.
Similar platelet phenotypes in CD148 and Csk conditional KO mice
After establishing the lineage specificity of Cre expression in hematopoietic cells, we sought to compare protein ablation efficiency and phenotypes of conditional KO mice generated with either Gp1ba-Cre or Pf4-Cre mice. To achieve this, we crossed both deleter strains to CD148-floxed,39 Csk-floxed,40 or Shp1-floxed mice,41 respectively, for direct comparison. CD148, Csk, and Shp1 are widely expressed in hematopoietic cells and have been implicated in regulating immune cell development and function.39-41,49
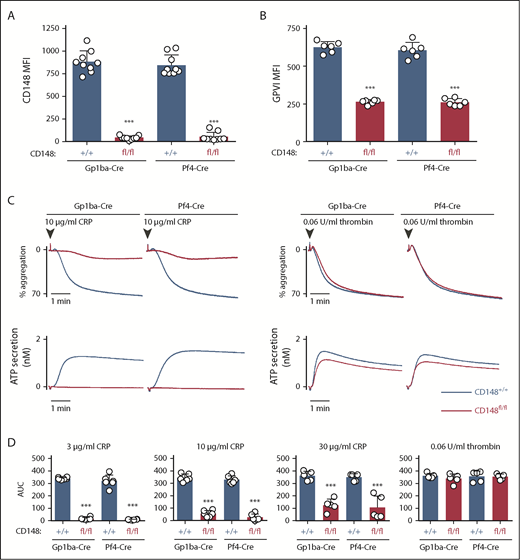
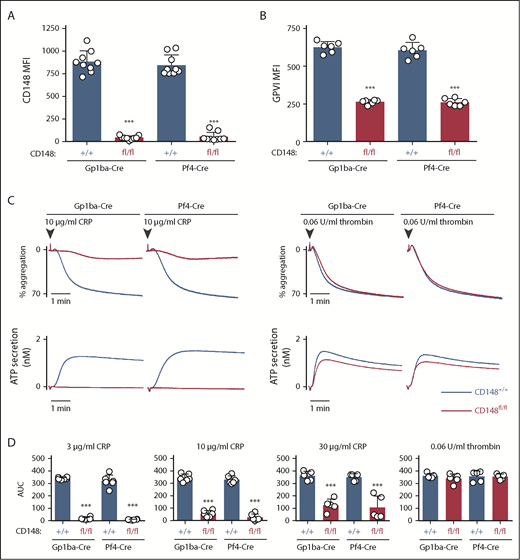
Surface receptor levels of CD148 on CD148fl/fl;Gp1ba-Cre+/KI and CD148fl/fl;Pf4-Cre+/KI platelets displayed 95.1% and 93.6% reduction, respectively, as determined by flow cytometry (Figure 5A). Surface levels of GPVI were reduced by 58% and 57% in the 2 mouse models, respectively (Figure 5B). CD148fl/fl;Gp1ba-Cre+/KI platelets showed similar aggregation responses to CD148fl/fl;Pf4-Cre+/KI platelets, including the previously described GPVI-specific aggregation defect in response to collagen-related peptide and normal aggregation in response to thrombin (Figure 5C).42,50 Highly similar aggregation responses were observed to a range of collagen-related peptide concentrations (Figure 5D), demonstrating that CD148fl/fl;Gp1ba-Cre+/KI platelets phenocopy their Pf4-Cre-generated counterparts.
Efficient protein ablation and similar platelet phenotypes in CD148;Gp1ba-Cre mice. (A) Platelet surface receptor expression of CD148 was measured by flow cytometry of the indicated genotypes and shown as MFI, n = 9 mice per genotype. (B) Platelet surface receptor expression of GPVI was measured by flow cytometry and shown as MFI, n = 6 mice per genotype. (C) Mean platelet aggregation and secretion traces in response to the indicated agonists, n = 5 to 6 mice per condition per genotype. (D) Platelet aggregation in response to the indicated agonists shown as area under the curve (AUC). (A-B,D) Asterisks refer to significant difference in CD148fl/fl;Gp1ba-Cre+/KI or CD148fl/fl;Pf4-Cre+/KI mice compared with CD148+/+;Gp1ba-Cre+/KI or CD148+/+;Pf4-Cre+/KI mice, respectively (***P < .001; 1-way ANOVA with Sidak’s test); data represent mean ± SD.
Efficient protein ablation and similar platelet phenotypes in CD148;Gp1ba-Cre mice. (A) Platelet surface receptor expression of CD148 was measured by flow cytometry of the indicated genotypes and shown as MFI, n = 9 mice per genotype. (B) Platelet surface receptor expression of GPVI was measured by flow cytometry and shown as MFI, n = 6 mice per genotype. (C) Mean platelet aggregation and secretion traces in response to the indicated agonists, n = 5 to 6 mice per condition per genotype. (D) Platelet aggregation in response to the indicated agonists shown as area under the curve (AUC). (A-B,D) Asterisks refer to significant difference in CD148fl/fl;Gp1ba-Cre+/KI or CD148fl/fl;Pf4-Cre+/KI mice compared with CD148+/+;Gp1ba-Cre+/KI or CD148+/+;Pf4-Cre+/KI mice, respectively (***P < .001; 1-way ANOVA with Sidak’s test); data represent mean ± SD.
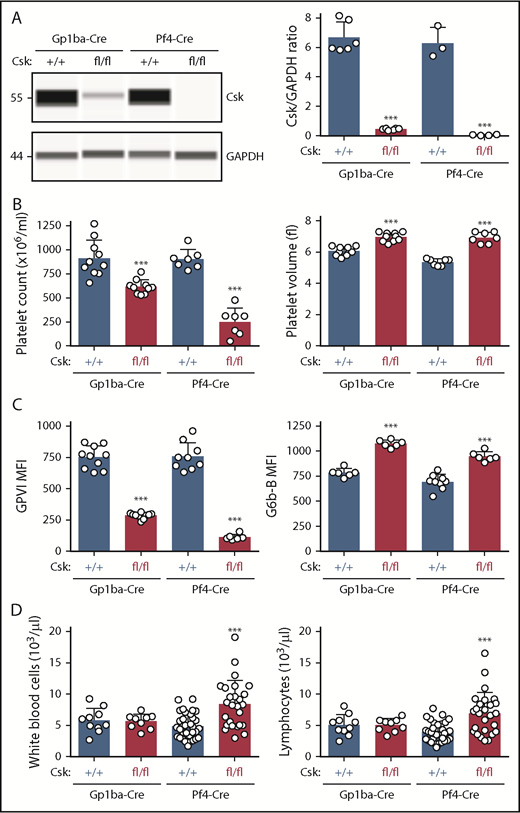
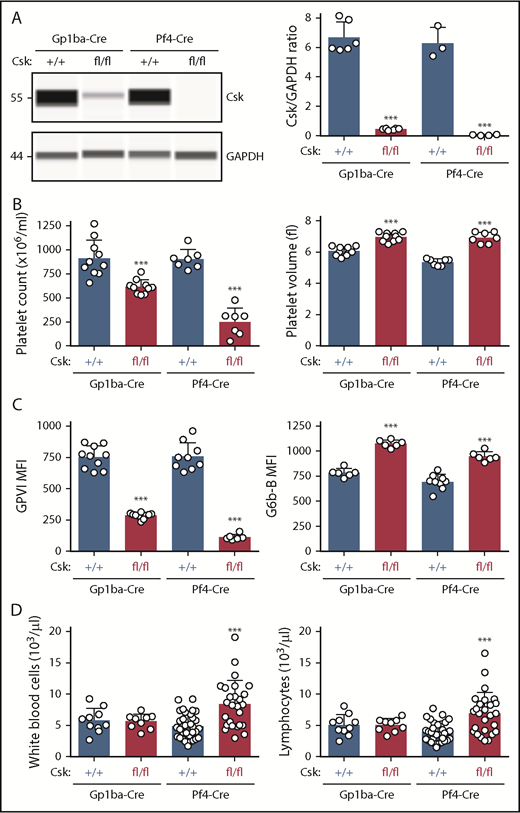
Protein levels of Csk from Cskfl/fl;Gp1ba-Cre+/KI and Cskfl/fl;Pf4-Cre+/KI platelets were analyzed by quantitative capillary-based immunoassays (ProteinSimple Wes) and compared with their corresponding controls, Csk+/+;Gp1ba-Cre+/KI and Csk+/+;Pf4-Cre+/KI platelet lysates, respectively. We found efficient protein ablation in Cskfl/fl;Gp1ba-Cre+/KI platelets revealing a 93% reduction of Csk levels, whereas Cskfl/fl;Pf4-Cre+/KI platelets showed a 99% reduction (Figure 6A). Cskfl/fl;Pf4-Cre+/KI mice showed a 71% reduction in platelet count, a 35% increase in platelet volume, an 85% reduction in GPVI, and a 42% increase in G6b-B levels (Figure 6B-C). Importantly, Cskfl/fl;Gp1ba-Cre+/KI mice exhibited a similar but less severe platelet phenotype, including a 32% reduction in platelet count, a 15% increase in platelet volume, a 62% decrease in GPVI, and a 37% increase in G6b-B levels (Figure 6B-C). Differences between the 2 models likely reflect incomplete Csk ablation in Cskfl/fl;Gp1ba-Cre+/KI platelets and can be explained by the later onset of Gp1ba-Cre transgene expression (Figure 4B). Notably, we found that white blood cell and lymphocyte counts were significantly elevated in the Cskfl/fl;Pf4-Cre+/KI mice, but unaltered in the Cskfl/fl;Gp1ba-Cre+/KI mice (Figure 6D), suggesting Csk deletion in lymphocytes in Cskfl/fl;Pf4-Cre+/KI mice.
Efficient protein ablation, similar platelet phenotypes, but no altered lymphocyte counts in Csk;Gp1ba-Cre mice. (A) Protein levels of Csk and GAPDH in platelet lysates of the indicated genotypes were determined by capillary-based immunoassays with the respective antibodies. Platelet Csk protein levels were determined by normalizing Csk peak areas by GAPDH peak areas (Csk/GAPDH ratio), n = 3 to 6 mice per genotype. (B) Platelet counts and platelet volumes, n = 7 to 10 mice per genotype. (C) Platelet surface receptor expression of GPVI and G6b-B were measured by flow cytometry and shown as MFI, n = 6 to 10 mice per genotype. (D) White blood cell and lymphocyte counts, n = 9 to 33 mice per genotype. (A-D) Asterisks refer to significant difference in Cskfl/fl;Gp1ba-Cre+/KI or Cskfl/fl;Pf4-Cre+/KI mice compared with Csk+/+;Gp1ba-Cre+/KI or Csk+/+;Pf4-Cre+/KI mice, respectively (***P < .001; 1-way ANOVA with Sidak’s test); data represent mean ± SD.
Efficient protein ablation, similar platelet phenotypes, but no altered lymphocyte counts in Csk;Gp1ba-Cre mice. (A) Protein levels of Csk and GAPDH in platelet lysates of the indicated genotypes were determined by capillary-based immunoassays with the respective antibodies. Platelet Csk protein levels were determined by normalizing Csk peak areas by GAPDH peak areas (Csk/GAPDH ratio), n = 3 to 6 mice per genotype. (B) Platelet counts and platelet volumes, n = 7 to 10 mice per genotype. (C) Platelet surface receptor expression of GPVI and G6b-B were measured by flow cytometry and shown as MFI, n = 6 to 10 mice per genotype. (D) White blood cell and lymphocyte counts, n = 9 to 33 mice per genotype. (A-D) Asterisks refer to significant difference in Cskfl/fl;Gp1ba-Cre+/KI or Cskfl/fl;Pf4-Cre+/KI mice compared with Csk+/+;Gp1ba-Cre+/KI or Csk+/+;Pf4-Cre+/KI mice, respectively (***P < .001; 1-way ANOVA with Sidak’s test); data represent mean ± SD.
No associated inflammatory complications in Shp1fl/fl;Gp1ba-Cre+/KI mice
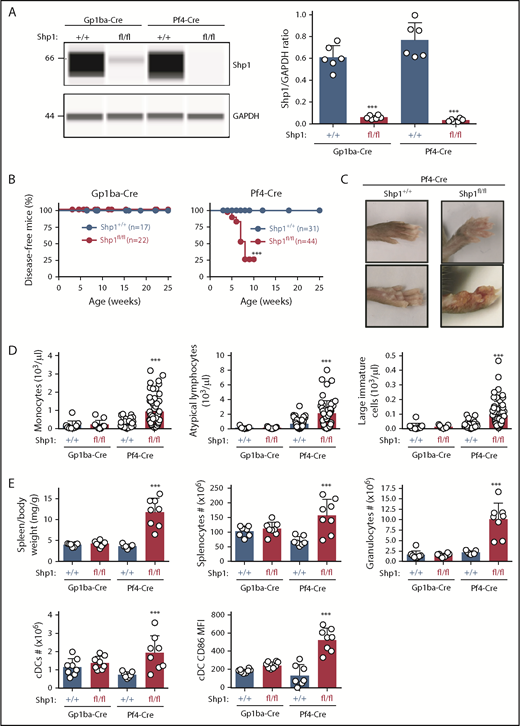
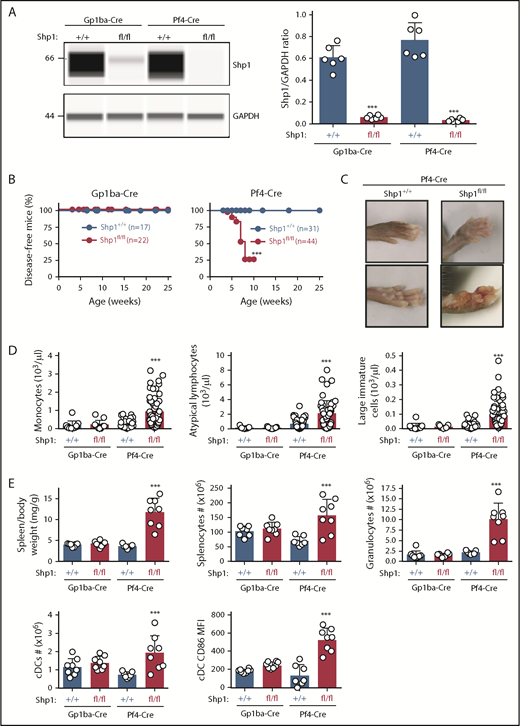
Shp1 levels were reduced by 90% and 96% in platelets from Shp1fl/fl;Gp1ba-Cre+/KI and Shp1fl/fl;Pf4-Cre+/KI, respectively (Figure 7A). Intriguingly, 75% of Shp1fl/fl;Pf4-Cre+/KI mice developed a motheaten-like inflammatory phenotype by 8 weeks of age, culminating in the inflammation of the paws, ears, or nose (Figure 7B-C). The motheaten phenotype arises from a spontaneous mutation in Shp1, leading to loss of Shp1 expression.49 It is characterized by hyperinflammatory and autoimmune responses, whereby different hematopoietic cell subsets contribute to different aspects of the phenotype.51 In contrast, none of the Shp1fl/fl;Gp1ba-Cre+/KI mice showed signs of the motheaten-like phenotype (Figure 7B), suggesting it is unlikely to be driven by the lack of platelet Shp1. To better understand the etiology of the inflammatory phenotype, we analyzed leukocyte counts in peripheral blood and spleen of these mice. Shp1fl/fl;Pf4-Cre+/KI mice showed significantly elevated monocyte, atypical lymphocyte, and large immature cell counts in the peripheral blood, suggesting aberrant development of multiple leukocyte subsets. This was not the case in Shp1fl/fl;Gp1ba-Cre+/KI mice, which had normal blood cell counts (Figure 7D). Shp1fl/fl;Pf4-Cre+/KI mice displayed significantly increased spleen/body weight ratio, total splenocyte numbers, granulocyte (Gr-1hiF4/80−CD11c−B220−) and conventional dendritic cell (cDC; CD11chiB220−) counts, and activated cDCs (CD86+; Figure 7E). All these parameters were unaltered in Shp1fl/fl;Gp1ba-Cre+/KI mice (Figure 7E), highlighting the lack of transgene expression in these other lineages. These results strongly suggest that Shp1 is ablated outside the MK lineage in the Shp1fl/fl;Pf4-Cre+/KI mice, supporting findings from mT/mGfl/+;Pf4-Cre+/KI mice (Figure 4A-C).
Hyperinflammatory phenotype and altered leukocyte counts are absent in Shp1;Gp1ba-Cre mice. (A) Protein levels of Shp1 and GAPDH in platelet lysates of the indicated genotypes were determined by capillary-based immunoassays with the respective antibodies. Platelet Shp1 protein levels were determined by normalizing Shp1 peak areas by GAPDH peak areas (Shp1/GAPDH ratio), n = 6 mice per genotype. (B) Mice were monitored every week for signs of motheaten-like phenotype (paw, nose, or ear inflammation). Percentage of disease-free mice for the indicated period was calculated using Kaplan-Meier survival analysis with a log-rank (Mantel-Cox) test. ***P < .001, n values are indicated in the figure. (C) Representative photographs of inflamed paws in Shp1;Pf4-Cre mice. (D) Monocyte, atypical lymphocyte, and large immature cell counts, n = 11 to 58 mice per genotype. (E) Spleen/body weight ratio, total splenocyte counts, granulocyte (Gr-1hiF4/80−CD11c−B220−) counts; cDC (CD11chiB220−) counts, and activated CD86+ cDC MFI of the indicated genotypes, n = 7 to 8 mice per genotype. (A,D-E) Asterisks refer to significant difference in Shp1fl/fl;Gp1ba-Cre+/KI or Shp1fl/fl;Pf4-Cre+/KI mice compared with Shp1+/+;Gp1ba-Cre+/KI or Shp1+/+;Pf4-Cre+/KI mice, respectively (***P < .001; 1-way ANOVA with Sidak’s test); data represent mean ± SD.
Hyperinflammatory phenotype and altered leukocyte counts are absent in Shp1;Gp1ba-Cre mice. (A) Protein levels of Shp1 and GAPDH in platelet lysates of the indicated genotypes were determined by capillary-based immunoassays with the respective antibodies. Platelet Shp1 protein levels were determined by normalizing Shp1 peak areas by GAPDH peak areas (Shp1/GAPDH ratio), n = 6 mice per genotype. (B) Mice were monitored every week for signs of motheaten-like phenotype (paw, nose, or ear inflammation). Percentage of disease-free mice for the indicated period was calculated using Kaplan-Meier survival analysis with a log-rank (Mantel-Cox) test. ***P < .001, n values are indicated in the figure. (C) Representative photographs of inflamed paws in Shp1;Pf4-Cre mice. (D) Monocyte, atypical lymphocyte, and large immature cell counts, n = 11 to 58 mice per genotype. (E) Spleen/body weight ratio, total splenocyte counts, granulocyte (Gr-1hiF4/80−CD11c−B220−) counts; cDC (CD11chiB220−) counts, and activated CD86+ cDC MFI of the indicated genotypes, n = 7 to 8 mice per genotype. (A,D-E) Asterisks refer to significant difference in Shp1fl/fl;Gp1ba-Cre+/KI or Shp1fl/fl;Pf4-Cre+/KI mice compared with Shp1+/+;Gp1ba-Cre+/KI or Shp1+/+;Pf4-Cre+/KI mice, respectively (***P < .001; 1-way ANOVA with Sidak’s test); data represent mean ± SD.
Taken together, these findings provide evidence that the Gp1ba-Cre mouse strain is highly efficient at ablating floxed genes in MKs, leading to an almost complete deletion of corresponding proteins in platelets. Reporter mouse experiments, together with the lack of leukocyte-related phenotypes in Gp1ba-Cre-generated mice, support MK lineage-specific transgene expression, which offers a powerful alternative strategy to generate MK/platelet-specific conditional KO mice.
Discussion
In this study, we report a new Cre deleter mouse strain that allows MK/platelet lineage-specific recombination of floxed genes. We demonstrate that 98.7% of platelets undergo recombination in Gp1ba-Cre+/KI mice. We show that this transgene is highly efficient at deleting proteins in platelets and results in similar platelet phenotypes to Pf4-Cre-generated KO mice. To test the utility of this new model, we have directly compared it with the Pf4-Cre mouse strain in fluorescent reporter mouse experiments revealing highly specific expression in the MK lineage with the Gp1ba-Cre transgene and wider expression with Pf4-Cre. We show that the motheaten-like inflammatory phenotype exhibited by Pf4-Cre-generated Shp1 conditional KO mice is not observed in Gp1ba-Cre-generated KO mice. These results, along with widespread recombination of mT/mG+/fl;Pf4-Cre+/KI leukocytes, strongly support that the inflammatory phenotype is driven by ablation of Shp1 in multiple leukocyte lineages. Platelets and leukocytes are often involved in the same processes, and recombined leukocytes can contribute toward the overall phenotype of Pf4-Cre-generated KO mice, as demonstrated in the present study. The Gp1ba-Cre mouse will advance the platelet and immunology fields by enabling the separation of platelet and leukocyte contributions to in vivo phenotypes in an array of pathological conditions.
Gp1ba was selected to drive Cre expression because it is highly specific and drives high expression of GPIbα from an early stage during megakaryopoiesis.44,52 Although its specificity to the MK lineage is well established, a limited number of early studies reported GPIbα expression in cultured endothelial and smooth muscle cells.53,54 However, these findings were never validated by other groups or in vivo.44 In our targeting strategy, we used the endogenous Gp1ba locus; thus, Cre is controlled by the native promoter with its intact enhancer and repressor elements. In contrast, the Pf4-Cre mouse contains 4 additional chemokine genes as a consequence of the bacterial artificial chromosome construct used for its generation; namely, Cxcl3, Cxcl5, Cxcl7, and Cxcl15.8 Additional copies of these genes may explain the significantly lower platelet counts in the Pf4-Cre mouse,9 but can also confound inflammatory and immune responses, as they are involved in modulating leukocyte recruitment and activation,55-57 which is not an issue with our targeting strategy.
The T2A sequence used to couple Cre to GPIbα expression allows the equimolar expression of proteins from a single multicistronic transcript.45 The T2A-based insertion results in the addition of a short peptide sequence to the C-terminus of GPIbα, which may explain the observed reduction in GPIbα surface expression levels in Gp1ba-Cre mice. Alternatively, reduced GPIbα expression might arise from less efficient chimeric mRNA transcription or reduced translation. The modest decrease in GPIbα likely underlies the marginal increase in platelet volume in Gp1ba-CreKI/+ mice, which is well established in humans and mice with reduced GPIbα expression.46,47,58 Although minor changes in platelet volume and receptor levels did not translate into defects in platelet function or hemostatic complications, we advocate using Gp1ba-Cre+/KI mice as controls for conditional KO mice generated with this strain, as is the case with Pf4-Cre-generated or any other conditional KO mouse models.
We demonstrate that 73.9% GPIbαhi MKs undergo recombination in the mT/mG;Gp1ba-Cre mice, pointing to high Cre expression in mature MKs. A possible reason for incomplete EGFP labeling of MKs is the short lag between GPIbα and Cre expression, recombination, and EGFP accumulation.38 GPIbα was recently reported in unipotent MK progenitors, further supporting the lineage specificity of Gp1ba.52 In contrast, CD41 is not exclusively MK-specific, but is also expressed on hematopoietic progenitors,59-61 providing an explanation for the observed lower recombination of 55.3% of CD41hi BM cells compared with GPIbαhi MKs. As a consequence, CD41-Cre mice are not suitable for generating MK/plt-specific KO mice.59
A limitation of the Gp1ba-Cre mouse is that the onset of recombination occurs later than Pf4-Cre during megakaryopoiesis, resulting in later gene deletion and a shorter timeframe for the remaining mRNA and protein to be degraded. This is demonstrated by a higher tdTomato signal in mT/mG+/fl;Gp1ba-Cre+/KI MKs and platelets compared with mT/mG+/fl;Pf4-Cre+/KI. As a consequence, we found that 5% to 10% of targeted proteins persisted in Gp1ba-Cre-generated platelets, whereas only 1% to 6% protein levels remained in Pf4-Cre-generated platelets. Several factors contribute to these differences in protein expression, including the onset of transgene and floxed gene expression, mRNA, and protein stability. This is exemplified by Shp1 and Csk conditional KO models, which have more residual protein in platelets than CD148 conditional KO mice as a result of earlier onset of target gene expression. An advantage of slightly higher residual protein levels is less severe developmental defects in MKs and platelets, providing more opportunity to investigate functional consequences of reduced protein expression. Dosing studies can also be designed by knocking out the same gene, using the Pf4-Cre and Gp1ba-Cre transgenes, and comparing phenotypes of heterozygous and homozygous KO models in parallel. This can be particularly useful in studying megakaryopoiesis. In addition, Gp1ba-Cre-generated mouse models may more closely model hemizygosity in human patients. Incomplete protein ablation is not uncommon with other hematopoietic deleter strains. For example, the lifespan of circulating neutrophils in mice is less than a day,62 and the protein ablation efficiency of the neutrophil lineage-specific Cre model Ly6G-Cre is approximately 55%.63 With 2 complementary MK/platelet-specific deleter strains now at our disposal, we can address the issues of leakiness and dosing effects of gene expression in the MK lineage.
In addition, we provide further evidence of the wide tissue expression of the Pf4-Cre transgene by analyzing mT/mG+/fl;Pf4-Cre+/KI mice. We observed recombination of 23% to 48% myeloid and 14% lymphoid cells from the spleen. These values are similar to previously reported data, including 32% to 38% recombination of different subsets of freshly isolated splenocytes,10 or 20% recombination of cultured splenocytes.9 Furthermore, wider recombination can also be detected in the spleen of R26R-lacZ;Pf4-Cre mice (www.informatics.jax.org/allele/MGI:3764698).64 Of note, we found an average of 26% EGFP+ peripheral blood leukocytes in contrast with the 0.3% reported by Pertuy et al.9 We suspect this discrepancy is primarily a result of methodological differences. Considering steady state leukocyte mobilization from and recruitment to the spleen,65,66 a similar level of recombination is expected in blood and spleen leukocytes, which is what we observed. Of note, we detected 6.2% TER-119+ BM cell population in the mT/mG+/fl;Gp1ba-Cre+/KI mice that underwent recombination, suggesting expression of GPIbα in a common MK-erythroid progenitor. We are currently actively engaged in developing a more detailed expression pattern of Gp1ba-Cre, covering a range of cell and tissue types under normal and inflammatory conditions. This will be further advanced as the Gp1ba-Cre mouse becomes more widely used, as was the case of Pf4-Cre.9-11
In summary, the Gp1ba-Cre mouse exhibits improved specificity toward the MK lineage compared with the existing Pf4-Cre mouse commonly used by platelet and leukocyte biologists. As proof of concept, we demonstrate that the motheaten-like hyperinflammatory phenotype observed on ablation of Shp1 with the Pf4-Cre transgene is absent in the Shp1 conditional KO mice generated with Gp1ba-Cre. These results discount the potential role of platelets in this phenotype and highlight the anomalies that arise from nonspecific ablation in Pf4-Cre-generated KO mice. The more lineage-specific gene ablation provided by the Gp1ba-Cre mouse will make it possible to delineate the relative contribution of platelets to a variety of pathophysiological processes and refine the involvement of leukocytes in thromboinflammatory conditions.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge all members of the Birmingham Biomedical Sciences Unit for maintenance of mouse colonies.
Z.N. and J.M. are British Heart Foundation (BHF) postdoctoral research associates (RG/15/13/31673) and A.M. is a BHF Intermediate Basic Science Research Fellow (FS/15/58/31784). This work was supported by BHF Senior Basic Science Research Fellowship FS/13/1/29894 and BHF Programme Grant RG/15/13/31673 (Y.A.S.). T.V. was supported by the Deutsche Forschungsgemeinschaft postdoctoral fellowship VO 2134/1-1. M.J.G. was funded by a Medical Research Council studentship GBT1564.
Authorship
Contribution: Y.A.S. provided conceptualization; Z.N., T.V., M.J.G., R.G., G.E.D., A.M., and Y.A.S. provided methodology; Z.N., T.V., M.J.G., J.M., S.H., G.D.N., G.E.D., A.M., and Y.A.S. provided the investigation; Y.A.S. provided resources; A.T., A.W., B.G.N., and Y.A.S. provided mouse models and reagents; Z.N. and Y.A.S. wrote the original draft; Z.N., T.V., A.M., and Y.A.S. performed review and editing; Y.A.S. provided supervision; and Y.A.S. provided funding acquisition.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yotis A. Senis, Institute of Cardiovascular Sciences, College of Medical and Dental Sciences, University of Birmingham, Birmingham, B15 2TT, United Kingdom; e-mail: y.senis@bham.ac.uk.