TO THE EDITOR:
Clonal evolution to myelodysplastic neoplasms/syndromes (MDSs) or acute myeloid leukemia represents the most serious late complication among patients with aplastic anemia (AA) and paroxysmal nocturnal hemoglobinuria (PNH).1,2 Most post-AA/PNH MDS cases present high-risk features, with chromosome 7 abnormalities and mutations in ASXL1, RUNX1, SETBP1, and RAS genes as the most frequent molecular lesions.3 However, other cases have more favorable outcomes and are mainly driven by low-risk cytogenetic alterations (eg, del[13q] and del[20q]).4 We previously showed that MDS arising after AA and PNH constitute a genetically distinct MDS subentity with cytogenetic and molecular features that are typical of myeloid evolution from a previous acquired bone marrow (BM) failure syndrome.1 Indeed, monosomy 7 as well as ASXL1 and RUNX1 mutations were significantly enriched in secondary AA and PNH cases when compared with a cohort of primary MDS pair-matched for other characteristics (age, gender, and BM blasts). Although some of the observed characteristics (eg, monosomy 7) may be similar to those of therapy-related MDS, cases arising from a prior acquired AA or PNH syndrome lack a specific enrichment in TP53 mutations and/or accumulation of complex karyotype abnormalities.5,6 Therefore, the specific genomic makeup, rarity of the disease, and information scarcity make the prognostication of MDS arising from AA and PNH challenging for routine clinical practice.
In recognition of the growing role of gene mutations, the Molecular International Prognostic Scoring System (IPSS-M) aims to improve the precision of prior classifications by adding molecular markers.7,8 Although validated for de novo and secondary/therapy-related cases,9-11 its applicability to post-AA/PNH MDS cases has not yet been explored. Here, we apply the IPSS-M to 69 post-AA/PNH MDS cases collected from a large multicenter data set of 1008 patients with AA/PNH, using a comparator cohort of primary MDS (n = 1281).
Complete clinical and molecular details of both AA/PNH1 and primary MDS9 cohorts have been previously described. A total of 69 patients developing MDS were selected for this study based on the availability of complete molecular annotations and clinical information. Diagnosis of MDS was based on the WHO 2016 criteria and substantiated with the new acquisition of cytogenetic abnormalities or significant dysplastic changes in previously healthy BMs.12 The IPSS-M R package tool indicated in the original study by Bernard et al8 (https://github.com/papaemmelab/ipssm) was used to compute IPSS-M, and the average score was selected to define the risk categories for subsequent outcomes analysis. Overall survival (OS) was defined as the time from MDS onset to last follow-up or death from any cause and was calculated using Kaplan-Meier estimation.13 Leukemia-free survival (LFS) was defined as the time from MDS onset to acute myeloid leukemia evolution. A 1:1 matched-pair analysis based on the nearest neighbor propensity score matching (R package MatchIt) was used to compare MDS cases arising from AA and PNH with matches from a cohort of de novo MDS using age, gender, and treatment type as relevant baseline covariates (confounders). Comparison between IPSS-M and IPSS-R was assessed via Harrell concordance index (C-index), using resampling with replacement. All statistical tests were 2-sided, and a P value <.05 was considered statistically significant. Review and collection of clinical and molecular data were performed in accordance with the protocols and written consent approved by the institutional review boards of each participating institutions and guidelines set forth by the Declaration of Helsinki.
Evolution to MDS occurred at a median time of 4.9 years (range, 1.9-8.3 years) from the initial diagnosis of AA and PNH. The median age at MDS onset was 63 years (interquartile range [IQR], 34-70 years), and most patients were male (Male:Female ratio, 1.65) (Table 1). Despite low BM blast percentages (median, 2; range, 0-7), 71% of the patients showed high-risk features per the IPSS-R (scores >3.514), mainly because of enrichment in poor-risk cytogenetics, most frequently monosomy 7. Overall, 48% of the patients were treated either with hypomethylating agents (28%), hematopoietic stem cell transplantation (60%), or both (12%). Leukemia progression occurred in 14% of cases after a median of 5.4 (1-14) months from post-AA/PNH MDS onset. With a median follow-up of 55.6 months (95% confidence interval, 42.5-75.8), the 60-month OS and LFS were 44% (32%-60%) and 42% (31%-58%), respectively.
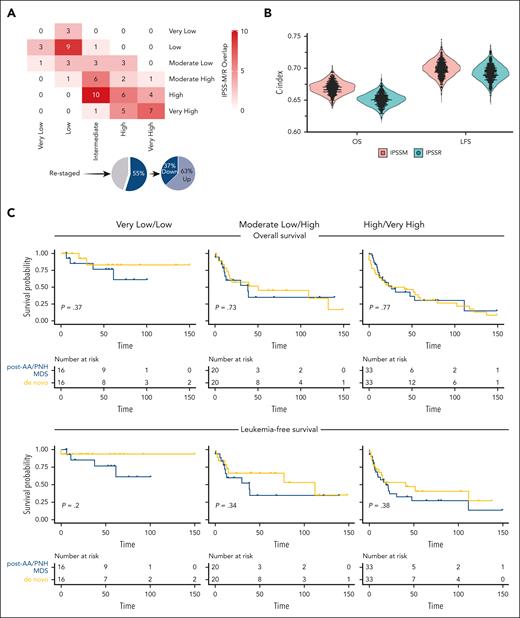
The IPSS-R distribution of our study cohort was as follows: very low (6%), low (23%), intermediate (30%), high (23%), and very high (18%). By applying IPSS-M, patients were redistributed in very low (5%), low (19%), moderate low (14%), moderate high (14%), high (29%), and very high (19%) risk categories. When comparing 5-to-5 (merging the moderate low/high IPSS-M categories for comparison purposes as previously done in the original study8), 55% of cases were restratified, of which 63% were up- or 37% down-staged (Figure 1A; supplemental Figure 1; supplemental Table 1). Indeed, 77% of patients with post-AA/PNH MDS harbored at least 1 mutation, with 43% presenting >2, thereby substantiating the differences observed between the 2 prognostication systems. Although according to IPSS-M, the majority (62%) of post-AA/PNH MDS cases still clustered in higher-risk categories, a nonnegligible fraction (38%) was assigned to lower-risk groups.
Application of IPSS-M to myelodysplastic neoplasms arising from AA and PNH. (A) A cross heatmap shows the distribution of patients with MDS based on the IPSS-R (x-axis) and the IPSS-M (y-axis) risk categories. Pie charts at the bottom illustrate the percentages of cases up- and down-staged. (B) C-indexes across bootstrapped samples (5000 runs) using risk groups as the covariate in OS and LFS modeling, showing a slight improvement of IPSS-M compared with IPSS-R. (C) OS and LFS of primary and secondary to aplastic anemia/PNH MDS cases within same IPSS-M risk categories after application of propensity score matching (1:1) for confounders (age, gender, and type of treatment received). From left to right, curves show very low/low, moderate low/high, and high/very high IPSS-M risk categories. Time (x-axis) is expressed in months.
Application of IPSS-M to myelodysplastic neoplasms arising from AA and PNH. (A) A cross heatmap shows the distribution of patients with MDS based on the IPSS-R (x-axis) and the IPSS-M (y-axis) risk categories. Pie charts at the bottom illustrate the percentages of cases up- and down-staged. (B) C-indexes across bootstrapped samples (5000 runs) using risk groups as the covariate in OS and LFS modeling, showing a slight improvement of IPSS-M compared with IPSS-R. (C) OS and LFS of primary and secondary to aplastic anemia/PNH MDS cases within same IPSS-M risk categories after application of propensity score matching (1:1) for confounders (age, gender, and type of treatment received). From left to right, curves show very low/low, moderate low/high, and high/very high IPSS-M risk categories. Time (x-axis) is expressed in months.
We then tried to assess the added benefit of IPSS-M for outcome predictions by computing Harrell c-statistics. The C-index was 0.67 (0.64-0.69) and 0.65 (0.63-0.67) in IPSS-M and IPSS-R for OS and 0.70 (0.67-0.72) and 0.69 (0.66-0.71) for LFS, respectively (Figure 1B).
To further explore whether IPSS-M could be used to also predict OS and LFS outcomes in post-AA/PNH MDS cases, we analyzed survival outcomes by taking advantage of a control cohort of de novo MDS cases. However, approximately half of the patients with post-AA/PNH MDS received disease-modifying treatments, potentially generating a bias when using survival as a surrogate for outcomes comparisons, even in cases falling into equivalent, molecularly defined risk categories. Therefore, we used a propensity score matching, accounting for age, gender, and hypomethylating agents and/or hematopoietic stem cell transplantation as relevant covariates to reduce possible confounders. As reported for other MDS subtypes in the original IPSS-M study,8 the OS and LFS outcomes for post-AA/PNH MDS paralleled those observed in the de novo group within the various IPSS-M risk categories (Figure 1C).
Altogether, to the best of our knowledge, we applied for the first time here the IPSS-M risk score in a cohort of MDS arising from AA and PNH and showed its differences from the prior IPSS-R prognostication system. It is important to notice that the actuarial survival of post-AA/PNH MDS cases was not distinct from that of de novo patients when matching for basic clinical features in equivalent IPSS-M risk groups. The genomic landscape of AA/PNH clonal evolution makes this MDS subtype a prototypic model, whereby the incorporation of molecular information may be important. However, to ensure adequate clinical management, some limitations must be noted.1,15 The majority of MDS cases arising from acquired BM failure syndromes typically show high-risk cytogenetic features (chiefly, monosomy 7), which already identify a specific prognostic subgroup within the IPSS-R, regardless of associated genomic makeup accounted for by the new IPSS-M. Thus, in our study IPSS-M did not add any further prognostic refinement. This does explain why we observed limited incremental benefit when assessing the predictive power of the new as compared with the older prognostication system. Indeed, we can envision IPSS-M to be more helpful in post-AA/PNH MDS cases lacking specific cytogenetic alterations whose prognostic role is already well established (eg, those without chromosome 7 abnormalities).
For all these reasons, the current efforts validating IPSS-M in a variety of MDS clinical settings9-11 are instrumental to define its clinical applicability and role in guiding both daily practice and patient allocation for clinical trials.16
Acknowledgments
The authors thank Elli Papaemmanuil and Elsa Bernard of the Memorial Sloan Kettering Cancer Center for their tremendous help in applying the IPSS-M tool in the study cohort.
This project was supported by National Institutes of Health, National Heart, Lung, and Blood Institute grant R35HL135795 (J.P.M.), grant AA&MDSIF (V.V., S.P., J.P.M.), VeloSano Pilot Award and Vera and Joseph Dresner Foundation–MDS (V.V.), Foundation For Rare Diseases and Association HPN France-Aplasie Médullaire, Italian Societies of Hematology and Experimental Hematology (S.P.), The Italian Foundation for Cancer Research 5 × 1000 project entitled “Metastatic disease: the key unmet need in oncology” to MYNERVA, award number 21267 (MYeloid NEoplasms Research Venture to M.T.V.), and Fondation Laurette-Fugain (J.S.). P.H.P. was supported by the American Society of Hematology Global Research Award and the Association HPN France-Aplasie Médullaire. C.G. was supported by a grant from the Edward P. Evans Foundation. R.T.C. was supported by the São Paulo Research Foundation.
Authorship
Contribution: C.G. collected, analyzed, interpreted clinical and molecular data, and wrote the manuscript; L.F.B.C. participated in patient recruitment, collected clinical and molecular data, and edited the manuscript; P.H.P. participated in patient recruitment, collected clinical and molecular data, and edited the manuscript; A.D. performed statistical analysis; S.P., L.L., M.C.B.I., F.S.d.F., M.S., and N.D. participated in patient recruitment and collected clinical and molecular data; A.L.P., V.A., L.H., E.C., and S.C.-Z. performed molecular analyses; M.T.V. and V.V. interpreted clinical and molecular data, gave helpful intellectual insights, and edited the manuscript; R.P.d.L., J.S., R.T.C., G.S., and J.P.M. designed the study, conceptualized, and sponsored the overall project and edited the manuscript; and all authors read and approved the final version of the manuscript and are accountable for all aspects of the work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jaroslaw P. Maciejewski, FACP Department of Translational Hematology and Oncology Research, Taussig Cancer Institute, 9620 Carnegie Ave, Building NE6-314, Cleveland, OH 44106; e-mail: maciejj@ccf.org.
References
Author notes
∗C.G., P.H.P., and L.F.B.C. contributed equally to this work.
†R.P.d.L., J.S., G.S., R.T.C., and J.P.M. contributed equally to this work.
Data are available upon request from the corresponding author, Jaroslaw P. Maciejewski (maciejj@ccf.org).
The online version of this article contains a data supplement.