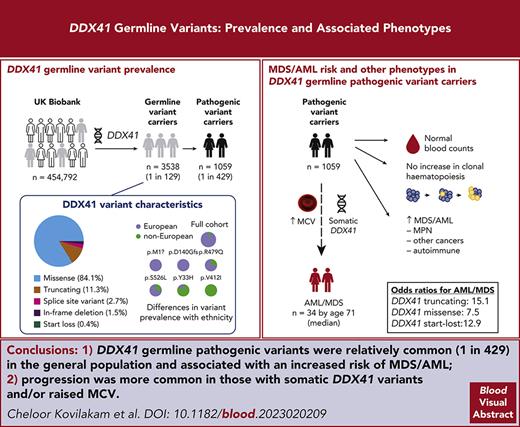
Key Points
We mapped DDX41 germ line variants in 454 792 volunteers and defined the risk of MDS/AML development associated with different variant types.
DDX41-mutant MDS/AML evolves differently from sporadic disease, but individuals at high risk often have somatic DDX41 mutations or a high MCV.
Abstract
Germ line variants in the DDX41 gene have been linked to myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML) development. However, the risks associated with different variants remain unknown, as do the basis of their leukemogenic properties, impact on steady-state hematopoiesis, and links to other cancers. Here, we investigate the frequency and significance of DDX41 variants in 454 792 United Kingdom Biobank (UKB) participants and identify 452 unique nonsynonymous DNA variants in 3538 (1/129) individuals. Many were novel, and the prevalence of most varied markedly by ancestry. Among the 1059 individuals with germ line pathogenic variants (DDX41-GPV) 34 developed MDS/AML (odds ratio, 12.3 vs noncarriers). Of these, 7 of 218 had start-lost, 22 of 584 had truncating, and 5 of 257 had missense (odds ratios: 12.9, 15.1, and 7.5, respectively). Using multivariate logistic regression, we found significant associations of DDX41-GPV with MDS, AML, and family history of leukemia but not lymphoma, myeloproliferative neoplasms, or other cancers. We also report that DDX41-GPV carriers do not have an increased prevalence of clonal hematopoiesis (CH). In fact, CH was significantly more common before sporadic vs DDX41-mutant MDS/AML, revealing distinct evolutionary paths. Furthermore, somatic mutation rates did not differ between sporadic and DDX41-mutant AML genomes, ruling out genomic instability as a driver of the latter. Finally, we found that higher mean red cell volume (MCV) and somatic DDX41 mutations in blood DNA identify DDX41-GPV carriers at increased MDS/AML risk. Collectively, our findings give new insights into the prevalence and cognate risks associated with DDX41 variants, as well as the clonal evolution and early detection of DDX41-mutant MDS/AML.
Introduction
Inherited variants in DDX41, the gene for DEAD-box RNA helicase 41, have been linked to an increased risk of myeloid neoplasia (MN), namely myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML).1 Insights into the type, prevalence, and clinical relevance of DDX41 variants have primarily come from studies of MDS/AML cases and their relatives.2-6 A recent study derived estimates of the risk of MDS/AML associated with different variants by comparing their prevalence in cases of MDS/AML with that observed among ∼20 000 healthy Japanese individuals.7 However, although valuable, risk estimates based on variant prevalence among MDS/AML cases vs the general population are influenced by the composition of the MDS/AML cohorts studied (eg, age, sex, driver mutations, etc) and cannot be used to estimate absolute risk in carriers of DDX41 germ line pathogenic variants (DDX41-GPV). Similarly, estimates of MDS/AML risk for relatives of patients that also carry DDX41-GPVs7 are likely to be higher than those of DDX41-GPV carriers in the general population. Here, to overcome these limitations and improve our understanding of the type, prevalence, and significance of DDX41 variants in the general population, we study 454 792 United Kingdom Biobank (UKB) participants for whom deep genetic, phenotypic, and clinical outcome data are available2 and analyze whole genome sequencing (WGS) data from 153 cases of AML with or without DDX41 variants from the United Kingdom’s 100,000 Genomes Project.8 Our findings give extensive new insights into the prevalence, clinical associations, MDS/AML risks, and leukemogenic processes associated with DDX41 variants.
Methods
DDX41 germ line variants in the UKB
Classification of germ line DDX41 gene variants
Germ line DDX41 variants predicted to result in protein truncation (namely stop-gained, frameshift, and essential splice) or start-lost (p.M1?) were considered pathogenic (DDX41-GPV). We also considered as DDX41-GPV all missense or in-frame deletion variants previously reported to cooccur with DDX41 somatic variants in MDS/AML samples3,4,6,7,11-14 or in pre-MDS/AML samples in the UKB (only 1 such sample was identified, carrying p.G313S together with the somatic DDX41 variant p.T227M that has been reported recurrently in DDX41-mutant MDS/AML). The remaining missense and in-frame deletion variants were classified as variants of unknown significance (VUS). Synonymous variants were considered nonpathogenic and grouped in a single set, with the exception of the common synonymous variant p.R400=, which was considered separately. For odds ratio estimates, splice site variants were grouped together with other truncating variants, and in-frame deletions were grouped together with missense variants. Participants without any DDX41 germ line variants were used as the control group for estimating odds ratios (OR) for different phenotypes.
Phenotypes
Phenotypic data were downloaded from UKB in April 2022. For each participant, the disease phenotypes were determined based on the presence of relevant traits (supplemental Table 1; available on the Blood website). For participants who had developed more than 1 myeloid neoplasm (myeloproliferative neoplasms [MPN], MDS, AML, or CMML), the first diagnosed disease was considered.
Statistical analyses
Multivariate logistic regression analysis using Python statsmodels (v.0.12.2)15 with sex (acquired from the central registry at recruitment or self-reported to the UKB), age, smoking status, and the first 10 principal components of genetic ethnicity as covariates were performed to detect associations between variables. For comparing the distribution of continuous variables between groups, we used the Mann-Whitney U test, and for associations between categorical variables, we used the Fisher exact test, both executed via the Python module Scipy (v.1.10.0).16 Bonferroni correction was applied to analyses involving multiple comparisons.
Somatic mutations in the UKB
Analysis of AML WGS data from Genomics England
Germ line DDX41 variants from WGS data of 153 AMLs were extracted from variant call format (VCF) files of germ line calls using BCFtools (v.1.11) and annotated using ANNOVAR (v.Nov2019)19 and Variant Effect Predictor (v.96).20 Somatic variants were extracted from tumor VCF files using BCFtools (v.1.11) and somatic single nucleotide variant (SNV) catalogs were generated using the signature.tools (v.2.1)21 package and used to calculate the somatic SNV burden of each AML.
The UKB resource was approved by the North West Multi-centre Research Ethics Committee under reference number 21/NW/0157 and all participants provided written, informed consent to participate. The 100,000 Genomes Project was approved by the NHS Health Research Authority, East of England–Cambridge South Research Ethics Committee (REC reference 14/EE/1112), and all participants provided written, informed consent to participate.
Results
DDX41 variants in the UK Biobank
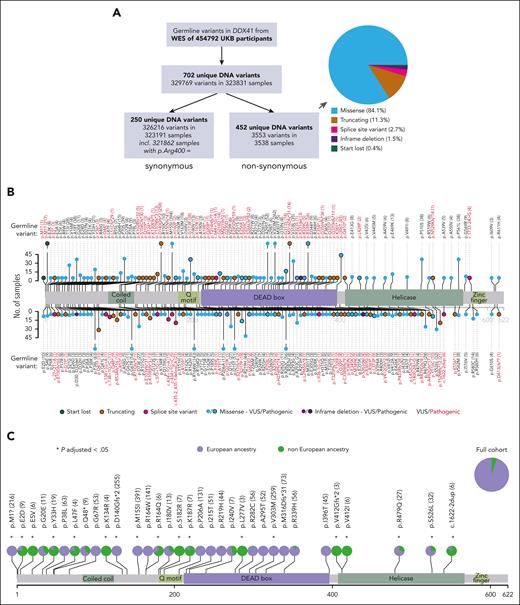
Analysis of blood DNA WES from 454 792 UKB participants (aged 37-73 years, median, 58 years) identified 3553 nonsynonymous DDX41 germ line variants in 3538 samples (Figure 1A-B), with 15 samples having 2 such variants (supplemental Table 3). Overall, we found 452 unique nonsynonymous variants distributed throughout DDX41 (Figure 1B; supplemental Table 4), with a lower density along the C-terminus. p.M155I was the most common nonsynonymous variant (n = 394), whereas 2 known pathogenic variants were also relatively common (p.M1?, n = 218 and p.D140Gfs∗2; n = 258). We next examined all 66 variants that had a frequency ≥0.0001 in Europeans or non-Europeans and found marked differences in prevalence between participants of European vs non-European ancestry, including p.M1? (start-lost) and p.D140Gfs∗2 that were present only in the former, as were several other nonsynonymous and synonymous variants (Figure 1C; supplemental Figure 1; supplemental Table 5). Overall, 55 nonsynonymous variants were unique to participants of non-European ancestry (supplemental Table 6), most of which have not been previously reported.
Prevalence of DDX41 germ line variants in the UKB and impact of ancestry. (A) Numbers and types of DDX41 germ line variants identified in the UKB. The pie chart subclassifies the 452 unique nonsynonymous variants. (B) Lolliplot depicting the location and frequency of nonsynonymous germ line DDX41 variants that are either known to be pathogenic (see “Methods”) or present in ≥3 participants. Variants are depicted above or below the protein sequence for clarity of representation only. The frequency of each variant in UKB is reflected in its lollipop height and is also entered numerically next to each variant. Different types of variants are distinguished by their color and the presence/absence of a black border, as per the key. (C) Differences in the prevalence of common DDX41 nonsynonymous variants (with a frequency ≥ 0.0001 in participants of European or non-European ancestry) between individuals with European (lavender) vs non-European (green) ancestry. Asterisks mark variants displaying a significantly different distribution among the 2 ancestries compared to the full cohort (P-adjusted < .05; P values adjusted according to Bonferroni correction for 66 tests).
Prevalence of DDX41 germ line variants in the UKB and impact of ancestry. (A) Numbers and types of DDX41 germ line variants identified in the UKB. The pie chart subclassifies the 452 unique nonsynonymous variants. (B) Lolliplot depicting the location and frequency of nonsynonymous germ line DDX41 variants that are either known to be pathogenic (see “Methods”) or present in ≥3 participants. Variants are depicted above or below the protein sequence for clarity of representation only. The frequency of each variant in UKB is reflected in its lollipop height and is also entered numerically next to each variant. Different types of variants are distinguished by their color and the presence/absence of a black border, as per the key. (C) Differences in the prevalence of common DDX41 nonsynonymous variants (with a frequency ≥ 0.0001 in participants of European or non-European ancestry) between individuals with European (lavender) vs non-European (green) ancestry. Asterisks mark variants displaying a significantly different distribution among the 2 ancestries compared to the full cohort (P-adjusted < .05; P values adjusted according to Bonferroni correction for 66 tests).
Risk of MN, other cancers, and common autoimmune diseases in carriers of DDX41 variants
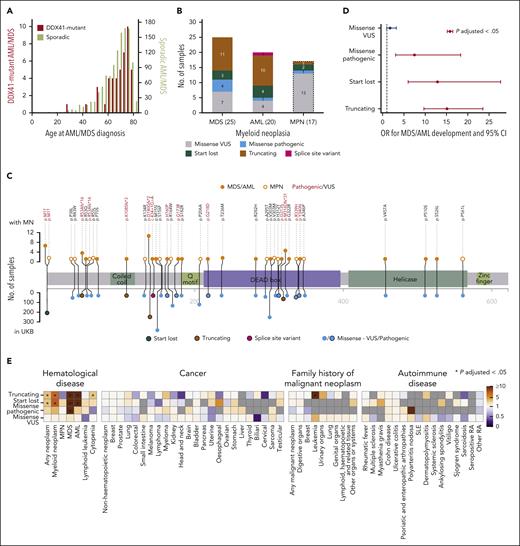
We next investigated the association between DDX41 germ line variants and MN risk and found that of 3538 DDX41 variant carriers, 25 developed MDS and 20 AML. The median age at MDS/AML onset was 71 years (Figure 2A), and there was a male predominance (male:female = 35:10 vs 1:1.2 in the UKB; OR, 4.15; P = 1.61 × 10–5; Fisher exact test), whereas carrier rates for DDX41 variants did not differ between males and females (supplemental Table 7). Of the 45 individuals who developed MDS/AML, 21 had truncating, 7 start-lost, 16 missense, and 1 had splice site variants (Figure 2B-C). Using logistic regression with age, sex, smoking status, and the first 10 principal components of genetic ethnicity as covariates, we found the OR for developing MDS/AML to be 15.12 (95% confidence interval [CI], 9.70-23.55; P = 3.30 × 10–33), 12.89 (95% CI, 5.99-27.73; P = 6.09 × 10–11) and 7.49 (95% CI, 3.06-18.34; P = 1.06 × 10–5), respectively, for truncating, start-lost, and pathogenic missense variants (Figure 2D; supplemental Table 8). Next, we evaluated the risk associated with individual variants identified in UKB participants with MDS/AML and also present in at least 10 UKB participants in total (supplemental Figure 2). Interestingly, the rare variant R53Afs∗16 was found to impart a very high risk, with 4 of 16 carriers developing MDS/AML (OR = 129.82; 95% CI, 38.80-434.32; P = 2.85 × 10–15) compared with odds ratios for more common variants, such as D140Gfs∗2 and start-lost. We did not identify any close relatives (third degree or closer) among the 45 DDX41 germ line variant carriers, proposing that leukemic progression was not substantially affected by coinherited genetic variation. However, there were a small number of close relatives among GPV carriers (48 relations among 1059 carriers).
Associations of DDX41 germ line variants with MDS/AML and other phenotypes. (A) Distribution of age at MDS/AML diagnosis of 45 DDX41 germ line variant carriers (red, left y-axis) and 1190 noncarriers (olive, right y-axis) who developed AML or MDS. (B) Frequencies of DDX41 variant types among carriers who developed AML, MDS, and MPN (variants in carriers who developed MPN are shown only for comparison and were not used for variant risk estimation). (C) Location and frequency of DDX41 variants present in participants who developed MN. The frequency of each variant in participants who developed MN is shown on the top and in the entire UKB on the bottom panel. Variants present in participants who developed MDS/AML are depicted as closed orange circles and MPN as open orange circles. Variant types are color-coded as per the figure key. (D) OR for developing MDS/AML with different types of germ line DDX41 variants compared with that of participants with no such variants. OR that are significantly different from 1 (P-adjusted < .05; P values adjusted for 173 tests, including the association tests for other phenotypes shown in Figure 2E, Bonferroni correction) are depicted in red. (E) Associations between different DDX41 variant types and other phenotypes. Gray color indicates that the analysis was not possible as no variant carrier developed the phenotype. An asterisk denotes significant associations after multiple testing correction (P-adjusted < .05; P values adjusted for 173 tests). SLE, systemic lupus erythematosus; RA, rheumatoid arthritis.
Associations of DDX41 germ line variants with MDS/AML and other phenotypes. (A) Distribution of age at MDS/AML diagnosis of 45 DDX41 germ line variant carriers (red, left y-axis) and 1190 noncarriers (olive, right y-axis) who developed AML or MDS. (B) Frequencies of DDX41 variant types among carriers who developed AML, MDS, and MPN (variants in carriers who developed MPN are shown only for comparison and were not used for variant risk estimation). (C) Location and frequency of DDX41 variants present in participants who developed MN. The frequency of each variant in participants who developed MN is shown on the top and in the entire UKB on the bottom panel. Variants present in participants who developed MDS/AML are depicted as closed orange circles and MPN as open orange circles. Variant types are color-coded as per the figure key. (D) OR for developing MDS/AML with different types of germ line DDX41 variants compared with that of participants with no such variants. OR that are significantly different from 1 (P-adjusted < .05; P values adjusted for 173 tests, including the association tests for other phenotypes shown in Figure 2E, Bonferroni correction) are depicted in red. (E) Associations between different DDX41 variant types and other phenotypes. Gray color indicates that the analysis was not possible as no variant carrier developed the phenotype. An asterisk denotes significant associations after multiple testing correction (P-adjusted < .05; P values adjusted for 173 tests). SLE, systemic lupus erythematosus; RA, rheumatoid arthritis.
The association between DDX41 mutations and MDS/AML is well established, but less is known about links to MPN. In the UKB, only 17 of 3538 participants with DDX41 germ line variants developed MPN (Figure 2B-C), and logistic regression analysis did not identify an increased risk of MPN in carriers of different types of DDX41 variants (Figure 2E; supplemental Figure 3). In addition, given reports of a possible association between DDX41 variants and lymphoid malignancies,22,23 we investigated possible links with lymphoma or other cancers but did not identify any significant associations (Figure 2E). Specifically, carriers of DDX41 p.R164W, a variant previously linked to familial lymphoma, did not confer a higher incidence of lymphoma (1/143) than the rest of UKB (1/102). As DDX41 is involved in the cyclic GMP-AMP synthase–stimulator of interferon genes (STING)-type I interferon pathway24 that is linked with various autoimmune diseases, we also looked for association of DDX41 germ line variants with 16 common autoimmune diseases but found no significant association (Figure 2E).
Blood cell parameters, DDX41 somatic variants, and early detection of MDS/AML
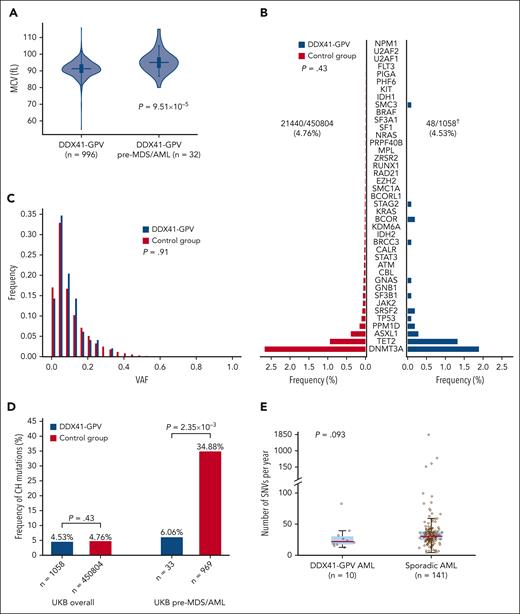
We first found that baseline blood cell parameters in carriers of different types of germ line DDX41 variants did not differ from controls (all UKB participants except those with nonsynonymous DDX41 variants and all participants who developed MN), with the exception of a marginally lower red blood cell count among carriers of variant p.D140Gfs∗2 (supplemental Figure 4; supplemental Table 8). We next investigated blood count results in the 32 individuals with available data who developed AML or MDS after sampling and found that their mean red cell volume (MCV) was higher compared with that of DDX41-GPV carriers who did not develop MDS/AML (median, 95.05 vs 91.3 fL; P = 9.51 × 10–5; Mann-Whitney U test, Figure 3A). In fact, of 25 pathogenic DDX41 variant carriers in the UKB with MCV >100.2 fL (=2 standard deviations above the mean), 4 (16%) developed MDS/AML within 1500 days (OR, 6.66; P = .006; Fisher exact test).
Early detection and leukemic evolution in DDX41-GPV carriers. (A) Distribution of MCV in DDX41-GPV carriers who did not develop any MN vs those who developed MDS/AML. (P = 9.51 × 10–5). (B) Prevalence and (C) variant allele fraction distribution of somatic variants associated with CH in individuals with DDX41-GPV vs controls (all UKB participants lacking nonsynonymous DDX41 germ line variants). There were no differences between the 2 group with regards to CH prevalence (OR, 0.89; 95% CI, 0.66-1.20; P = .43, multivariate logistic regression) or clone size (P = .91; Mann-Whitney U test). (D) Comparison of frequencies of CH somatic mutations in all DDX41-GPV carriers vs controls overall (left), and specifically in those UKB participants who later developed MDS/AML (right). (E) Total somatic SNV burden in the genomes of DDX41-GPV–associated (n = 10) vs sporadic (n = 141) AML cases. There was no significant difference between the 2 groups (P = .093, Mann-Whitney U test), indicating that there is no increased mutagenesis in the former. In panels A and E, the horizontal lines mark the medians, the box marks the interquartile range, and whiskers extend to the last data point lying within 1.5× the interquartile range (Tukey whiskers). † denotes somatic mutation calling with Mutect2 failed for 1 of 1059 DDX41-GPV samples.
Early detection and leukemic evolution in DDX41-GPV carriers. (A) Distribution of MCV in DDX41-GPV carriers who did not develop any MN vs those who developed MDS/AML. (P = 9.51 × 10–5). (B) Prevalence and (C) variant allele fraction distribution of somatic variants associated with CH in individuals with DDX41-GPV vs controls (all UKB participants lacking nonsynonymous DDX41 germ line variants). There were no differences between the 2 group with regards to CH prevalence (OR, 0.89; 95% CI, 0.66-1.20; P = .43, multivariate logistic regression) or clone size (P = .91; Mann-Whitney U test). (D) Comparison of frequencies of CH somatic mutations in all DDX41-GPV carriers vs controls overall (left), and specifically in those UKB participants who later developed MDS/AML (right). (E) Total somatic SNV burden in the genomes of DDX41-GPV–associated (n = 10) vs sporadic (n = 141) AML cases. There was no significant difference between the 2 groups (P = .093, Mann-Whitney U test), indicating that there is no increased mutagenesis in the former. In panels A and E, the horizontal lines mark the medians, the box marks the interquartile range, and whiskers extend to the last data point lying within 1.5× the interquartile range (Tukey whiskers). † denotes somatic mutation calling with Mutect2 failed for 1 of 1059 DDX41-GPV samples.
Somatic mutations affecting the second DDX41 allele are present in most cases of DDX41-mutant MDS/AML and most commonly take the form of the R525H substitution, which disrupts the adenosine triphosphate-dependent helicase function of the protein.1,7 To determine if such changes can be identified before MDS/AML onset, we searched for DDX41 somatic mutations in WES from all 454 335 UKB participants using Mutect2 (supplemental Methods). This identified 342 DDX41 somatic variants in 321 participants, including 5 of the 1059 carriers of pathogenic germ line DDX41 variants (OR vs controls, 5.51; 95% CI, 2.05-14.82; P = 7.15 × 10–4; logistic regression). Notably, of these 5 carriers of both a DDX41 germ line pathogenic and a somatic variant, 1 was diagnosed with MDS 201 days earlier, and 2 developed AML or MDS subsequently, including 1 with p.G313S (see “Methods”; supplemental Table 10). This suggests that the detection of somatic variants can herald subsequent progression to MN. However, Mutect2 did not identify any R525H somatic mutations in carriers of DDX41-GPVs, which proposes that R525H mutations may be associated with more rapid progression to MDS/AML. As the sequencing depth in WES data are relatively shallow (supplemental Table 2), we further investigated this by analyzing R525 codon sequencing pileups to look for low-level variants missed by Mutect2 in the 1059 carriers of DDX41-GPVs. This identified 2 participants with 2 reads reporting somatic R525 variants: 1 with R525H and 1 with R525S (which is not known to occur somatically in DDX41-mutant MDS/AML). Notably, the R525H carrier developed MDS within 2 years of blood sampling, whereas the R525S carrier did not. Notably, the identification of DDX41 somatic variants in individuals lacking germ line DDX41 was not associated with subsequent MDS/AML development (OR, 1.4; 95% CI, 0.19-9.93; P = .74; logistic regression).
DDX41 mutations and preleukemic evolution
Sporadic MDS/AML commonly arises from preexisting clones of CH; however, the clonal evolution of DDX41-mutant MDS/AML is not very well understood. To determine whether DDX41 germ line mutations are associated with an increased prevalence of CH overall or CH due to specific mutations, we searched for CH driver mutations in 454 335 UKB exomes using Mutect2 (supplemental Methods). This identified 22 987 CH driver mutations in 21 608 UKB participants, of whom 48 were carriers of a DDX41-GPV. The prevalence and clonal size of CH among carriers of pathogenic DDX41 germ line variants did not differ from those of controls (Figure 3B). Furthermore, the rate of CH in carriers of DDX41-GPV who subsequently developed MDS/AML was significantly lower than that in participants who subsequently developed sporadic MDS or AML (2/33 [6.06%] vs 338/969 [34.88%]; OR, 0.11; 95% CI, 0.03-0.45; P = 2.35 × 10–3; logistic regression, Figure 3D), suggesting that DDX41-mutant MDS/AML does not generally develop through progression of CH. Furthermore, we explored the possibility of an association between DDX41-GPV and mutations in genes other than known CH drivers using WGS data from 12 DDX41-mutant AML samples from the Genomics England cohort and found no gene to be recurrently mutated in these samples (supplemental Table 11).
Finally, given the links between DEAD-box RNA helicases and genomic stability,25 we investigated if the increased risk of MDS/AML in DDX41-GPV carriers may be due to an increased rate of somatic mutations. Analysis of WGS data of 153 adult AML samples from the Genomics England cohort did not show any significant differences in mutation rates (number of somatic SNVs/year) between DDX41-GPV–mutant (n = 10) and sporadic AML (n = 141) (P = .093; Mann-Whitney U test, Figure 3E and supplemental Table 11). Two AML cases with DDX41 as variants of unknown significance were excluded from this comparison.
Discussion
Pathogenic germ line mutations in the DDX41 gene are the leading cause of familial MDS/AML,5,26,27 yet their prevalence and associated risks remain incompletely understood, as does the basis of their leukemogenic properties.28 Here, we provide a comprehensive description of DDX41 mutations in >450 000 UKB participants, the largest such study to date. First, we show that DDX41 GPVs are relatively common in the general population (∼1 in 429), and this may need to be considered when devising guidelines for choosing unrelated adult donors for hematopoietic stem cell transplantation. We go on to show that the likelihood of developing AML or MDS (but not MPN) is several times higher in DDX41-GPV carriers than controls (OR, 12.33), with start-lost and truncating variants (OR, 12.89 and 15.12, respectively) conferring approximately twice the risk of missense ones (OR, 7.49). Nevertheless, a high OR could be used as a PS4 evidence criterion for the pathogenicity of DDX41 substitution variants. We also found that after a ∼13-year follow-up of the UKB population (median age 58 years at recruitment), the absolute risk of developing MDS/AML in DDX41-GPV carriers was 3.21% (5.50% in male and 1.37% in female carriers), compared with 0.26% for sporadic MDS/AML (0.32% in males vs 0.21% in females, calculated using the total numbers of AML + MDS in UKB participants lacking nonsynonymous DDX41 variants). These estimates are lower than those derived from relatives of DDX41-mutated MDS/AML,7,29 most likely because the latter group is enriched in higher-risk variants (eg, truncating) than the UKB/general population, although the possibility of coinheritance of risk-modifying alleles in trans cannot be excluded.
We also reveal significant differences in the prevalence of known DDX41 pathogenic variants among individuals of different ancestries and identify several variants that were not previously reported. Certain variants were common enough to expect that they would be identified by chance in sporadic MDS/AML, despite being likely nonpathogenic (eg, M155I), emphasizing that recurrence in MDS/AML should not be used as the sole criterion for pathogenicity. Furthermore, we performed the first large-scale analysis of associations of DDX41-GPV with hematological and other cancers and confirmed the strong association with AML and MDS, but found no association with MPN, lymphoma, or other cancers. Similarly, we found no association between DDX41-GPVs and 16 common autoimmune diseases.
Furthermore, we used data from the UKB and Genomics England to investigate the paths and mechanisms of progression to DDX41-mutant MN. This revealed that the prevalence of CH is not higher in DDX41-GPV carriers and that, unlike sporadic MDS/AML,30,31DDX41-mutant MDS/AML does not commonly evolve from preexisting CH but follows a distinct evolutionary path. This is reflected in observations that the mutational spectra in DDX41-mutant AML differ from those of sporadic AML.3 In addition, given the links between DEAD-box RNA helicases and DNA damage,25 we wanted to investigate the possibility that DDX41-GPV may drive leukemogenesis by increasing the rate of somatic mutations, but found this not to be the case by comparing total somatic SNV rates in WGS data from sporadic vs DDX41-mutant AML. Finally, our study reveals that, among carriers of DDX41-GPV, a raised MCV and the presence of somatic DDX41 mutations are biomarkers of increased risk of progression to MDS/AML and should be monitored in the context of early detection/prevention in families with these mutations. Precise recommendations for how DDX41-GPV carriers should be monitored will require input from panels of experts; however, it may be reasonable to propose that, after the age of ∼40 years, a rise in MCV should trigger additional investigations and that routine monitoring for DDX41 somatic mutations should be considered (particularly R525H).
Collectively, our study gives significant new insights into the population-wide prevalence, clinical significance, and leukemic evolution associated with germ line DDX41 gene mutations that will assist research into familial MDS/AML and help guide the clinical management of DDX41-GPV carriers, patients, and their families.
Acknowledgments
The authors thank the participants and investigators involved in the UK Biobank resource and Genomics England Limited who collectively made this research possible.
This work was funded by an Early Detection Project Grant from Cancer Research UK (EDDCPJT∖100010). W.G.D. is funded by a Clinical Research Fellowship from the Cancer Research UK Cambridge Centre (CTRQQR-2021∖100012). S.P.K. is supported by a United Kingdom Research and Innovation Future Leaders Fellowship (MR/T043202/1). S.N.-Z. is supported by a National Institute for Health and Care Research (NIHR) Research Professorship (NIHR301607). P.M.Q. is funded by the Miguel Servet Program (CP20/00130). G.S.V. is supported by a Cancer Research UK Senior Cancer Fellowship (C22324/A23015) and work in his laboratory is also funded by the European Research Council, Leukemia and Lymphoma Society, Rising Tide Foundation for Clinical Cancer Research, Kay Kendall Leukaemia Fund, Blood Cancer UK, and the Wellcome Trust. This research was also supported by the NIHR Cambridge Biomedical Research Centre (NIHR203312). This research was conducted using the UK Biobank resource under approved application 56844. This research was made possible through access to the data and findings generated by the 100,000 Genomes Project (RR239). The 100,000 Genomes Project is managed by Genomics England Limited (a wholly owned company of the Department of Health and Social Care). The 100,000 Genomes Project is funded by the National Institute for Health Research and NHS England. The Wellcome Trust, Cancer Research UK, and the Medical Research Council have also funded research infrastructure. The 100,000 Genomes Project uses data provided by patients and collected by the National Health Service as part of their care and support.
Authorship
Contribution: G.S.V. conceived, designed, and supervised the study; S.C.K. carried out data analyses and generated tables and figures; M.G. called somatic mutations; P.M.Q. managed UK Biobank data access, performed somatic mutation filtering, and helped with regression models; W.G.D. analyzed mutation rates in Genomics England AML samples with help from S.N.-Z. L.M., C.B., I.M., S.P.K., S.N.-Z., and M.A.F. contributed ideas and analytical tips/guidance during this project; S.C.K. and G.S.V wrote the manuscript with help from all coauthors; and all authors approved the final version of the manuscript.
Conflict-of-interest disclosure: G.S.V. is a consultant to STRM.BIO and holds a research grant from AstraZeneca for research unrelated to that presented here. M.A.F. is an employee and stockholder of AstraZeneca. The remaining authors declare no competing financial interests.
A complete list of the members of the Genomics England Research Consortium appears in the supplemental Appendix.
Correspondence: George S. Vassiliou, Department of Haematology, University of Cambridge, Cambridge Stem Cell Institute, Jeffrey Cheah Biomedical Centre, Cambridge CB2 0AW, United Kingdom; e-mail: gsv20@cam.ac.uk.
References
Author notes
Individual-level UK Biobank data, including DDX41 and CH DNA variants, can be requested via application to the UK Biobank (https://www.ukbiobank.ac.uk). Access to AML whole-genome sequencing data can be requested via application to Genomics England (https://www.genomicsengland.co.uk/research/research-environment).
The DDX41 call set has been returned to the UK Biobank to enable individual-level data linkage for approved UK Biobank applications. Primary data from the 100,000 Genomes Project, which are held in a secure research environment, are available to registered users. Refer to https://www.genomicsengland.co.uk/research/academic for further information.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.