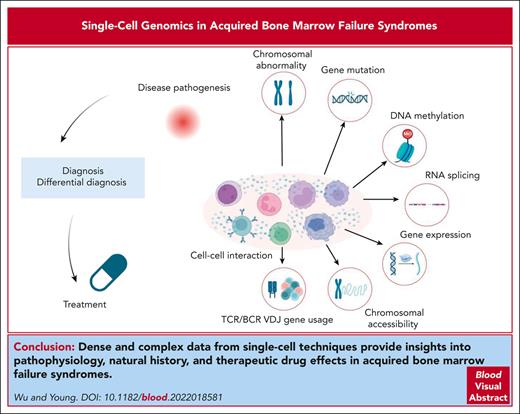
Abstract
Mechanistic studies of immune bone marrow failure are difficult because of the scarcity of residual cells, the involvement of multiple cell types, and the inherent complexities of hematopoiesis and immunity. Single-cell genomic technologies and bioinformatics allow extensive, multidimensional analysis of a very limited number of cells. We review emerging applications of single-cell techniques, and early results related to disease pathogenesis: effector and target cell populations and relationships, cell-autonomous and nonautonomous phenotypes in clonal hematopoiesis, transcript splicing, chromosomal abnormalities, and T-cell receptor usage and clonality. Dense and complex data from single-cell techniques provide insights into pathophysiology, natural history, and therapeutic drug effects.
Introduction
Inability to produce blood cells follows from chemical and physical damage (as with cytotoxic drug therapies and radiation), as a component of constitutional syndromes (classically Fanconi anemia and the telomere biology disorders), or as an acquired disease. Acquired bone marrow failure (BMF) diseases include aplastic anemia (AA), (hypoplastic) myelodysplastic syndrome (MDS), paroxysmal nocturnal hemoglobinuria, pure red blood cell aplasia and other single-lineage syndromes, and large granular lymphocytic leukemia (LGLL).1 Patients typically respond to immunosuppression: these diseases are more specifically categorized as immune BMF syndromes.1-6 Their pathogenesis is broadly understood from decades of laboratory studies using traditional methods of cell culture, functional assays, immunophenotyping, and molecular biology using blood and marrow samples, as well as animal models.7-25 However, scarcity of marrow cells, the heterogeneity of hematopoietic and immune system elements, and the complexity and variability of intrinsic cell behaviors and cell–cell interactions have been major limitations to deeper understanding of disease processes.
Conventional laboratory methods require in vitro manipulation: physical cell separation, exposure to nonphysiologic conditions of oxygen and temperature, concentrations of growth factors, culture media, other cell populations and cell densities, and regenerative stress. Laboratory experiments are optimized to provide measurable outputs in vitro, which are assumed to correlate with in vivo physiology and pathophysiology. Optimization also entails simplification, and conventional assays consequently generate low dimensional and single-layer data that cannot identify the complex parallel processes at play. Absence of specific cell-membrane markers of cytogenetically or genetically abnormal cells has hindered examination of dysregulated molecular mechanisms. Thus, familiar techniques, or gold standards, have considerable deficiencies.
Recent rapid, often startling, advances in single-cell methods are based on genomics, large-scale and deep sequencing of DNA and RNA, and associated protein detection. Single-cell studies coupled with bioinformatics generate extensive, multidimensional, and multiomic information from limited numbers of cells, an ideal approach to study BMF. Genomic approaches have encountered some skepticism, in part because of the massive amounts of data, reliance on mathematical calculations, “noise,” and the peculiarities of the workflow. Most of the experimenters’ involvement begins after the “wet” laboratory work is completed, the experimental designs may purposely avoid hypothesis testing, and cooperation must be achieved between (nonquantitative) biologists and computational analysts (untrained in biology). Single-cell RNA sequencing (scRNA-seq)-related data should be broadly consistent with results from conventional experiments and our understanding of pathophysiology, but new genomic methods should also reveal novel and unanticipated phenomena (Figure 1). A brief summary of background knowledge of BMF diseases is shown in Table 1.
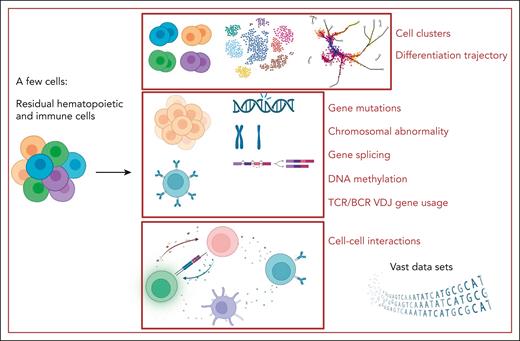
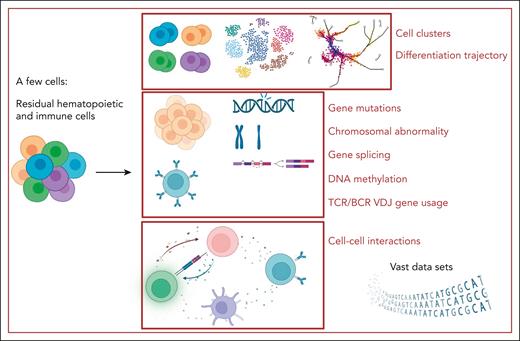
Single-cell multiomics study in BMF syndrome. By starting with just a few cells, single-cell multiomics studies can yield a wealth of information on various aspects of BMF syndrome diseases and treatment: vast data sets, cell clusters, hematopoietic differentiation trajectory, chromosomal abnormalities, gene mutations, gene splicing, DNA methylation, TCR/B-cell receptor (BCR) variable diversity joining (VDJ) gene usage, and cell–cell interactions. Figure created with BioRender.com.
Single-cell multiomics study in BMF syndrome. By starting with just a few cells, single-cell multiomics studies can yield a wealth of information on various aspects of BMF syndrome diseases and treatment: vast data sets, cell clusters, hematopoietic differentiation trajectory, chromosomal abnormalities, gene mutations, gene splicing, DNA methylation, TCR/B-cell receptor (BCR) variable diversity joining (VDJ) gene usage, and cell–cell interactions. Figure created with BioRender.com.
Immune AA
In acquired AA, the initiating antigenic targets of the immune response cells are unknown. Antigenic targets and potential viral infections can be inferred from T-cell receptor (TCR) usage: TCR clones that are individual-public (shared among patients) and/or disease specific (common among patients and absent in other populations) would implicate a common initiating antigen. However, in a study using scRNA-seq and scTCRαβ-seq (scRNA + TCRαβ-seq),64 the epitope specificities of individual-private (not shared among individuals) response clonotypes were unlikely to be a common viral antigen but a more homogenous target population within the clones of patients with AA.64 Most T cells with private response clonotypes had an activated CD8+ effector phenotype, characterized by expression of GZMH, GNLY, and PRF1. Private response clonotypes were suppressed in a patient responding to immunosuppression and increased in a patient who was not responding. In a screen for somatic mutations in AA, variants were found to be common in both patients and healthy controls but enriched in CD8+ T cells in AA, and they were located in the JAK-STAT and MAPK pathways.65 Mutation burden was associated with CD8+ T-cell clonality. Paired scRNA + TCRαβ-seq in patients with STAT3 or other mutations in CD8+ T cells linked clonotypes with phenotypes. In 2 index patients, somatic STAT3 mutations were restricted to a single CD8+ T-cell clone. Phenotypically, STAT3 mutations associated with CD8+ terminally differentiated effector memory T cells, which exhibited enhanced expression in pathways of immune response, cytotoxic, and lymphocyte activation. With immunosuppression, TCRVB clones carrying STAT3 mutations decreased in 1 case on normalization of blood counts but further increased in another patient during response and later relapse.
Subpopulations of CD8+ and CD4+ T cells in AA have been imputed from single-cell data to directly interact with hematopoietic stem and progenitor cells (HSPCs). In pediatric AA cases, mass cytometry (cytometry by time of flight) identified a subgroup of noncanonical CD4+ naive T cells with elevated expression of pNFkBS529 and pSTAT3Y705.66 Using 5′ scRNA-seq of CD3+ BM mononuclear cells (BMMNCs), activation of the JAK3/STAT3 pathway in Th17-polarized CD4+ naive T cells in patients with severe AA (SAA) was observed. Proteomics and metabolomics analyses of plasma and BM supernatants from patients with SAA were the basis for another scRNA-seq experiment.67 Differential proteins and metabolites in SAA were related to energy metabolism, the complement and coagulation cascades, and hypoxia-inducible factor (HIF)-1a signaling pathways. On reanalyzing scRNA-seq data, these pathways were enriched in T cells from patients with AA. A highly activated CD38+CD8+ T-cell subset, which was increased in AA and a murine model of AA, contained genes relating to T-cell activation (the glycolysis or gluconeogenesis pathway, HIF-1a signaling, and the complement-associated pathways). Zhu et al sequenced sorted single HSPCs and T cells from patients with AA.68 CD4+ T cells showed upregulation of genes associated with antigen presentation and cell death regulation, whereas CD8+ T cells displayed high expression of genes associated with cytokine production. There were increased interactions between HSPCs and T cells in AA, including Fas/Fas ligand and tumor necrosis factor (TNF) receptors/TNF-α, already implicated in immune-mediated disruption of hematopoietic cells; some of these ligand–receptor interactions were affected by treatment.
Immune cells do not function in isolation but operate within complex and dynamic networks, which can be recognized in single-cell data. Using high-dimensional mass cytometry and subcluster frequency correlation analyses of the BM, 2 cell networks were identified in AA.69 Network AA was composed of CD16+ myeloid cells, CCR6++ B-cells, Th17-like CCR6+ memory CD4+ T cells, and KLRG1+ terminally differentiated effector memory CD8+ T cells. These cells were increased in AA before immunosuppressive therapy, and with hematologic response, the immune cell compartment largely normalized, with reduced numbers of CD16+ myeloid cells. In a single-cell transcriptomic analysis focused on natural killer (NK) cells from the peripheral blood (PB) and the BM of patients with SAA,70 8 clusters of NK cells were identified, indicating remarkable cellular heterogeneity. NK cell numbers in both the PB and BM were reduced in SAA, and their cytotoxic function was downregulated.
Regulatory T cells downmodulate autoreactive T cells, a mechanism of central tolerance, and they are decreased in many autoimmune diseases, including AA.22,23 Recent studies have highlighted the immune suppressive functions of regulatory B cells (Bregs). CD19+CD24hiCD38hi Bregs can suppress cytotoxic T lymphocytes and Th1 responses and promote conversion of CD4+ T cells to regulatory T cells via interleukin-10, PDL1, CD80, CD86, and CD1d.71-75 Bregs are reduced in AA, particularly in very severe disease, but residual Bregs remain functional and produce interleukin-10.76 In scRNA-seq analysis of the BM cells of 2 patients with SAA,77 focusing on B-cell receptor and variable diversity joining genes, the highest pairing frequencies were between IGHV3-20-IGKJ2, IGHV3-20-IGKJ4, and IGHV3-20-IGHLJ2, and 3 V genes (IGHV3-7, IGHV3-33, and IGLV2-11) had elevated expression in B cells of patients with AA. The ligand–receptor pairs of B cells with hematopoietic cells involved antigen presentation, inflammation, apoptosis, and proliferation of B cells. This study was limited by the small number of patients and lack of correlation of B-cell receptor usage with B-cell phenotypes. Cell cross talk appears to have a crucial role not only in immune responses and inflammation in BMF syndrome but also in the pathogenesis of clonal hematopoiesis (CH).
Genes coding for RNA splicing factors are frequently mutated in CH of indeterminate potential (CHIP) and in myeloid malignancies.78,79 Spliceosome genes have been reported as differentially expressed in HSPCs from patients with AA; full-length scRNA-seq of HSPCs showed altered isoform usage for thousands of genes.68 When the splicing spectrums of aggregated HSPCs of patients with AA and MDS were compared, there were shared altered splicing events, and the affected genes were associated with DNA damage and repair response (FANCG, ATF2, and RFC1) and cell cycling signals. These results suggested a possible mechanism of AA progression to MDS. In addition, in AA there was downregulation of genes regulating poly-A tail shortening in AA HSPCs, and for many genes, altered poly-A tail usage associated with DNA repair signaling.68
MDSs
scRNA-seq has been applied to define the transcriptome in lineage-negative BMMNCs in MDS; features included upregulation of neutrophil granule genes and downregulation of ribosomal genes in MDS.80 In a large cohort of patients with MDS,81 scRNA-seq was used to validate the 2 distinct differentiation patterns in 2 representative patients from their lineage-negative CD34+ HSPC compartments. In common myeloid progenitor–pattern MDS, the cells atop the HSPC hierarchy maintained the transcriptional profile of the most immature long-term repopulating hematopoietic stem cells (HSCs), including expression of MLLT3, PBX1, and HLF. In granulocytic-monocytic progenitor–pattern MDS, these cells expressed myeloid-affiliated genes in the lymphoid-primed multipotent progenitor population, including CEBPA and CSF3R. Pseudotime analysis of HSPCs from patients with MDS showed trajectories that converged at the myeloid progenitor state, consistent with similar myelomonocytic differentiation potentials and the clinical phenotypes of the 2 groups of patients with MDS. These findings were extended using mouse models and ex vivo perturbations to identify molecular drivers in blast progression after failure of frontline hypomethylating agent treatment.
Spliceosome genes are more frequently mutated in MDS than in AA. Among these mutated genes, SF3B1, a core component of the spliceosome complex, remains most prevalent across hematologic malignancies and solid tumors.82-84 Splicing aberrations have been documented in a recent advanced multiomics single-cell approach in samples with mutated SF3B1.85 GoT-Splice, integrates genotyping of transcriptomes with enhanced efficiency for long-read single-cell transcriptome profiling, with proteogenomics (cellular indexing of transcriptomes and epitopes by sequencing). This new technique allows simultaneous profiling of gene expression, cell surface protein markers, somatic mutation status, and RNA splicing within individual cells, overcoming limitations of 3′- or 5′-biased short-read sequencing. SF3B1-mutated cells in the megakaryocytic-erythroid lineage showed increased fitness, as inferred from upregulation of genes involved in cell cycle and messenger RNA (mRNA) translation. SF3B1-mutated cells also had aberrant 3′ splicing site usage. Disruptive and pathogenic SF3B1 mutation–driven missplicing affected key mediators of hemoglobin synthesis and differentiation at all stages of erythroid maturation. The single-cell approach enabled the detection of erythroid lineage bias and cell type–specific cryptic 3′ splice site usage in SF3B1-mutated cells in patients with CH, preceding the development of overt MDS.
Complex clonal and molecular landscapes in MDS and other myeloid malignancies have been described from next-generation sequencing. As inferred from bulk DNA sequencing and bioinformatic analyses, mutations are acquired stepwise, but these methods cannot discriminate mutations in the same clone or define the sequence of mutation acquisition. Somatic mutations are common in patients with MDS, and many patients carry multiple mutations. Coexistence of splicing factor mutations in patients with myeloid malignancy can be inferred from both bulk and single-cell DNA sequencing (scDNA-seq) analyses.86 In the majority of cases, mutations in splicing factor genes were mutually exclusive, with <1% of patients carrying 2 concomitant mutations (∼50% of such double mutations were found in the same individual cells). Patients with double mutations showed selection against the most common alleles and selection for less common alleles, preserving 1 wild-type allele. A possible functional basis for the coexistence of splicing factor mutations is that SF3B1 and SRSF2 alleles, which are enriched in patients with double mutations, have a reduced impact on RNA splicing and/or binding compared with more common alleles.
In a study using scDNA-seq to evaluate the clonal dynamics of pathogenic mutations in 2 patients with MDS,87 clonal heterogeneity of pathogenic mutations, including FLT3-ITD, IDH2, EZH2, and GATA2, was associated with disease progression and resistance to hypomethylating agent therapy, and was accompanied by copy number loss in DNMT3A, TET2, and GATA2. scDNA-seq detected rare cell clones and mutations that were undetectable by bulk tumor sequencing. To further investigate the clonal framework of myeloid malignancies, scDNA-seq of 31 frequently mutated genes was performed in 146 samples from 123 patients with myeloid malignancies, including CHIP, myeloproliferative neoplasm, and acute myeloid leukemia (AML).88 AML was dominated by a small number of clones, which frequently harbored cooccurring mutations in epigenetic regulators. Mutations in signaling genes were often present more than once in distinct subclones, consistent with increasing clonal diversity. Simultaneous scDNA-seq and immunophenotyping revealed differential lineage contributions of DNMT3A R882 (myeloid bias) and DNMT3A R635Q (less in myeloid and B-cell lineages).
Chromosomal abnormalities are typical of cancer and hematologic neoplasms, and also occur in BMF.89-92 The presence of complex cytogenetics and monosomy 7 is prognostic of refractory cytopenia, clonal evolution to MDS/AML, and an adverse prognosis in AA.3 Assessing the functional implications of these cytogenetic abnormalities at the cellular level is difficult because of the absence of markers to distinguish abnormal cells from diploid cells. scRNA-seq allows identification of aneuploid cells, by analysis of relative global gene expression levels, copy-number variation, and loss of heterozygosity. Monosomy 7 cells in patients with MDS, including 2 cases of de novo MDS and 3 cases with clonal evolution from AA, had diverse differentiations pattens and showed downregulation of genes involved in immune response, DNA damage checkpoints, and apoptosis pathways.93 Monosomy 7 cells also displayed downregulated long noncoding RNAs associated with immune response, cell apoptosis and cell death, and DNA modification,94 suggesting coordinated mRNA and long noncoding RNA transcription in the regulation of cellular functions. Monosomy 7 and trisomy 8 are frequent chromosomal abnormalities in GATA binding protein 2 (GATA2) deficiency, a constitutional disease with immunologic and hematologic manifestations, and they corelate with disease prognosis and malignant transformation. scRNA-seq of HSPCs in patients with GATA2 deficiency provided molecular signatures of monosomy7, trisomy 8 cells, and complex cytogenetic abnormalities.95
T-cell LGLL
scRNA + TCRαβ-seq profiling was used to analyze sorted CD45+ PB mononuclear cells and CD3+ T cells from patients with T-cell LGLL (T-LGLL) in 2 recent companion studies from the National Institutes of Health and the University of Helsinki. Their data are complementary: the American team examined patients before and after effective therapy, and the Finnish group examined a wider range of cell types and also included other autoimmune diseases and hematopoietic malignancies.96,97 In both studies, bioinformatics was conducted in conjunction with bulk RNA-seq and TCRβ-seq data; flow cytometry, serum protein profiling, and ex vivo validations provided complementary data. TCRαβ-seq analysis provided high-resolution profiling of individual clones and allowed for flexibility in adjusting the threshold for comparisons. TCR clones (at least 2 cells with identical TCRs) and expanded clones (at least 10 cells with identical TCRs) were identified in patient samples. T-LGLL has been hypothesized to be driven by chronic antigen exposure, and efforts have been made to identify shared antigens imputed from common TCR sequences. However, in both studies T-LGLL clonotypes were restricted to individual patients (and therefore private), and no structural amino acid–level similarities were identified (no disease-specific clones), even when the analysis focused only on 43 HLA-A∗02+ T-LGLL clones.96 These results imply a lack of common clonotypes in T-LGLL.96,97 TCR clones in T-LGLL are present in healthy donors but at very low frequencies,97 and antigen-driven clonotypes are even more frequently observed in healthy controls than among nonantigen-driven clonotypes in T-LGLL.96 Antigen-driven clonotypes defined in T-LGLL might recognize commonly encountered antigens, including cytomegalovirus pp65.
Single-cell analysis has directly linked phenotypes to clonotypes. In our study, T cells belonging to the same clonotype (with identical TCR sequences) had similar transcriptional phenotypes and they occupied a confined region in a t-distributed stochastic neighbor embedding projection: TCR use may affect T-cell phenotypes.97 On diffusion mapping, T-cell activation and TCR usage were the main components contributing to T-cell phenotypes, with the most expanded clones being effector memory or activated T cells. In the Finnish study, in contrast to the positive correlation of TCR clonality with an effector memory phenotype observed in AA65 and in our study of T-LGLL,97 clonally expanded T cells appeared more phenotypically diverse than in healthy donors, but they had higher expression of proliferation, activation, and exhaustion genes, and lower expression of antiapoptosis genes.96 We classified clone dynamics (based on clone size changes) after treatment with alemtuzumab into 3 patterns: increasing, decreasing, and unchanged. Clones with increasing sizes showed upregulation of genes enriched in immune response and cell activation, whereas these genes were downregulated in unchanged and decreased clone groups.97 Expanding, antigen-driven wild-type STAT3 clones had higher cytotoxic gene expression than in the decreasing STAT3 mutated clones.96
Various cytokines and chemokines were elevated in T-LGLL, likely produced by monocytes and dendritic cells rather than by T cells, and these cytokines can remain elevated after treatment, despite suppression of T cells and a hematologic response.97 The nonleukemic immune cell repertoire likely also has a role in T-LGLL pathogenesis; T cell–derived interferon gamma (IFN-γ) may drive activation of nonleukemic immune cells. T-LGLL clones had elevated predicted cell–cell interactions and many costimulatory interactions with other immune cells.96 scRNA + TCRαβ-seq has expanded our understanding of TCR usage, T-cell clonality with phenotype and activation, and clonal dynamics with treatment.
CH
Experiments in human CH can be problematic because of phenotypic and transcriptomic similarities between mutated and wild-type cells, which make it difficult to isolate an abnormal population by morphology or cell surface markers. Mutated clones at low variant allele frequency are diluted in bulk cell experiments. Although the exact mechanisms by which somatic mutations disturb hematopoietic homeostasis are not yet fully understood, both cell-intrinsic and non–cell-intrinsic effects of mutations, including interactions between mutated cells and the BM environment, likely are important. Multimodal single-cell sequencing techniques can simultaneously detect gene mutations, gene expression, and proteins, and, thus, integrate genotype–phenotype correlations, differentiation bias, and associations among different types of cells.
Direct comparison of mutated clones with wild-type cells at the single-cell level in the same individual has shown proliferative advantages and lineage bias of mutated cells.98,TET2-mutated HSCs were shifted toward a more differentiated state in pseudotime, with downregulated long-term HSC signatures; these mutated clones expanded further in multiple downstream progenitors. Upregulation of myeloid lineage–affiliated transcription factors (CEBPD and IRF8) in TET2-mutated granulocytic-monocytic progenitors may be the basis of myeloid skewing early in differentiation. The clone size of DNMT3A-mutated cells was maintained throughout differentiation, without differentiation delay or lineage bias. Multimodal single-cell sequencing capturing genotype, transcriptomes, and methylomes in HSPCs, was applied to individuals with DNMT3A R882-mutated CH and multiple myeloma.99 DNMT3A mutations resulted in myeloid bias and an expansion of immature myeloid progenitors primed toward a megakaryocytic-erythroid fate, with dysregulated expression of lineage and leukemia stem cell genes. Mutated DNMT3A cells displayed preferential hypomethylation of polycomb repressive complex 2 targets and a specific CpG flanking motif. Notably, the hypomethylation motif was enriched in binding motifs of key hematopoietic transcription factors, a potential mechanistic link between DNMT3A mutations and aberrant transcription. Single-cell studies also facilitated identification of cooccurring mutations within the same cell. Cells with double mutations of DNMT3A/TET2 or biallelic TET2 mutations tended to have a higher variant allele frequency, and these mutations together may confer an enhanced advantage.100
An inflammatory phenotype is frequently associated with CH, especially with TET2 mutations: evidence includes skewing to proinflammatory tissue-resident macrophages, and clinical associations of atherosclerotic cardiovascular disease, obstructive pulmonary disease, and gout. Supporting CHIP as a secondary phenomenon are observations of clonal expansion of DNMT3A-mutated cells in mouse models of infection and inflammation.101-106 Inflammation might be driven by primary cell-autonomous effects of the CH mutant cells, or preexisting CH mutations may be adaptive and expand secondary to an inflammatory environment. Gene signatures associated with previous exposure to inflammation are upregulated in HSCs from individuals with CH, supporting a role of inflammation in the development of CH.98 In addition, wild-type HSCs in DNMT3A- and TET2-mutated samples showed enhanced expression of inflammatory, quiescence, and chemokine gene signatures, and enhanced cell proliferation, compared with wild-type HSCs in non-CH samples. Similarly, wild-type TET2 HSCs from CH individuals had aberrant IFN-response signatures compared with wild-type HSCs from healthy controls. In a mouse model, transplantation of heterozygous Tet2-hKO cells resulted in an enhancement of IFN-response signatures in recipient wild-type cells.100 Overall, these observations suggest that the CH clones affect wild-type cells and may alter the BM environment to promote further positive selection of CH clones.
Other BMF syndromes
Marrow failure is a recognized complication of immunotherapy for cancer and, like other autoimmune toxicities (hepatitis and colitis), is assumed to be secondary to off-target immune effects.107,108 Cytopenias are frequent after chimeric antigen receptor T-cell (CAR T) infusions: local inflammation, CH, and MDS have been hypothesized as mechanisms (MDS has been diagnosed in ∼5% of patients).109-114 scRNA-seq of the BM aspirates from 16 patients with diffuse large B-cell lymphoma treated with axicabtagene ciloleucel, of whom 11 had a grade 3 to 4 cytopenia at day 30,115 revealed GZMH+ FGFBP2+ CD8+ T cells with a cytotoxic signature, and IFN signaling and inflammatory pathways were elevated in multiple immune cells and hematopoietic cells. Compared with CD8+ T cells, from patients without toxicity CD8+ T cells in patients with CART-associated cytopenia had more clonal expansion but they did not express the CAR transcript. Using scRNA + TCRαβ-seq, we obtained similar results in a diffuse large B-cell lymphoma case treated with tisa-cel116; T cells with oligoclonal expansion and a CD8+CD57+ phenotype were observed, as in AA and T-LGLL.
In vacuoles, E1 enzyme, X-linked, autoinflammatory, somatic (VEXAS) syndrome, somatic UBA1 mutations in HSPCs and myeloid cells in the PB cause decreased ubiquitylation, which triggers cellular stress responses that upregulate the unfolded protein response, and activate multiple innate immune pathways.42 We, and others, have begun to investigate VEXAS syndrome using integrated analyses of single-cell immunophenotyping, bulk RNA-seq, cytokine profiling, scRNA-seq of peripheral cells and skin in patients with VEXAS syndrome, and overlapping phenotypes (VEXAS syndrome–like autoinflammatory disease, low-risk MDS, and healthy controls).117 Circulating monocytes are quantitively decreased with features of inflammatory activation and exhaustion. Migration of monocytes to tissues also contributes to monocytopenia: CD16+CD163+ monocytes and M1 macrophages localize in skin lesions of patients with VEXAS syndrome. Analysis of PB mononuclear cells using scRNA-seq confirmed dysregulated proinflammatory and cell death signatures in VEXAS monocytes. In our study of BMMNCs and HSPCs, there was early activation of inflammatory pathways (in particular TNF-α and both IFN-α and IFN-γ) in the HSC compartment, likely intrinsic to UBA1-mutated cells; hematopoiesis markedly biased toward myeloid (particularly granulocytic) differentiation of VEXAS syndrome HSCs; and increased apoptosis of UBA1-mutated lymphoid progenitors, all potential mechanisms of clonal dominance of UBA1-mutated myeloid cells and for lymphocytopoiesis.118 Ongoing single-cell multiomic approaches of VEXAS syndrome BM should reveal early events in HSPCs and the cell-autonomous and nonautonomous immune activations of UBA1-mutated cells.
Frequent events and molecular pathways involved in BMF syndromes are summarized in Figure 2. Main findings from key studies using single-cell genomics techniques on immune BMF diseases are in Table 2.
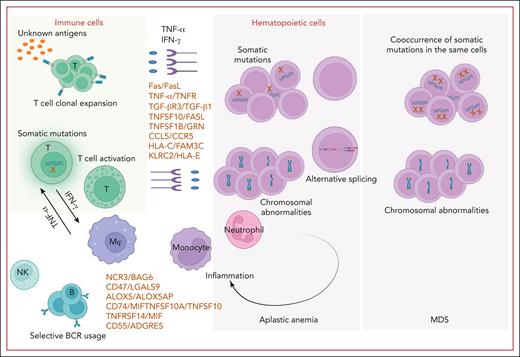
Frequent events and molecular pathways involved in immune BMF syndrome. In immune BMF syndrome, T cells, with or without somatic mutations (eg, on STAT3) undergo clonal expansion and activation. This occurs after recognizing unknown antigens, causing the T cells to polarize to type 1 T helper cell response and stimulate cytotoxic T cells. Excessive production of proinflammatory cytokines like IFN-γ and TNF-α and cytotoxic T lymphocytes lead to HSPC destruction via Fas/Fas ligand (FasL) and the TNF-α and receptor apoptosis pathway. Newly identified (by scRNA-seq) ligand–receptor pairs such as TGF-βR3/TGF-β1, TNFSF10/FasL, and CCL5/CCR5 also contribute to cell–cell interactions in this process. B cells exhibit distinct BCR usage and interact with other cell types through ligand–receptor pairs such as NCR3/BAG6, CD47/LGALS9, and ALOX5/ALOX5AP. NK cells play an immunomodulatory role in AA. Somatic mutations and chromosomal abnormalities are prevalent and participate not only in CH but also contribute to an inflammatory phenotype through cell-autonomous and non–cell-autonomous effects. Alternative splicing is observed in AA and MDS, contributing to lineage bias. Figure created with BioRender.com.
Frequent events and molecular pathways involved in immune BMF syndrome. In immune BMF syndrome, T cells, with or without somatic mutations (eg, on STAT3) undergo clonal expansion and activation. This occurs after recognizing unknown antigens, causing the T cells to polarize to type 1 T helper cell response and stimulate cytotoxic T cells. Excessive production of proinflammatory cytokines like IFN-γ and TNF-α and cytotoxic T lymphocytes lead to HSPC destruction via Fas/Fas ligand (FasL) and the TNF-α and receptor apoptosis pathway. Newly identified (by scRNA-seq) ligand–receptor pairs such as TGF-βR3/TGF-β1, TNFSF10/FasL, and CCL5/CCR5 also contribute to cell–cell interactions in this process. B cells exhibit distinct BCR usage and interact with other cell types through ligand–receptor pairs such as NCR3/BAG6, CD47/LGALS9, and ALOX5/ALOX5AP. NK cells play an immunomodulatory role in AA. Somatic mutations and chromosomal abnormalities are prevalent and participate not only in CH but also contribute to an inflammatory phenotype through cell-autonomous and non–cell-autonomous effects. Alternative splicing is observed in AA and MDS, contributing to lineage bias. Figure created with BioRender.com.
Constitutional BMF syndromes
Single-cell methods have been applied to inherited disorders including pure red blood cell aplasia in its congenital form, Diamond-Blackfan anemia,45-53 GATA2 deficiency,54-58 deficiency of adenosine deaminase 2 (DADA2),59-62 and telomere biology disorders63 (Table 1). Transcriptomics of single erythroid progenitors in patients with Diamond-Blackfan anemia demonstrated shortened cell cycle in erythroid progenitors, and IFN-α–inhibited cell cycle progression in patients responding to glucocorticoid treatment.120 In GATA2 deficiency, HSCs with increased erythroid/megakaryocytic priming contribute to aberrant lymphoid/myeloid differentiation.95 By sequencing single monocytes, increased nonclassical monocytes and activation of IFN pathways were implicated in DADA2; previously unsuspected cross talk of monocytes with T cells appeared to drive upregulation of STAT1 and activation and cytotoxicity of T cells in DADA2.121,122 Transcriptome and chromatin accessibility assays of single HSPCs in telomerase-deficient mice and patients with heterozygous pathogenic germ line TERT mutations showed that cell-intrinsic upregulation of the innate immune signaling response directly compromised self-renewal in HSCs and led to their exhaustion; targeting the IFI16 signaling axis of a cytosolic DNA sensor overcame IFN activation and skewed differentiation toward the megakaryocytic lineage in telomere-dysfunctional HSCs.123
Conclusions and future directions
For investigators who have learned and used single-cell genomics, the method can appear revolutionary, as remarkable as the invention of the microscope with regard to the quality, quantity, and interrelatedness of the data generated. In acquired AA, cytotoxic lymphocyte destruction of stem cells has been implicated experimentally for decades, but no technique could provide the detailed resolution of scRNA-seq or cytometry by time of flight, with which relatively superficial analysis many unsuspected features of the immune pathophysiology and the status of target cells have been revealed. Like other remarkable inventions, single-cell genomics developed from a simple, if challenging, combination of technologies, that is, single-cell separation and sequencing and the associated algorithms are critical. The vast amount of information generated by single-cell experiments is susceptible to deep mining and reanalysis. As with much in the digital revolution, the “black boxes” containing the equations can be discomfiting, and the computational team generating the data may be strangers to biologic and medical research questions. Mitigating concerns regarding the black box, sequence and code are available for most single-cell sequencing experiments to allow reanalysis, reinterpretation, and interlaboratory comparisons and reconciliations.
Single-cell data are different quantitatively and qualitatively from the historical results acquired in hematology laboratories to address blood diseases: experiments based on cell culture and phenotyping, cell biology, biochemistry, and molecular biology. Compared with conventional experiments, the amount of data from a single scRNA-seq run is orders of magnitude greater: hundreds to thousands of individual cells, hundreds to thousands of transcripts from thousands of individual genes as the initial data set, which is then subject to digital quality control and deep analysis of mRNA length, splicing, and concomitant mutations detectable in complementary DNA or mRNA and now also simultaneous proteomics and epigenetics of individual cells. Analysis of raw data involves filtering, accounting for missing data, mathematical and statistical adjustments, and a wide variety of algorithms to assess ligands and their receptors and other cell–cell relationships, much of which is novel to the biologically oriented experimentalists. As with any powerful new technology, the method itself can impel novel approaches to biology that would not be suggested in conventional experiments.
Methods of validation of scRNA-seq are not well established, universally accepted, or necessarily rational; for example, quantitative gene amplification and measures of alternatively spliced transcript sizes almost always replicate sequence data. “Orthogonal” studies, poorly defined in theory but commonly requested by reviewers and editors, may be of an uncertain value because of the biologic complexity of transcription regulation, and RNA processing, protein translation, transport, and degradation. Whether results based on isolated and manipulated cells in strikingly unphysiologic conditions under artificial laboratory conditions should be privileged as "gold standards", should retain interpretive value, or could be predicated to capture the complex interactions of single-cell genomics, is arguable. The sheer number of such hypothetical experiments that might follow on a scRNA-seq result is so large as to be impractical, nor is it obvious how such correlative studies should be assessed statistically to validate a sequencing experiment. Finally, single-cell genomics is sufficiently novel that, in the absence of simple and universally applied platforms, pipelines of data processing, and analytical methods, comparison of results among laboratories remains problematic. Exactly how single-cell genomics, as well as associated proteomics and immunogenic studies, will be integrated into historical data and routine research laboratory approaches is unclear. Nonetheless, the amount, depth, and dimension of data generated by single-cell methods cannot be replicated or replaced by other methods.
Over time, single-cell genomics have evolved from a monolayer of data of gene expression or mutation to integration with protein markers and, ultimately, combined for examination of transcript splicing, DNA methylation, and other epigenetic modifications. Combined with advanced and improving bioinformatics, single-cell genomics can be applied to nearly every aspect of hematopoiesis: cell identity and abundance, differentiation potential and trajectories, chromosomal abnormalities, gene mutations, gene splicing, and epigenetics alterations, all with or without disease or perturbations. Due to high cost and scarcity of samples, current single-cell studies in BMF diseases have focused on elucidating disease pathogenesis in small patient cohorts, characterizing disease features, and only a few have explored treatment mechanisms and effects. Future studies on larger patient numbers in diseases with overlapping manifestations, and on longitudinal samples before and after treatment should facilitate identification of biomarkers for diagnosis, differential diagnosis, and predictors of response to treatment. We anticipate the appearance of single-cell studies of many human diseases; most immediately, single-lineage BMF syndromes and immune cell compositions in MDS, as well as mechanistic studies in animal models and with in vitro perturbations with growth factors, cytokines, and specific cell populations. Comparison and collation of these large, but also enormously rich, data sets are the challenges for the future.
Acknowledgments
The authors thank Sachiko Kajigaya, Emma Groarke, Bhavisha Patel, Jichun Chen, Diego Quinones Raffo, Juan Coelho Da Silva, and Ash Lee Manley (the National Heart, Lung, and Blood Institute/National Institutes of Health) for their critical reading of the manuscript.
This research was supported by the intramural research program of the National Heart, Lung, and Blood Institute. National Institutes of Health.
Authorship
Contribution: Z.W. and N.S.Y. reviewed the relevant literature, wrote the manuscript, and drew the figures.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Zhijie Wu, Hematology Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, 10 Center Dr, Bethesda, MD 20892-1202; e-mail: zhijie.wu@nih.gov.