Abstract
One of the most challenging aspects of stem cell research is the reliance on retrospective assays for ascribing function. This is especially problematic for hematopoietic stem cell (HSC) research in which the current functional assay that formally establishes its HSC identity involves long-term serial transplantation assays that necessitate the destruction of the initial cell state many months before knowing that it was, in fact, an HSC. In combination with the explosion of equally destructive single-cell molecular assays, the paradox facing researchers is how to determine the molecular state of a functional HSC when you cannot concomitantly assess its functional and molecular properties. In this review, we will give a historical overview of the functional and molecular assays in the field, identify new tools that combine molecular and functional readouts in populations of HSCs, and imagine the next generation of computational and molecular profiling tools that may help us better link cell function with molecular state.
Introduction
As a modern-day experimental hematologist, it is difficult to imagine a world where methods now considered essential for the study of single cells, such as fluorescence-activated cell sorting (FACS), single-cell transcriptomics, genetic barcoding, and single-cell transplantations, weren’t readily available. Indeed, clonal assays have been part of the very fabric of hematopoietic stem cell (HSC) research, with the first clonal assays dating back almost as far as the discovery of HSCs.1 The now classical colony forming unit spleen (CFU-S) assay demonstrated the clonal origin of individual hematopoietic colonies by microscopically examining the unique radiation-induced karyotypes of cells extracted from spleen colonies.2,3 Similarly, knowledge about the initial progeny of stem and progenitor cells was largely derived from in vitro methods in the 1970s and 1980s that assessed divisional symmetry (or asymmetry) and cellular morphology.4,5 Although the tools available to researchers have certainly developed since these initial experiments, it is humbling to think that much of our fundamental understanding of HSC biology stems from a time when relatively simple and crude assays were available. In this review, we will chart the history of such assays up to present day, move onto approaches that tackle the nature of retrospective assays (Figure 1) by combining cell function with molecular profiling, and look forward to a time when we can simultaneously monitor the initial cell state of functional HSCs and obtain a better understanding of the fundamental molecular wirings of these critical units of cell and gene therapy.
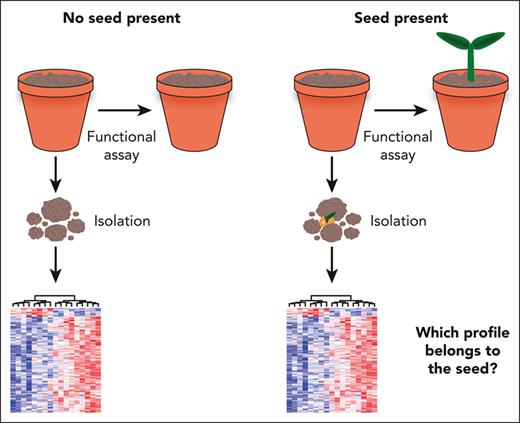
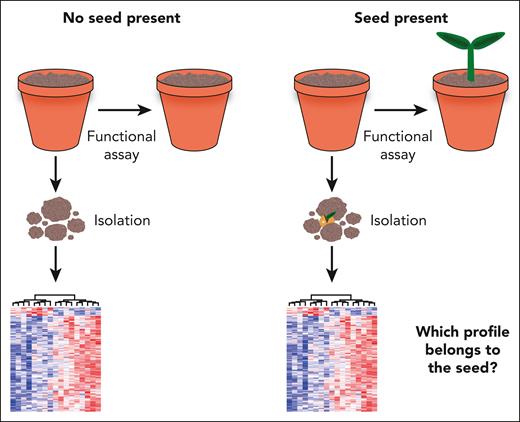
The fundamental challenge of retrospective assays. The vast majority of HSC assays require destruction of the initial cell state to perform either a functional or a molecular assay. Establishing that a cell population, being the “soil,” contains an HSC, the “seed,” necessitates lengthy functional assays but also destroys the seed in the process. Once the presence of an HSC has been established, the cell is no longer present, and the molecular program of similar cells isolated at the same time cannot easily be distinguished from other non-HSCs profiled at the same time.
The fundamental challenge of retrospective assays. The vast majority of HSC assays require destruction of the initial cell state to perform either a functional or a molecular assay. Establishing that a cell population, being the “soil,” contains an HSC, the “seed,” necessitates lengthy functional assays but also destroys the seed in the process. Once the presence of an HSC has been established, the cell is no longer present, and the molecular program of similar cells isolated at the same time cannot easily be distinguished from other non-HSCs profiled at the same time.
In vitro single-cell assays
In terms of clonal assays aimed at probing single hematopoietic stem and progenitor cell (HSPC) function, cultures performed in semisolid media (predominantly agar6 or methylcellulose7) followed by microscopy-based morphological analysis have been a robust and consistent tool. Semisolid culture systems, such as the colony forming cell (CFC) assay provide a simple yet elegant approach for tracking the cellular output of a single cell by physically restricting its progeny to grow in isolation from other progenitors in the dish. As such, these assays allow for robust identification of multipotent and self-renewing HSPCs from populations of largely unfractionated cells derived from various hematopoietic organs, something that is particularly useful in the absence of FACS. Cultures in semisolid media have been used in several landmark discoveries, including the identification of single HSPCs with multilineage potential in fetal liver, neonatal liver, and adult bone marrow (BM)8,9 and the demonstration of the dual ability of single HSPCs to self-renew and differentiate during serial replating; the latter representing early evidence for the existence of a hematopoietic hierarchy.10,11 In addition, CFC assays combined with sophisticated micromanipulation of the paired progeny of single hematopoietic progenitors were used in a series of studies examining differentiation behavior of HSPCs, demonstrating the stochastic nature of this process4,5: an idea pioneered already during the field’s infancy as the “hematopoiesis engendered randomly” hypothesis (recently reviewed by us elsewhere12). In vitro clonal HSPC assays on stromal feeder layers13 have also been instrumental in early as well as modern-day studies. One such example of a stroma-dependent in vitro assay with the ability to distinguish short- and long-term HSCs is the cobblestone area–forming cell (CAFC) assay,14 supported by the fact that the frequency of in vivo long-term repopulating cells in a BM cell suspension is strongly correlated with the ability to form cobblestone areas in coculture with stromal cells.15 Shortly after, a slightly less complicated long-term culture-initiating cell (LTC-IC) assay was introduced as another surrogate for in vivo assessment of stem cell activity in which a period of long-term culture is followed by a CFC assay to quantify the number of CFCs in a given cell suspension.16,17 Moreover, although many early clonal in vitro HSPCs assays, such as the CFC and CAFC culture systems described earlier were limited to assessing the cells’ ability to form myeloerythroid colonies, modification of the stromal layer used in LTC-IC assays enabled robust production of lymphoid cells.18 By transferring clonal cultures initiated in a CAFC assay to Whitlock-Witte conditions, researchers could then demonstrate combined myeloerythroid and lymphoid potential of HSPCs in vitro,17 thus, further expanding the toolbox of ex vivo assays capable of reliably identifying and quantifying primitive, multipotent hematopoietic progenitors in the BM. The most commonly used feeder-based assays in use today include the OP919,20 and OP9-DL121 cell lines, capable of supporting lympho-myeloid and T-cell differentiation, respectively, as well as the MS-5 stromal cell line, which supports the generation of B and myeloid cells from single human HSPCs.22 In particular, clonal analysis of index-sorted human HSPCs on the OP9, OP9-DL1, and MS-5 cell lines has led to identification of an isolation strategy for human lymphoid-restricted progenitors (multilymphoid progenitor).23 More recently, elegant combinations of feeder-based and feeder-free cultures with index sorting and single-cell RNA sequencing (seq) have been used to define lineage branching points in human cord blood HSPCs.24
In vivo single-cell assays
As described in “Introduction,” the inclusion of radiation-induced random chromosomal aberrations has been used to track the clonal progeny of HSPCs since the early 1960s2,25,26 and represents the earliest form of genetic barcoding experiments. Several decades later, another important breakthrough emerged when the significantly more efficient system of retroviral barcoding of HSCs was introduced. After the transplantation of retrovirally transduced BM cells (in which the individual integration site patterns could be tracked), the presence of mature cells in the BM, spleen, and thymus harboring viral integration sites provided formal evidence for a stem cell hierarchy in the hematopoietic system, with the most potent cells (HSCs) being capable not only of producing mature progeny of the B, T, and myeloid lineages but also of downstream, unilineage hematopoietic progenitors at the single-cell level in vivo.27-29
These assays made it increasingly clear that assays such as the CFU-S significantly overestimated the number of bona fide HSCs because of the inability of the CFU-S assay to distinguish HSCs from more downstream progenitors based on the reconstitution of mature cell lineages alone. In addition, concerns were raised about missing HSCs owing to recipients perishing because of myeloablation before sufficiently robust donor-derived reconstitution.30,31 This prompted a need for a new assay to assess HSCs more quantitatively, and the competitive repopulation unit assay was introduced as an elegant solution to the issue of HSC quantification, with the limiting dilution analysis becoming the gold standard for quantifying HSCs. In this initial work, Szilvassy et al tracked the ability of limiting numbers of male donor BM cells to reconstitute female recipients who underwent cotransplantation using compromised female BM cells, which retained the capacity for rapid production of mature blood cells but lacked durable self-renewal. Their results indicated a frequency of 1 in 1200 murine BM cells that met the definition of a competitive repopulation unit capable of sustaining hematopoiesis for 10 weeks.31 The true breakthrough in directly measuring in vivo clonal HSC function occurred a few years later when Osawa et al undertook transplantation of single FACS–purified HSCs and identified long-term (>3 months) lymphomyeloid reconstitution in a proportion of recipient animals.32 Subsequently, single-cell transplantations were used to formally show that all bona fide HSCs have the capacity to be engrafted into host animals33 (albeit with varying efficiency, depending on the developmental stages of the donor and recipient34) and to demonstrate that individual fetal,35 adult,36-38 and aged39 mouse HSCs exhibit sustained, heterogeneous behavior in vivo: properties that are today considered to be fundamental traits of HSCs, but these would have been impossible to prove without the use of single-cell transplantations. The combination of classical distinction of myeloid and lymphoid contribution based on differential expression of CD45.1 or CD45.2 between donor and recipient with fluorescent reporters for the megakaryocytic and erythroid lineages represents an even more comprehensive tool for assessing the lineage output of single HSCs in transplantation settings.40 Single-cell transplantation into immunodeficient recipient mice has also been used to formally prove that human HSC activity is contained within a fraction of cells expressing CD49f, providing an enrichment strategy for human HSCs from pools of progenitors for subsequent molecular and functional assays,41 although relative purities still lag behind those of mouse purification strategies.
Gene expression analysis coupled with functional assays
Although clonal functional assays reporting on single HSCs have been in place, essentially, since the discovery of HSCs, quantitative global molecular assays required substantial technological advances before being applicable to single cells. The desire to have such information at the single-cell level prompted rapid application of gene expression analysis in single cells using polymerase chain reaction (PCR), which was published just a few years after the initial invention of PCR.42,43 Methods enabling amplification of the global messenger RNA in a single cell were developed and applied first to mature hematopoietic cell types44 and subsequently to the clonal progeny of single hematopoietic progenitor cells,45,46 thus making the probing of rare populations of cells such as HSCs a tangible possibility.
With single-cell gene expression assays becoming more widely available and clonal functional assays growing increasingly sophisticated, the first attempts to integrate the 2 technologies in HSCs were reported. Using the knowledge about the heritable nature of daughter cell behavior after the first 2 or 3 divisions of immature hematopoietic precursors in vitro,4,5 Brady et al performed an elegant study in which some of the clonal progeny of HSPCs were collected for PCR analysis, and the remainder of the clone was left for subsequent characterization of the cells it would produce (plus an additional sampling for PCR).45 This approach, the first to link HSPC function with a molecular profile, enabled the identification of genes specific to particular hematopoietic lineages as well as distinct progenitor stages. Later works extended this assay to more highly enriched fractions of primitive HSPCs and genes with less well-defined roles in hematopoiesis.46 Since these pioneering studies, the field of gene expression analysis in HSCs has progressed tremendously in terms of throughput, from single-cell microarrays for specific target genes47 to the global characterization of the transcriptome in highly purified, single HSCs,48 shaping our view of hematopoiesis and its hierarchical organization.
FACS coupled to in vitro and in vivo assays
At this stage, it is important to highlight flow cytometry and FACS in the context of single-cell molecular assays that can be linked to HSC function. Since its invention in the late 1960s and subsequent commercialization approximately a decade later, FACS has had a tremendous impact on our ability to molecularly characterize single cells and continues to be one of the technological backbones in the field of experimental hematology.49 Although it is restricted to molecules on the cell surface, a FACS experiment coupled to a downstream functional assay is an excellent example of how one can simultaneously integrate the molecular and functional properties of single cells.
The first strategies for enriching primitive mouse hematopoietic precursors using FACS were reported as early as 1982.50 Building upon previous work identifying putative hematopoietic progenitors by using density centrifugation and morphological analysis,51 the De Leuuw group examined the CFU-S capacity of FACS-purified cells from the low-density fraction expressing wheat germ agglutinin50 and, later, wheat germ agglutinin in addition to the protein H-2K,52,53 resulting in an isolation strategy yielding >100-fold enrichment of cells with CFU-S capacities and radioprotective properties. Soon after, the work from Weissman et al showed that clonogenic hematopoietic progenitors, which lacked the expression of mature lineage markers (Lin– cells), could be further enriched via FACS purification of cells and had low expression of Thy-154 and positively expressed both the Sca-155 and cKit56 antigens. Using downstream in vitro and in vivo assays, their work demonstrated that the population of cells immunophenotypically defined as Lin– Sca-1+ cKit+ (LSK) contains the vast majority of the HSC activity in the mouse BM.
The next leap forward was the discovery of the signaling lymphocytic activation molecule (SLAM) markers, which allowed further discrimination between HSCs and their downstream progeny. They were identified via comparative gene expression analysis of Thy-1lo LSK cells and their nonstem cell progeny,57 further demonstrating the power of integrating molecular profiling with functional assays. Similarly, gene expression analyses in combination with clonal assays were also used to identify endothelial protein C receptor (EPCR) as a cell surface marker almost exclusively associated with robust HSC activity,58-60 as well as CD49b as a marker that separates long- and short-term HSCs.61 Collectively, the combined efforts of many research groups coupling multilevel molecular analyses and functional assays over several decades has led to FACS-purification strategies enabling the isolation of populations with an HSC content in >50% of the sorted cells,62 paving the way for virtually all of the current approaches aimed at simultaneously assessing molecular and functional cell states. It is important to note that many of the markers used to identify HSCs via flow cytometry show fluctuations in their expressions of response to external stimuli that alter parameters such as cell-cycle status and motility in HSCs.63,64 However, expression of more recently identified HSC markers (including Fgd5,65 EPCR,59,60 and α-catulin66) have been shown to be less sensitive to such changes in the cellular state66,67 and, as such, represent potentially more reliable markers for HSC identification across different perturbation scenarios in a similar manner to that of CD34 expression on human HSCs across the development and tissue types.
The application of coupled molecular and functional assays, such as FACS, in human HSPCs has historically been hampered by the low availability of human tissues for analysis as well as the added difficulties of establishing robust in vitro and in vivo assays to probe human HSPC functionality. In the mid-1980s, researchers established that the large majority of LTC-IC in human BM express the antigen CD34,68,69 and the LTC-IC assay was subsequently used to estimate the HSC content of the human CD34+ population.70 Using the severe combined immunodeficient mouse model, labs began to show that human Thy-1+ CD34+ cells were capable of maintaining hematopoiesis in vivo, 71 thus, further refining purification strategies for the population of cells containing HSC activity in human BM. Since then, several research groups have applied the approach of letting high-parameter assays such as RNA-seq72 and mass cytometry73,74 inform subsequent lower-parameter assays (ie, FACS) to refine immunophenotypes of human HSPCs. These studies have resulted in FACS gating strategies for human tissues that now yield an HSC frequency of 1 in 5 cells, as recently reviewed elsewhere.75 Although this purity still remains below what can be achieved in the mouse,62 the ability to more precisely resolve the pool of human HSPCs has certainly paved the way for a multitude of assays integrating molecular and functional profiling.
Index sorting coupled to downstream assays
The advent of index sorting,76,77 that is, the ability to link the immunophenotype of a single FACS–purified cell to its position in a multiwell plate, has accelerated studies focused on rare cell types, the HSC field included. Index sorting coupled with downstream functional or molecular assays have enabled researchers to perform large-scale experiments that directly link cell surface phenotypes to the molecular profile or cellular output in a retrospective manner. The first example of large-scale integrated analysis of surface, molecular, and functional phenotypes in HSCs was reported by Wilson et al in 2015.62 In this work, 4 commonly used gating strategies for FACS-based purification of HSCs were used to isolate cells for single-cell gene expression analysis, in addition to in vitro and in vivo functional assays. Importantly, the cell surface markers for the other strategies were included in a multicolor panel without being sorted upon, thereby, permitting retrospective assessment of how HSCs with a different phenotype would behave in the same assay. Integration of gene expression data from single-cell quantitative PCR and genome-wide single-cell RNA-seq with the in vivo behavior of the different sorted HSC populations, allowed for the derivation of a molecular signature that spanned multiple HSC subsets in which durable self-renewal capacities were present (termed as the molecular overlap signature). Integration of the molecular overlap signature with the index sorting data revealed a refined surface marker profile for FACS-isolation of a cell population containing at least 60% bona fide HSCs, as validated via single-cell transplantation experiments.62 Using a similar strategy of integration of cell surface phenotypes with functional analyses, Schulte et al correlated differences in surface marker expression with the capacity of HSCs to form stem and progenitor enriched clones in vitro.78 These studies launched the field of combining index sorting with single-cell RNA-seq, chromatin profiling, and functional assays and has also been successfully used to link states of lineage priming with the transcriptional and epigenetic profile of Lin– Sca-1– cKit+ myeloid progenitors, as in the report by Wilson et al,62 leading to refined FACS-purification strategies for progenitor populations with differential lineage potential.79 Taken together, the integration of multilevel cell-state analyses as described in the aforementioned studies has proven to be an immensely powerful approach for resolving heterogeneous cell populations and providing strategies for the prospective isolation of highly homogeneous pools of cells for further functional and molecular profiling.
Despite these advantages, index sorting is limited to the protein-level molecular phenotype of a cell, and for functional assays, which require a viably sorted cell, this is further limited to those proteins on the cell surface. Moreover, FACS is often limited by available detectors, with only a handful of studies reporting >20 color panels. Here, cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) in which antibodies are coupled to oligonucleotides rather than fluorophores, represents a substantially higher throughput approach for combined transcriptional profiling and surface phenotyping than FACS and single-cell RNA-seq.80 CITE-seq has been usefully applied for both human81 and mouse82 HSCs to further resolve the hematopoietic hierarchy; however, unlike FACS, the CITE-seq workflow is destructive and cannot be linked to single-cell functional assays.
Sample splitting to capture molecular and functional states
Although index sorting coupled with downstream assays can link molecular and functional phenotypes via cell surface marker expression, each sorted HSC can only be assessed using a single assay (Figure 1), making it impossible to directly measure function while capturing the global molecular profile of a specific cell. Remarkably, the Iscove laboratory pioneered a potential solution to this issue, several decades ago, in the form of temporal quantitative PCR sampling of clonally derived progeny of hematopoietic progenitor cells,45,46 with the idea of allowing a single HSC to divide in vitro into roughly equivalent progeny, thereby permitting sampling of some progeny for functional assays and some for molecular assays. However, there were some challenges associated with such an approach, including data from multiple groups using paired daughter assays indicating that most wild-type HSCs divide asymmetrically in culture to give 1 HSC and 1 non-HSC.4,5,58,83 In 2019, a major breakthrough in this space occurred when Wilkinson et al developed a serum-free, optimized culture system that allows for up to 900-fold expansion of multipotent, self-renewing HSCs.84 This system allowed us to undertake an Iscove-inspired set of assays in which we performed single HSC expansion cultures and split the clonal progeny into functional and molecular assays.67 After identifying that all HSC activity was contained in the EPCR+ LSK (ELSK) fraction of the clone, we split the non-ELSK and ELSK progeny of single ex vivo expanded HSCs for molecular profiling via RNA sequencing and functional in vivo analysis. By integrating the transcriptional and functional profiles of each clonally derived population of cells, we derived a molecular signature that robustly marks cells with repopulation capacity (termed as RepopSig). Critically, because of its direct connection with the range of different cellular functions in transplantation assays, RepopSig proved capable of not only identifying HSCs after culture but also freshly isolated cycling as well as dormant HSCs from mouse BM and fetal liver, and, even more remarkably, it identifies HSCs from transcriptome profiles of a broad range of human stem and progenitor cells.67 The ability to undertake computational analyses in which the different functional outputs are used to define functional HSC clusters allows for much more refined strategies, with higher likelihoods of finding molecular programs of the function being assessed.
In addition to transcriptional profiling, HSC expansion conditions with an accompanying HSC sorting strategy after ex vivo expansion has substantial implications for future work aimed at directly coupling function and molecular profile. Specifically, it will greatly aid the field in moving beyond the transcriptome, enabling the correlation of functional outcomes with, for example, proteome and chromatin state, protein complex, and other assays currently not applicable to HSCs because of the large numbers of cells required. However, further optimization of the culture system introduced by Wilkinson et al is required in order to tackle the current heterogeneity in the expansion ability of individual HSCs, and to translate the protocol for human setting.
Keeping pace with new bioinformatic tools
As alluded to the previously stated explanation, we now have the capacity to generate much more sophisticated data sets involving multiple types of data inputs. However, the pace at which single-cell molecular assays have emerged has been much quicker than that of the emergence of clonal functional assays (Figure 2). Likewise, the range and type of tools developed to assess and integrate data have far outstripped the assay development, leading to a complex landscape from which biologists must select their analysis tools. To illustrate this rapid development, it is worth recalling that the SmartSeqII approach for single-cell RNA-seq was only published in 2014,85 and only a few years later, the variety and scale of methods developed to analyze single-cell gene expression data is mind-boggling.86 Bioinformatic workflows, nowadays, have multiple steps and use various software packages developed at different institutes. Furthermore, running bioinformatic pipelines often requires substantial computational power, thereby requiring high performance computing clusters and cloud-based computing services. Although this all sounds daunting, it is also an incredibly exciting time for those involved in single-cell informatics. Some tools have already been developed and shared to integrate multiple single-cell-omics data sets,87 and many more are on the horizon.
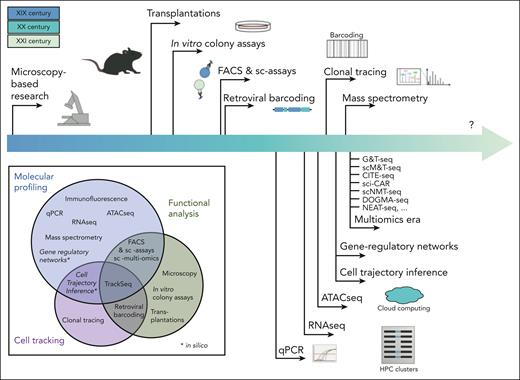
A broad timeline of single-cell functional, molecular, and computational techniques. Although clonal functional assays have developed slowly over many decades, global single-cell molecular profiling (and its associated analytical tools) have been largely developed in the last 10 years. The pace of these developments and the breadth of competing/complementary tools makes it more challenging to navigate. The Venn diagram (bottom-left) depicts each tool and identifies the aspects of HSC biology that can be assessed via each technique (cellular function, molecular profile, or clonal tracking). In addition to these individual limitations, it is important to note that most multiomic techniques are not as robust as singleomic techniques optimized to capture 1 molecular aspect, and these combinatorial technologies should be used with great caution. ATAC-seq, assay for transposase-accessible chromatin sequencing; G&T-seq, genome and transcriptome sequencing; NEAT-seq, sequencing of nuclear protein epitope abundance, chromatin accessibility and the transcriptome in single cells; qPCR, quantitative polymerase chain reaction; sc-assays, single-cell assays; sci-CAR, single-cell combinatorial indexing-based coassay that jointly profiles chromatin accessibility and mRNA; scM&T-seq, single-cell methylome and transcriptome sequencing; scNMT-seq, single-cell nucleosome, methylation and transcription sequencing.
A broad timeline of single-cell functional, molecular, and computational techniques. Although clonal functional assays have developed slowly over many decades, global single-cell molecular profiling (and its associated analytical tools) have been largely developed in the last 10 years. The pace of these developments and the breadth of competing/complementary tools makes it more challenging to navigate. The Venn diagram (bottom-left) depicts each tool and identifies the aspects of HSC biology that can be assessed via each technique (cellular function, molecular profile, or clonal tracking). In addition to these individual limitations, it is important to note that most multiomic techniques are not as robust as singleomic techniques optimized to capture 1 molecular aspect, and these combinatorial technologies should be used with great caution. ATAC-seq, assay for transposase-accessible chromatin sequencing; G&T-seq, genome and transcriptome sequencing; NEAT-seq, sequencing of nuclear protein epitope abundance, chromatin accessibility and the transcriptome in single cells; qPCR, quantitative polymerase chain reaction; sc-assays, single-cell assays; sci-CAR, single-cell combinatorial indexing-based coassay that jointly profiles chromatin accessibility and mRNA; scM&T-seq, single-cell methylome and transcriptome sequencing; scNMT-seq, single-cell nucleosome, methylation and transcription sequencing.
Single-cell transcriptome studies have been a massive boon to the HSC field, with 2 of the most popular tools being Seurat88 and Scanpy,89 although a quick scan of the literature reveals thousands of task-specific tools available. With a seemingly infinite list of new tools published every year, the critical role of comparative benchmarking studies becomes rapidly apparent.90 The results of such studies are databases such as the single-cell RNA-seq tool database (www.scrna-tools.org) and well-controlled benchmarking studies (eg, Tian et al for single-cell RNA-seq pipelines91). Moreover, analysis pipelines in the literature can be studied to determine the direction in which the field is moving, such as in the work from Zappia et al90 illustrating that researchers have moved away from methods to order cells into continuous trajectories (eg, pseudotime) and toward methods for integrating multiple data sets and public reference data sets to undertake cell classification.
Ordering cells into differentiation landscapes
Inferring the trajectory of an HSC via the hematopoietic differentiation cascade is a much sought-after goal of advanced transcriptomic analysis. Early studies that inferred cell differentiation trajectories used diffusion maps to delineate the journey of a start cell (eg, HSC) and an end cell (specific progenitor or mature cell) that effectively charts the distribution of expression across high dimensionality coordinates and orders cells in pseudotime.92 Such studies allow us to correlate selected gene expression along the trajectories, construct gene regulatory networks, and simulate gene expression in silico.92-94 Numerous tools now exist to undertake trajectory inference and are usefully summarized by Saelens et al.95 Another approach for extracting information about the cell state is RNA velocity. Here, computational modeling principles are underpinned by assumptions about splicing kinetics (ie, comparing spliced and nonspliced reads) to determine future cell phenotype,96-98 with exciting potential to extrapolate the identity of a cell, in the future.99 Useful reference data sets in hematopoiesis now exist92,100 that can be used to infer linkages between gene expression and cell-state changes. Perhaps, most critically, these tools, and new tools in the future, can make predictions in silico about cell fate, differentiation regulation, and transcription factor networks that can be tested experimentally. These types of studies also set the stage for more complex modeling exercises that use bioinformatic approaches to build network models of disease (eg, as recently reported by Talarmain et al101) and generate new hypotheses such as continuous differentiation landscapes and the fluid/cloud state of HSCs.102,103
Adding new layers: the multiomics era
Single-cell RNA-seq is a fantastic tool, but it is widely recognized as an incomplete snapshot of a current cellular state captured at a particular time.103 More recently, the multiomics age has been initiated in an attempt to address this shortcoming by profiling other aspects of the cell of interest.104,105 From simple layers, such as chromatin accessibility (eg, ATAC-seq) and G&T-seq,106 to complex perturbation/profiling applications (eg, multi-CUT&Tag,107 spatial CUT&Tag,108 spear-ATAC,109 and snNMTseq110 among others), the list of new ways to simultaneously profile cells continues to grow.105,111
Integrating these newly expanded molecular catalogs of cells requires new data integration tools (reviewed by Forcato et al112). Through combinations of dimensionality reduction, pattern recognition, machine learning,113 and statistical modeling (and others), these strategies attempt to bring together multiple data points from different data platforms. One feature that can aid this is if the same cell is assayed via multiple techniques (as opposed to populations of single cells ran separately and then compared afterwards). Integration tools, at present, are regularly plagued with different sensitivities of different technologies, especially when biological noise and technical noise are not easily disentangled.114 Usefully though, as with transcriptomic data described earlier, reference data sets are now emerging in which multiomic landscapes are now published for numerous HSPCs.115
With all of these “one-stop shop” techniques, however, it is important to also consider what is lost from an individual assay compared with the multiomic one (eg, how deeply surveyed is the transcriptome? And what is the sensitivity of mutational calling?).116 This becomes a large problem in upscaling single- and multiomic approaches, as costs inflate, to try to address the core problems of dropping out and data sparsity.117 Moreover, each technology has its own limitations, including signal to noise ratio, different sensitivities and resolution, and statistical power, etc.118 As the tools and technologies grow, it becomes more imperative to ensure that reproducibility in computational analyses remains paramount119 to avoid problems such as undocumented assumptions, data filtering, and different computational environments among other things.120
New horizons in single-cell and clonal analysis
Several new technologies are emerging as potential tools to solve our inability to track molecular and cellular state simultaneously. One approach, the application of heritable, genetic barcoding of HSPCs coupled to genome-wide single-cell RNA-seq has aided substantially in deepening our understanding of heterogeneity within the HSC pool (reviewed by Kester and Oudenaarden121). A recent study from Weinrab et al used temporal sampling of cells labeled with single-cell RNA-seq–compatible barcodes and assayed the cells both in vitro and in vivo.122 Although this study was potentially hampered by the same issue of HSCs undergoing asymmetric division in culture, as described in “Sample splitting to capture molecular and functional states,”58,83 the work did show that the transcriptional state of a single cell does not always explain its ultimate mature cell production, highlighting the need to develop other global -omics or tracking approaches. Another interesting area of development is in multiparameter timelapse imaging in which continuous imaging of live cells allows morphology and cell behavior to be integrated with downstream molecular or functional assays. An early example included video monitoring of HSC clones over a period of 4 days followed by transplantation to integrate the cell-cycle status and uropod formation of HSCs with their potential to sustain long-term, multilineage hematopoiesis among recipients who received transplantation.123 Remarkably, in 2019, temporal video monitoring was used to capture the asymmetric cell division of HSCs for the very first time and to couple HSC daughter cell fate to the unequal segregation of organelles during the earliest cell divisions.124 More recently, Wehling et al introduced trackSeq,125 a platform that integrates microscopy-based monitoring of the fate of 2 daughter cells arising from single HSCs with a downstream single-cellomic assay, enabling the generation of molecular profiles of cells with known historical behavior. Other recent advances in the imaging field include systems that enable repeated fluorescence-based imaging of the behavior of clonally derived hematopoietic cell types, for example, in response to cytokine stimulation.126
Also on the horizon, but less developed as of now, are new tools such as high-speed image-enabled cell sorting, in which image analysis can be integrated with FACS (similar to the current ImageStream technology but with a sorting of live cells option available),127 high throughput analysis of signaling/secretion at the single-cell level,128 and mass spectrometry-based proteomics (eg, cytometry by time of flight [CyTOF]) that can profile ∼100 targets simultaneously at the protein level but destroy the cell in the process. Moreover, unbiased characterization of the cellular proteome of single human leukemia stem cells using mass spectrometry was also recently reported.129 Even more appealing are tools such as LiveSeq130 that uses fluidic force microscopy to sample a cell without killing it, and can, thus, be used to capture a picture of molecular state while also assessing its function.
In summary, this is a very exciting time to ponder the linkage of cell function with molecular profile as new tools for HSC expansion,67,84 clonal tracking,121,122 and gene therapy all become available for experimental manipulation. Although the questions might well have existed many decades ago when the first clonal assays were being devised, the opportunity to match them to a molecular program and to potentially direct that molecular program for cell-state change to avert disease or improve transplantation/transfusion medicine is only becoming available now. These tools should also advance clinical sciences by identifying and assisting development of novel drug targets that are simply not discoverable through standard differential gene expression analyses that have been used to date.
Acknowledgments
The authors thank all members of the Kent laboratory for helpful discussion.
M.J. was supported by an international postdoctoral grant from the Swedish Research Council (2021-00185), M.G. was the recipient of a Biotechnology and Biological Sciences Research Council White Rose DTP PhD Studentship (BB/T007222/1), and research in the laboratory of D.G.K. is supported by an ERC Starting Grant (ERC-2016-STG–715371), an MRC-AMED joint award (MR/V005502/1), a Bill and Melinda Gates Foundation award (INV-038816), and a Cancer Research UK Programme Foundation Award (DCRPGF\100008).
Authorship
Contribution: M.J. and M.G. researched and wrote the initial draft of the paper; D.G.K. supervised the research and edited the manuscript; and all authors contributed to the figures and final version of the text.
Conflict-of-interest disclosure: The Kent laboratory receives research funding from Strm.bio. D.G.K. is a former board member of the International Society for Experimental Hematology and currently serves in several committees for the European Hematology Association. The remaining authors declare no competing financial interests.
Correspondence: David G. Kent, York Biomedical Research Institute, Department of Biology, University of York, York, United Kingdom; e-mail: david.kent@york.ac.uk.