In this issue of Blood, Barajas et al report that acute myeloid leukemia (AML) with UBTF internal tandem duplications (UBTF-ITDs) is dependent on the interaction between menin and KMT2A.1 This adds UBTF-ITD AML to an expanding list of hematologic malignancies that depend on menin and opens up a new therapeutic avenue for a high-risk leukemia.2
There is considerable excitement for menin inhibition in acute leukemias. Several different menin inhibitors are currently in clinical trials as a single agent or in combination. Emerging clinical data suggest excellent single-agent activity in highly pretreated patients with KMT2A-rearranged (KMT2A-r) or NPM1-mutant AML. Based on these encouraging results, menin inhibitors are being tested in combination and rapidly moved into earlier lines of therapy.
The molecular rationale for menin inhibition in KMT2A-r leukemias is strong. The interaction between KMT2A and menin is critical for the recruitment of KMT2A-fusions to key target loci and essential for the survival of KMT2A-r leukemia. In contrast, the effects of menin deletion on normal hematopoiesis are subtle, despite the fact that KMT2A itself is essential for hematopoiesis. Although these characteristics nominated menin as a potential therapeutic target, protein-protein interactions pose substantial medicinal chemistry challenges. The groundbreaking work of Grembecka and colleagues provided evidence for the feasibility and therapeutic potential of menin inhibition in KMT2A-r leukemias,3 sparking the development of today’s range of agents.
A parallel strand of investigations forms the mechanistic basis for the report from Barajas et al. Initial work on menin inhibitors focused solely on KMT2A-r leukemias, which represent 10% of acute lymphoblastic leukemia (ALL) and AML. However, almost 60% of AML, and a substantial portion of B- and T-ALL, express many of the KMT2A-fusion target genes in the absence of a KMT2A fusion. A role for unmutated KMT2A in these leukemias was first shown for AML with fusions or overexpression of meningioma-1.4 This was quickly followed by a report that menin-inhibition had preclinical efficacy in NPM1-mutant AML.5 Currently, menin-sensitive leukemias include those with KMT2A-fusions, KMT2A-partial tandem duplications, MN1-fusions,6NPM1 mutations,5NUP98-fusions,7 and now UBTF-ITDs. All of these leukemias share expression of HOXA (and sometimes HOXB, including UBTF-ITD) cluster genes and Meis1. Overexpression of HOXA9 and Meis1 are sufficient to induce AML in mice, documenting the central importance of these target genes.
A number of critical and interesting questions remain, both for UBTF-ITD specifically and menin inhibition in general. Menin binds KMT2A-, KMT2B-, and KMT2A-fusions. In KMT2A-r leukemias, transcriptional targets of the KMT2A-fusion but not those of wild type KMT2A (transcribed from the second, unmutated allele) or KMT2B are most affected by menin inhibition.8 In contrast, menin inhibition in non–KMT2A-r HOX-high leukemias affects the transcriptional targets of wild type KMT2A. Why this difference? And why are leukemias such as UBTF-AML that rely on wild type KMT2A more sensitive to menin inhibition than normal hematopoiesis, when KMT2A is needed for both? What is the molecular reason for the therapeutic window that clearly exists? In addition, it has been shown that only some KMT2A-fusion targets are sensitive to menin inhibition. For example, menin inhibition prevents KMT2A-fusion binding to the Meis1 locus but not the HOXA cluster.9 The molecular basis for this selectivity is ultimately not understood.
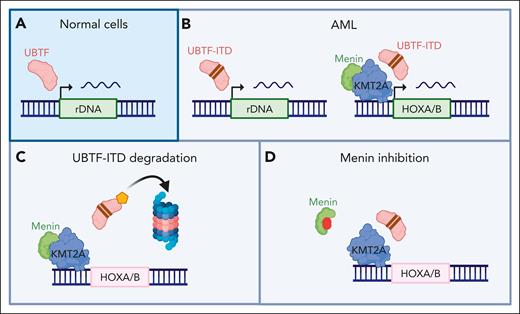
The molecular mechanism of how UBTF-ITD acts on KMT2A:menin is even more mysterious. Cut&Run experiments by Barajas et al show aberrant recruitment of UBTF-ITD but not wild type UBTF to sites of KMT2A:menin binding (see figure). However, degradation of UBTF-ITD did not alter recruitment of KMT2A or menin, but it did result in loss of gene expression at these sites. How exactly UBTF-ITDs alter the effect of KMT2A:menin on gene expression therefore remains unresolved. One may speculate that future insights into the effects of UBTF-ITDs on menin and KMT2A may help solve some of the remaining mysteries around target specificity and the mechanistic basis for the therapeutic window in non–KMT2A-r AML.
Menin inhibition in UBTF-ITD AML. (A) UBTF is recruited to ribosomal DNA (rDNA) loci in normal cells. (B) In AML with UBTF-ITD, the mutated UBTF is recruited to both rDNA loci and sites of KMT2A/menin binding, including the HOXA and HOXB cluster, resulting in aberrant expression of these genes. (C) Degradation of UBFT-ITD results in transcriptional downregulation of HOXA and HOXB cluster genes, despite maintained KMT2A and menin binding. (D) Inhibition of the menin:KMT2A interaction also results in downregulation of HOXA and HOXB cluster genes and induces differentiation in UBTF-ITD leukemia cells. Created with BioRender.com.
Menin inhibition in UBTF-ITD AML. (A) UBTF is recruited to ribosomal DNA (rDNA) loci in normal cells. (B) In AML with UBTF-ITD, the mutated UBTF is recruited to both rDNA loci and sites of KMT2A/menin binding, including the HOXA and HOXB cluster, resulting in aberrant expression of these genes. (C) Degradation of UBFT-ITD results in transcriptional downregulation of HOXA and HOXB cluster genes, despite maintained KMT2A and menin binding. (D) Inhibition of the menin:KMT2A interaction also results in downregulation of HOXA and HOXB cluster genes and induces differentiation in UBTF-ITD leukemia cells. Created with BioRender.com.
The study also raises the interesting question of how to best identify leukemias with menin sensitivity in a clinical setting. So far, all leukemias with high HOXA expression that have been tested were reported to be sensitive to menin inhibition. Is this a universal feature or due to publication bias? Would HOXA (and/or HOXB) expression be the most straightforward way to identify patients who might benefit from menin inhibition? As RNA sequencing enters the realm of CLIA–approved clinical testing, should clinical translation be focused on a gene signature for eligibility rather than an expanding list of DNA mutation? Currently used standard next generation sequencing pipelines are less well suited to pick up indels such as UBTF-ITDs2 or enhancer hijacking events such as MN1-fusions.10 Expression-based screening might more comprehensively capture patients who could benefit from menin inhibition. Future studies will also need to address resistance mutations and determine the best combinatorial therapies.
UBTF-ITD leukemias have an intermediate to poor prognosis.2 Despite residual mechanistic questions, this report is an important step forward in understanding the role of KMT2A and menin in leukemia and offers hope for a patient population that has not done well with conventional therapy.
Conflict-of-interest disclosure: K.M.B. has consulted for Agios and Novartis and has received research funding from Syndax Pharmaceuticals Inc.