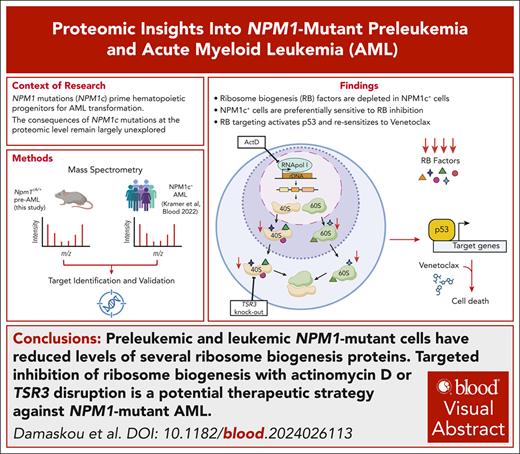
Key Points
Preleukemic and leukemic NPM1-mutant cells have reduced levels of several ribosome biogenesis proteins.
Targeted therapeutic disruption of ribosome biogenesis is a potential strategy against NPM1-mutant AML.
Visual Abstract
NPM1 is a multifunctional phosphoprotein with key roles in ribosome biogenesis among its many functions. NPM1 gene mutations drive 30% of acute myeloid leukemia (AML) cases. The mutations disrupt a nucleolar localization signal and create a novel nuclear export signal, leading to cytoplasmic displacement of the protein (NPM1c). NPM1c mutations prime hematopoietic progenitors to leukemic transformation, but their precise molecular consequences remain elusive. Here, we first evaluate the effects of isolated NPM1c mutations on the global proteome of preleukemic hematopoietic stem and progenitor cells (HSPCs) using conditional knockin Npm1cA/+ mice. We discover that many proteins involved in ribosome biogenesis are significantly depleted in these murine HSPCs, but also importantly in human NPM1-mutant AMLs. In line with this, we found that preleukemic Npm1cA/+ HSPCs display higher sensitivity to RNA polymerase I inhibitors, including actinomycin D (ActD), compared with Npm1+/+ cells. Combination treatment with ActD and venetoclax inhibited the growth and colony-forming ability of preleukemic and leukemic NPM1c+ cells, whereas low-dose ActD treatment was able to resensitize resistant NPM1c+ cells to venetoclax. Furthermore, using data from CRISPR dropout screens, we identified and validated TSR3, a 40S ribosomal maturation factor whose knockout preferentially inhibited the proliferation of NPM1c+ AML cells by activating a p53-dependent apoptotic response. Similarly, to low-dose ActD treatment, TSR3 depletion could partially restore sensitivity to venetoclax in therapy-resistant NPM1c+ AML models. Our findings propose that targeted disruption of ribosome biogenesis should be explored as a therapeutic strategy against NPM1-mutant AML.
Introduction
Nucleophosmin, coded by the NPM1 gene, is a highly abundant phosphoprotein involved in diverse cellular processes.1-6 It can shuttle between the nucleus and cytoplasm, but at a steady state, it is primarily located at the nucleolus, the main site of ribosomal RNA (rRNA) transcription and early ribosome biogenesis.7 More specifically, nucleophosmin is located at the outer, granular component of the nucleolus,8 contributes to maintaining the organelle’s structure,9,10 and is involved in rRNA 2′-O-methylation.11
Somatic mutations affecting the NPM1 gene are found in ∼30% of cases of acute myeloid leukemia (AML)12,13 and define the most common AML subtype. AML-associated NPM1 mutations, usually 4-base pair duplications/insertions within the gene’s final exon, disrupt a nucleolar localization signal and create a novel nuclear export signal. As a result, mutant nucleophosmin is aberrantly localized to the cytoplasm (NPM1c) through interaction with XPO1, a major effector of nucleocytoplasmic traffic.14 The current standard-of-care treatment for patients with NPM1c-mutant AML is 3+7 chemotherapy, whereas for patients carrying FLT3 comutations, such as internal tandem duplications (ITDs), a FLT3 inhibitor is incorporated into the first-line treatment.15,16 Most patients enter complete remission, but approximately half relapse and most succumb to their disease, highlighting the need for new therapies.17
Despite its high prevalence in AML, the molecular consequences of the NPM1c mutation are incompletely understood, which poses an obstacle to the design of effective therapeutic strategies.18 Recent advances have given insights into the transcriptomic and epigenetic effects of NPM1c mutations,19 but less is known about its global posttranscriptional and proteomic consequences.
In this study, we determined the global proteomic consequences of isolated NPM1c mutations in preleukemic hematopoietic progenitors from knockin Npm1c-mutant mice, which faithfully recapitulate the human mutations.19-21 We then investigated which changes were also detected in full-blown NPM1-mutant AML22 and evaluated these against CRISPR knockout screens23-25 to identify potential therapeutic targets. Notably, we found significant decreases in abundance of multiple ribosome biogenesis factors in NPM1c+ preleukemia which were also observed in human NPM1c+ AML and were associated with increased sensitivity to the RNA polymerase (pol) I inhibitor actinomycin D (ActD) alone and particularly in combination with the Bcl-2 inhibitor venetoclax. Focusing on individual targets, we discovered that NPM1c-mutant cells are dependent on TSR3, a normally nonessential maturation factor of the 40S ribosomal subunit. Finally, we demonstrated that both low-dose ActD treatment and TSR3 knockout induce resensitization to venetoclax in previously resistant NPM1c+ leukemia cells.
Methods
Also see the supplemental Methods (available on the Blood website).
Mouse model
Mx1-Cre; Npm1flox−cA/+ mice have been described previously.19,20 Cre expression was induced in 8 to 10 weeks old Mx1-Cre; Npm1flox−cA/+ mice by administration of polyinosinic:polycytidylic acid (catalog no. P1530; Sigma-Aldrich). Blood counts were calculated by a SciI Vet abc hematology analyzer. All mice were kept in a pathogen-free environment, and all procedures were performed according to the regulation of the UK Home office, under project license PP3797858, in accordance with the Animal Scientific Procedures Act 1986.
Proteomic analysis of preleukemic murine hematopoietic progenitors
A total of 7 Npm1cA/+ and 5 control Npm1+/+ mice were collected 8 weeks after polyinosinic:polycytidylic acid, and the bone marrow lineage-negative (lin–) cells were subjected to proteomic analysis (supplemental Table 1). Sample preparation, mass spectrometry analysis, and data processing are described in detail in the supplemental Methods.
Proteomic and RNA expression analysis of samples from patients with AML
Experimental procedures and data sets can be found in Kramer et al.22 t test was used to calculate P values for differences between the NPM1wt and NPM1c+ AML groups, followed by multiple hypothesis correction using the Benjamini-Hochberg method.
Study of patient-derived NPM1-mutant AML cells
For patient-derived xenograft (PDX) studies, 1 × 106 cells of a luciferase-expressing NPM1-mutant AML PDX were injected IV into NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice, and animals were treated with intraperitoneal ActD or vehicle from day 35 after transplantation and monitored for AML tumor volume and survival. Detailed experimental procedures are provided in the supplemental Methods.
NPM1-mutant normal karyotype AML cells were obtained from a 33-year-old man with the patient's informed consent as part of the Hémopathies Inserm Midi-Pyrénées collection (BB-0033-00060). According to French law, the Hémopathies Inserm Midi-Pyrénées collection has been declared to the Ministry of Higher Education and Research (DC 2008-307 collection 1) and a transfer agreement (AC 2008-129) was obtained after approbation by the Comité de Protection des Personnes Sud-Ouest et Outremer II (ethical committee). The study was performed in accordance with the Declaration of Helsinki.
Results
Npm1cA/+ hematopoietic progenitors have reduced levels of ribosome biogenesis proteins
We collected bone marrow lineage-depleted (lin–) hematopoietic stem/progenitor cells (HSPCs) 8 weeks after activation of the conditional Npm1cA allele in Mx1-Cre; Npm1flox-cA/+ mice.20 At that stage, the mice displayed normal hematological parameters and no signs of leukemia (supplemental Figure 1A-D), but the characteristic Npm1c-associated HoxA and HoxB gene upregulation was clearly evident in HSPCs (supplemental Figure 1E; supplemental Table 2). In line with this, Npm1cA/+ HSPCs demonstrated enhanced serial replating in colony-forming unit assays, compared with their wild-type counterparts (supplemental Figure 1F-G).
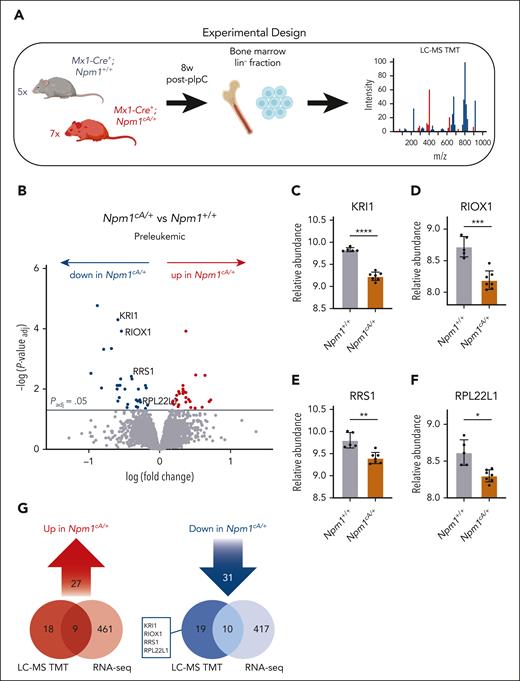
To capture the global impact of isolated Npm1c mutations at the protein level, we performed tandem mass tag (TMT)-based quantitative proteomics in HSPCs from Npm1cA/+ mutant (n = 7) vs Npm1+/+ (n = 5) mice (Figure 1A). Of 7409 proteins detected overall, we found 58 high-confidence proteins whose abundance was significantly different (Padj < .05) between Npm1cA/+ and Npm1+/+ HSPCs (Figure 1B; supplemental Table 3). Notably, NPM1 protein levels were found to be significantly reduced in Npm1cA/+ mice. However, evaluation of the identified peptide spectra revealed that this was because the only NPM1-derived peptide detected was unique to the wild-type protein (amino acids 276-289). With no mutant-specific peptides identified, this resulted in an apparent reduction in NPM1 protein levels in heterozygous mice.
Reduced abundance of ribosomal proteins in Npm1cA/+ hematopoietic progenitors. (A) Workflow for global quantitative proteomic study of lineage-negative (lin–) bone marrow–derived hematopoietic progenitors using TMT. Preleukemic Npm1cA/+ (n = 7) vs Npm1+/+ (n = 5) murine hematopoietic progenitors were collected 8 weeks after Mx1-Cre induction with pIpC. (B) Volcano plot revealing relative protein abundances in preleukemic Npm1cA/+ vs Npm1+/+ lin– progenitors. (C-F) Relative abundance of selected ribosome-related proteins in Npm1cA/+ vs Npm1+/+ lin– progenitors. (G) Overlap between messenger RNAs (RNA-seq) and proteins (LC-MS) that were significantly over- or under-expressed in preleukemic Npm1cA/+ vs Npm1+/+ lin– cells. Panel B, unpaired t test with multiple-hypothesis–testing correction (FDR; Benjamini-Hochberg). (C-F) Unpaired t test with multiple-hypothesis–testing correction (FDR; Benjamini-Hochberg). ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001; ∗∗∗∗P ≤ .0001. Data represent n = 7 (Npm1cA/+) or n = 5 (Npm1+/+) biological replicates (mean ± SD). FDR, false discovery rate; LC-MS TMT, liquid chromatography mass spectrometry, tandem mass tagging; pIpC, polyinosinic:polycytidylic acid. Panels A and G were created with BioRender.com. Vassiliou, G. (2025) https://BioRender.com/9tt8kww.
Reduced abundance of ribosomal proteins in Npm1cA/+ hematopoietic progenitors. (A) Workflow for global quantitative proteomic study of lineage-negative (lin–) bone marrow–derived hematopoietic progenitors using TMT. Preleukemic Npm1cA/+ (n = 7) vs Npm1+/+ (n = 5) murine hematopoietic progenitors were collected 8 weeks after Mx1-Cre induction with pIpC. (B) Volcano plot revealing relative protein abundances in preleukemic Npm1cA/+ vs Npm1+/+ lin– progenitors. (C-F) Relative abundance of selected ribosome-related proteins in Npm1cA/+ vs Npm1+/+ lin– progenitors. (G) Overlap between messenger RNAs (RNA-seq) and proteins (LC-MS) that were significantly over- or under-expressed in preleukemic Npm1cA/+ vs Npm1+/+ lin– cells. Panel B, unpaired t test with multiple-hypothesis–testing correction (FDR; Benjamini-Hochberg). (C-F) Unpaired t test with multiple-hypothesis–testing correction (FDR; Benjamini-Hochberg). ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001; ∗∗∗∗P ≤ .0001. Data represent n = 7 (Npm1cA/+) or n = 5 (Npm1+/+) biological replicates (mean ± SD). FDR, false discovery rate; LC-MS TMT, liquid chromatography mass spectrometry, tandem mass tagging; pIpC, polyinosinic:polycytidylic acid. Panels A and G were created with BioRender.com. Vassiliou, G. (2025) https://BioRender.com/9tt8kww.
The top 3 proteins with the most significantly increased levels were the polyol pathway enzyme SORD (sorbitol dehydrogenase), the centrosomal protein CEP44 (centrosomal protein 44), and the F13A1 (coagulation factor XIII A chain) blood coagulation factor (supplemental Figure 2A-C). In addition, among proteins having increased abundance were several members of the Karyopherin family (supplemental Figure 2D), mirroring recent findings in human NPM1c-mutant AML.22 Interestingly, NPM1c+ OCI-AML3 cells were found to be particularly sensitive to KPNA2 knockout (DepMap24,25; supplemental Figure 2F-G).
Among the 31 proteins having decreased abundance in Npm1cA/+ HSPCs, there were several ribosome-related proteins, including ribosome biogenesis factors and structural constituents of ribosomes.7 Significantly depleted proteins included KRI1 (KRI1 homolog),26 RIOX1 (ribosomal oxygenase 1),27 RRS1 (ribosome biogenesis regulator 1 homolog),28,29 and RPL22L1 (ribosomal protein L22 like 1)30 (Figure 1C-F), none of which demonstrated reduction at the messenger RNA level (Figure 1G). Gene ontology term analysis of all depleted proteins (n = 270) among the top 500 differentially abundant proteins demonstrated enrichment for the preribosome and biological processes involving rRNA processing and ribosome biogenesis (supplemental Figure 3A; supplemental Table 4). Polysome profiling of whole bone marrow cells from Npm1cA/+ and control Npm1+/+ mice revealed no major differences, although there was a trend toward higher abundance for the 40S ribosomal subunit in the mutant cells (supplemental Figure 3B-E).
Ribosome biogenesis factors are depleted in human NPM1c+ AML
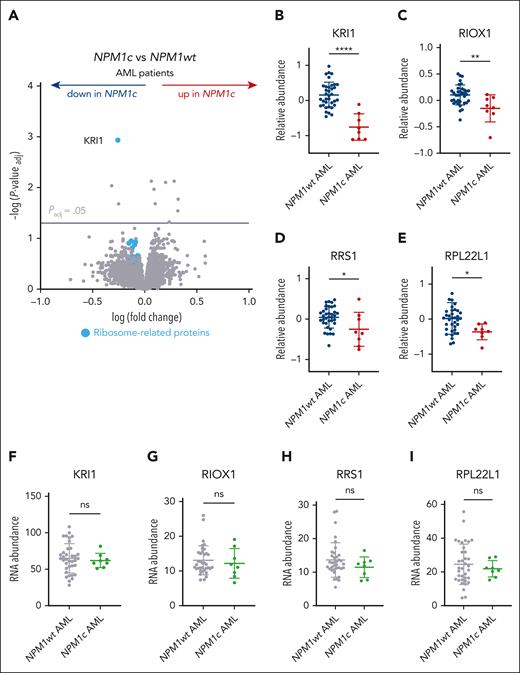
To pinpoint which changes identified in NPM1c+ preleukemic cells can also be detected after progression to AML, we evaluated recently reported TMT proteomics data from bone marrow samples of 44 patients with de novo AML, including 8 with NPM1c+ AML.22 By comparing protein levels between NPM1c+ and NPM1wt AMLs, we identified only 11 significantly differentially abundant proteins (Padj < .05), among which KRI1 was the most depleted (Figure 2A-B; supplemental Table 5). Importantly, gene ontology analysis of all depleted proteins (n = 191) among the top 500 differentially abundant proteins revealed an enrichment for ribosome biogenesis factors, rRNA processing enzymes, and maturation factors of rRNA precursors (supplemental Figure 4A; supplemental Table 6), similar to the murine preleukemic cells. RIOX1, RRS1, and RPL22L1, all of which were significantly depleted in preleukemic Npm1cA/+ cells, also demonstrated reduced abundance in NPM1c+ AML (Figure 2C-E), albeit without reaching significance after correcting for multiple testing. Other ribosome-related proteins that were depleted in NPM1c+ AML included KRR1 (KRR1 small subunit processome component homolog), an important interactor of KRI1,26 RPS9 (ribosomal protein S9), and RBIS (ribosomal biogenesis factor) (supplemental Figure 4B-D). Notably, none of the abovementioned candidates demonstrated downregulation in RNA-seq data from the same patients, suggesting that these genes/proteins are regulated posttranscriptionally (Figure 2F-I; supplemental Figure 4E-G). Finally, no significant differences were observed in the abundance of the abovementioned proteins between NPM1c+, FLT3-ITD+, and all other NPM1c+ AMLs (supplemental Figure 4H), suggesting that these changes were not related to FLT3-ITD, the most common comutation of NPM1c in AML.
Depletion of ribosome processing factors in human NPM1c+ AML. (A) Volcano plot of protein abundance in bone marrow samples from NPM1c+ (n = 8) vs NPM1wt (n = 36) human de novo AMLs.22 (B-I) Relative protein (B-E) or RNA abundance (F-I) of selected ribosome-related candidates in NPM1c+ (n = 8) vs NPM1wt (n = 36) AML patients (mean ± SD). Panel A, unpaired t test with multiple-hypothesis–testing correction (FDR, Benjamini-Hochberg method). (B-I) Unpaired t test was used to calculate P values between groups. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗∗P ≤ .0001. ns, nonsignificant. Data available in Kramer et al.22
Depletion of ribosome processing factors in human NPM1c+ AML. (A) Volcano plot of protein abundance in bone marrow samples from NPM1c+ (n = 8) vs NPM1wt (n = 36) human de novo AMLs.22 (B-I) Relative protein (B-E) or RNA abundance (F-I) of selected ribosome-related candidates in NPM1c+ (n = 8) vs NPM1wt (n = 36) AML patients (mean ± SD). Panel A, unpaired t test with multiple-hypothesis–testing correction (FDR, Benjamini-Hochberg method). (B-I) Unpaired t test was used to calculate P values between groups. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗∗P ≤ .0001. ns, nonsignificant. Data available in Kramer et al.22
Preleukemic Npm1cA/+ HSPCs demonstrate enhanced sensitivity to RNA pol I inhibitors
In light of the mislocalization of mutant and the reduced levels of NPM1wt, targeting ribosome homeostasis has been previously proposed as a therapeutic strategy against NPM1-mutant AML.31-33 Our data suggest that, in addition to the reduced levels of NPM1wt in the nucleolus, mutant cells are depleted of several ribosome biogenesis factors. We hypothesized that this depletion creates a vulnerability that can be therapeutically exploited to specifically target mutant cells. To test this, we compared the responses of preleukemic Npm1cA/+ vs control Npm1+/+ HSPCs to drugs that modulate ribosome biogenesis, including ActD and other RNA pol I inhibitors. Indeed, we found that compared with Npm1+/+ cells, preleukemic Npm1cA/+ lin– cells were significantly more sensitive to ActD treatment (Figure 3A-C), particularly at lower concentrations where the drug effects are known to be on-target (supplemental Figure 5A).34 Following treatment with low-dose ActD (1 nM), both mutant and wt HSPCs exhibited characteristic features of nucleolar stress, including a shift in nucleolar shape from irregular to round conformation (Figure 3D).35,36 This suggests that both cell types activate a nucleolar stress response, although impact on survival is greater for NPM1c+ cells. Nucleoplasmic dislocation of NPM1wt, another indicator of nucleolar stress,33,36,37 was observed in preleukemic Npm1cA/+ lin– cells treated with higher drug doses (5 nM) (supplemental Figure 5B). No DNA damage was observed in Npm1cA/+ lin– cells after treatment with 2.5 nM ActD for 48 hours (supplemental Figure 5C).
Preleukemic Npm1cA/+ cells demonstrate enhanced sensitivity to RNA pol I inhibitors. (A) Relative proliferation of preleukemic Npm1cA/+ vs Npm1+/+ lin– cells after 4 days of treatment with ActD at the indicated doses (MTS assay). (B) Representative CFU assays from preleukemic Npm1cA/+ vs Npm1+/+ lin– cells after 24 hours pretreatment with 2.5 nM ActD. (C) Total number of colonies in CFU assays of preleukemic Npm1cA/+ vs Npm1+/+ lin– cells after 24 h pretreatment with 2.5 nM of ActD. (D) Nucleolar morphology of Npm1+/+ (top panel) or Npm1cA/+ (bottom panel) lin– cells after treatment with 1 nM ActD for 18 hours, highlighted using fluorescent staining for wild-type NPM1 (NPM1WT) and nucleolin (NCL) (scale, 10 μm). (E-G) Relative proliferation of preleukemic Npm1cA/+ vs Npm1+/+ lin– cells after 4 days of treatment with BMH-21 (E), CX-5461 (F), or cycloheximide (G) at the indicated doses (MTS assay). (A,C,E-G) Unpaired t test was used to calculate P values between groups. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001. (A,E-G) Absorbance values were normalized to DMSO-treated cells. Data represent 4 to 7 biological replicates per group (mean ± SD). (C) Values represent 4 biological replicates per group. (D) Representative example from 1 of 2 to 4 biological replicates. DAPI, 4′,6-diamidino-2-phenylindole.
Preleukemic Npm1cA/+ cells demonstrate enhanced sensitivity to RNA pol I inhibitors. (A) Relative proliferation of preleukemic Npm1cA/+ vs Npm1+/+ lin– cells after 4 days of treatment with ActD at the indicated doses (MTS assay). (B) Representative CFU assays from preleukemic Npm1cA/+ vs Npm1+/+ lin– cells after 24 hours pretreatment with 2.5 nM ActD. (C) Total number of colonies in CFU assays of preleukemic Npm1cA/+ vs Npm1+/+ lin– cells after 24 h pretreatment with 2.5 nM of ActD. (D) Nucleolar morphology of Npm1+/+ (top panel) or Npm1cA/+ (bottom panel) lin– cells after treatment with 1 nM ActD for 18 hours, highlighted using fluorescent staining for wild-type NPM1 (NPM1WT) and nucleolin (NCL) (scale, 10 μm). (E-G) Relative proliferation of preleukemic Npm1cA/+ vs Npm1+/+ lin– cells after 4 days of treatment with BMH-21 (E), CX-5461 (F), or cycloheximide (G) at the indicated doses (MTS assay). (A,C,E-G) Unpaired t test was used to calculate P values between groups. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001. (A,E-G) Absorbance values were normalized to DMSO-treated cells. Data represent 4 to 7 biological replicates per group (mean ± SD). (C) Values represent 4 biological replicates per group. (D) Representative example from 1 of 2 to 4 biological replicates. DAPI, 4′,6-diamidino-2-phenylindole.
To further test whether the preferential sensitivity of Npm1cA/+ cells to ActD was due to RNA Pol I inhibition, and not due to off-target effects, we then tested BMH-21 and CX-5461, 2 more recently developed small-molecule inhibitors with different modes of RNA pol I inhibition.38-40 Similarly to ActD, we found that Npm1cA/+ HSPCs were significantly more sensitive to both agents compared with wild-type Npm1+/+ cells (Figure 3E-F; supplemental Figure 5D-G). Although off-target effects cannot be entirely excluded, the consistent phenotype observed across the 3 chemically distinct compounds suggests that Npm1c specificity is primarily driven by on-target RNA pol I inhibition. By contrast, we did not observe differential sensitivity between Npm1cA/+ and Npm1+/+ HSPCs to cycloheximide, a drug that inhibits translational elongation (Figure 3G).
Single-agent ActD inhibits expansion of an NPM1-mutant AML PDX
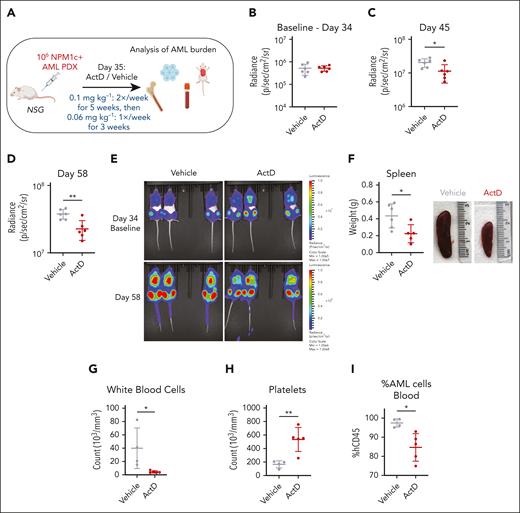
To investigate the effects of ActD treatment in vivo using a clinically relevant AML model, we used a luciferase-expressing NPM1-mutant PDX harboring DNMT3A R882H, FLT3-ITD, and RAD21 R146G comutations. One million NPM1c+ AML PDX cells were injected IV into 12 NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice (Figure 4A; supplemental Figure 6A). After 35 days, mice were allocated into 2 groups of similar AML burden (Figure 4B) to receive ActD (0.1 mg kg−1) or vehicle twice weekly for 5 weeks. Repeat bioluminescent imaging revealed a significant reduction of AML burden in ActD-treated animals at 45 and 58 days after IV injection (Figure 4C-E). Dosing was reduced to 0.06 mg kg−1 once weekly for another 3 weeks, and animals were monitored until they developed leukemia symptoms. ActD-treated animals had a significantly longer survival at 90 days (P = .031), but this became nonsignificant soon after stopping the treatment and until the end of the study (median survival ActD 84.5 days vs vehicle 99 days) (supplemental Figure 6B). Notably, at collection, ActD-treated mice exhibited milder AML-related pathological findings, including significantly smaller spleens, lower white blood cell, and higher platelet counts (Figures 4F-H). The antileukemic effect of ActD was also confirmed by the significant reduction of circulating human CD45+ AML cells in the blood (Figure 4I) and trends toward lower levels in the bone marrow and spleen (supplemental Figure 6C-D). Overall, these data reveal that single-agent ActD has antileukemic activity in vivo against human NPM1-mutant AML. Nonetheless, further studies will be needed to assess the selectivity of this treatment for patients with NPM1c+ AML vs patients with NPM1wt AML.
Actinomycin D reduces AML burden in a xenograft model. (A) Schematic representation of PDX experiment. Mice transplanted with 106 luciferase-expressing NPM1c+ AML PDX cells were treated with 0.1 mg kg−1 ActD or vehicle twice weekly for 5 weeks, followed by treatment with 0.06 mg kg−1 ActD or vehicle once weekly for another 3 weeks. Leukemia burden was monitored by bioluminescence imaging and quantified in tissues after animal collection. (B-D) Bioluminescence radiance on day 34 (baseline) (B), day 45 (C), and day 58 (D) in ActD vs vehicle-treated animals. (E) Bioluminescence imaging of animals treated with ActD vs vehicle at indicated time points. (F) Spleen weights and representative spleen pictures in ActD vs vehicle-treated animals. (G-H) Counts of white blood cells (G) and platelets (H) in ActD vs vehicle-treated animals. (I) Percentage of cells expressing human CD45 (hCD45) of total CD45 in blood in ActD vs vehicle-treated animals. (C-D,F-I) Unpaired t test was used to calculate P values between groups. ∗P ≤ .05; ∗∗P ≤ .01. Data represent 4 to 6 biological replicates per group (those with available tissues on disease presentation) (mean ± SD). Panel A was created with BioRender.com. Vassiliou, G. (2025) https://BioRender.com/9tt8kww.
Actinomycin D reduces AML burden in a xenograft model. (A) Schematic representation of PDX experiment. Mice transplanted with 106 luciferase-expressing NPM1c+ AML PDX cells were treated with 0.1 mg kg−1 ActD or vehicle twice weekly for 5 weeks, followed by treatment with 0.06 mg kg−1 ActD or vehicle once weekly for another 3 weeks. Leukemia burden was monitored by bioluminescence imaging and quantified in tissues after animal collection. (B-D) Bioluminescence radiance on day 34 (baseline) (B), day 45 (C), and day 58 (D) in ActD vs vehicle-treated animals. (E) Bioluminescence imaging of animals treated with ActD vs vehicle at indicated time points. (F) Spleen weights and representative spleen pictures in ActD vs vehicle-treated animals. (G-H) Counts of white blood cells (G) and platelets (H) in ActD vs vehicle-treated animals. (I) Percentage of cells expressing human CD45 (hCD45) of total CD45 in blood in ActD vs vehicle-treated animals. (C-D,F-I) Unpaired t test was used to calculate P values between groups. ∗P ≤ .05; ∗∗P ≤ .01. Data represent 4 to 6 biological replicates per group (those with available tissues on disease presentation) (mean ± SD). Panel A was created with BioRender.com. Vassiliou, G. (2025) https://BioRender.com/9tt8kww.
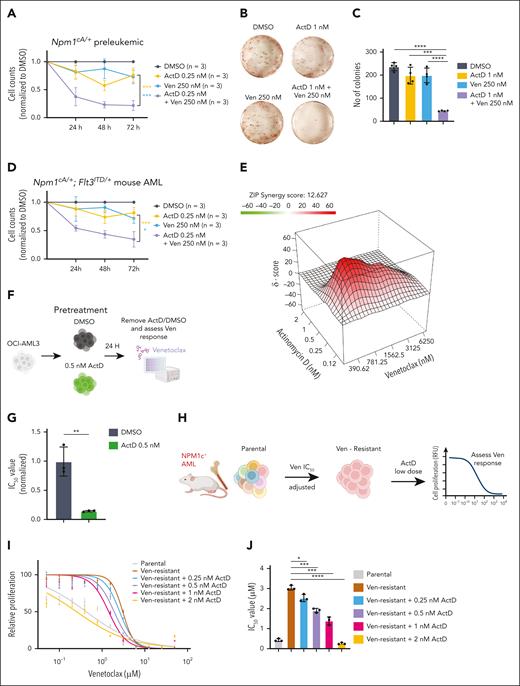
ActD potentiates the effects of venetoclax against preleukemic and leukemic NPM1c+ cells
Because venetoclax has become the standard of care for the older/unfit patients with AML and is particularly effective against NPM1-mutant AML,41,42 we next wanted to evaluate whether the ActD-venetoclax combination could synergistically suppress the growth of NPM1c+ hematopoietic cells. We found that doses of ActD or venetoclax that had mild effects on cell proliferation in isolation significantly inhibited the growth of preleukemic Npm1cA/+ hematopoietic progenitors in liquid culture when combined (Figure 5A). Moreover, the combination of ActD and venetoclax synergistically reduced the number of total colony-forming unit colonies in semisolid media at doses that had minimal anticlonogenic effect when each drug was used in isolation (Figure 5B-C).
Actinomycin D enhances cytotoxicity and reverses resistance to Ven in NPM1c+ preleukemic and leukemic cells. (A) Counts of preleukemic Npm1cA/+ lin– cells treated with DMSO, ActD, venetoclax (Ven), or a combination (mean ± SD, n = 3 independent experiments). (B) Representative CFU assays of preleukemic Npm1cA/+ lin– cells treated with DMSO, ActD, Ven, or a combination. (C) Total CFU colony numbers of preleukemic Npm1cA/+ lin– cells treated with DMSO, ActD, Ven, or a combination (n = 2 biological replicates, each performed in duplicate). (D) Counts of murine Npm1cA/+; Flt3ITD/+AML cells treated with DMSO, ActD, Ven, or a combination (mean ± SD, n = 3 independent experiments). (E) Representative synergy plot revealing the interaction between ActD and Ven in inhibiting the proliferation of murine Npm1cA/+; Flt3ITD/+AML cells after 4 days of drug incubation (MTS assay) (representative examples from 1 of 3 independent experiments). (F) OCI-AML3 cells were pretreated with 0.5 nM ActD vs DMSO for 24 hours and then tested for sensitivity to venetoclax. (G) IC50 values of DMSO or ActD pretreated OCI-AML3 cells after 4 days of incubation with Ven at the indicated doses (MTS assay). (H) Outline of approach to generate Ven-resistant murine Npm1c-mutant AML cells by treatment with escalating concentrations of Ven. Ven-resistant cells were then pretreated with ActD for 24 hours and their sensitivity to Ven was assessed using the MTS assay. (I) Relative proliferation of DMSO or ActD pretreated murine NPM1c+ AML cells after 4 days incubation with Ven at the indicated doses. Absorbance values were normalized to DMSO-treated cells (mean ± SD, data from 1 of 2 biological replicates, performed in triplicate). (J) IC50 values of DMSO or ActD pretreated murine NPM1c+ AML cells after 4 days of incubation with Ven (mean ± SD, data from 1 of 2 biological replicates, performed in triplicate). (A,C-D,G,J) t test was used to calculate P values between groups. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001; ∗∗∗∗P ≤ .0001. ZIP, zero interaction potency. Panels F and H were created with BioRender.com. Vassiliou, G. (2025) https://BioRender.com/9tt8kww.
Actinomycin D enhances cytotoxicity and reverses resistance to Ven in NPM1c+ preleukemic and leukemic cells. (A) Counts of preleukemic Npm1cA/+ lin– cells treated with DMSO, ActD, venetoclax (Ven), or a combination (mean ± SD, n = 3 independent experiments). (B) Representative CFU assays of preleukemic Npm1cA/+ lin– cells treated with DMSO, ActD, Ven, or a combination. (C) Total CFU colony numbers of preleukemic Npm1cA/+ lin– cells treated with DMSO, ActD, Ven, or a combination (n = 2 biological replicates, each performed in duplicate). (D) Counts of murine Npm1cA/+; Flt3ITD/+AML cells treated with DMSO, ActD, Ven, or a combination (mean ± SD, n = 3 independent experiments). (E) Representative synergy plot revealing the interaction between ActD and Ven in inhibiting the proliferation of murine Npm1cA/+; Flt3ITD/+AML cells after 4 days of drug incubation (MTS assay) (representative examples from 1 of 3 independent experiments). (F) OCI-AML3 cells were pretreated with 0.5 nM ActD vs DMSO for 24 hours and then tested for sensitivity to venetoclax. (G) IC50 values of DMSO or ActD pretreated OCI-AML3 cells after 4 days of incubation with Ven at the indicated doses (MTS assay). (H) Outline of approach to generate Ven-resistant murine Npm1c-mutant AML cells by treatment with escalating concentrations of Ven. Ven-resistant cells were then pretreated with ActD for 24 hours and their sensitivity to Ven was assessed using the MTS assay. (I) Relative proliferation of DMSO or ActD pretreated murine NPM1c+ AML cells after 4 days incubation with Ven at the indicated doses. Absorbance values were normalized to DMSO-treated cells (mean ± SD, data from 1 of 2 biological replicates, performed in triplicate). (J) IC50 values of DMSO or ActD pretreated murine NPM1c+ AML cells after 4 days of incubation with Ven (mean ± SD, data from 1 of 2 biological replicates, performed in triplicate). (A,C-D,G,J) t test was used to calculate P values between groups. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001; ∗∗∗∗P ≤ .0001. ZIP, zero interaction potency. Panels F and H were created with BioRender.com. Vassiliou, G. (2025) https://BioRender.com/9tt8kww.
In a similar fashion, ActD treatment synergistically enhanced the antileukemic effects of venetoclax in murine Npm1cA/+; Flt3ITD/+ AML cells (Figure 4D). In the same model, high zero interaction potency synergistic scores were obtained in 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) proliferation assays when venetoclax was combined with low concentrations of ActD (Figure 5E; supplemental Figure 7A).
Low-dose ActD treatment resensitizes resistant NPM1-mutant AML cells to venetoclax
NPM1c-mutant OCI-AML3 cells carry biallelic loss-of-function BAX mutations and are resistant to venetoclax (supplemental Figure 7B-C). We pretreated OCI-AML3 cells for 24 hours with low-dose ActD (0.5 nM), a concentration that did not affect cell viability (supplemental Figure 7D). We, then, removed ActD and exposed the pretreated cells to a range of doses of venetoclax (Figure 5F). We found that ActD pretreated OCI-AML3 were significantly more sensitive to venetoclax compared with dimethyl sulfoxide (DMSO)-pretreated cells, with a dramatic fivefold reduction in 50% inhibitory concentration (IC50) (Figure 5G; supplemental Figure 7E). In addition, Annexin V staining demonstrated a significant increase in apoptosis induction in NPM1-mutant cells after combination treatment with low-dose ActD and venetoclax (supplemental Figure 7F).
To test whether this observation held true for acquired venetoclax resistance outside immortalized cell lines, we turned to our Sleeping Beauty transposon-driven NPM1c+ AML model.20 In brief, we exposed murine NPM1c+ AML cells harboring active (jumping) transposons to gradually increasing doses of venetoclax, until they developed resistance. The derivative resistant AML cells displayed a 4 to 7 times higher IC50 value for venetoclax compared with the parental cells. However, as with the OCI-AML3 cells, pretreatment with low concentration of ActD restored their sensitivity to venetoclax in a dose-dependent manner (Figure 5H-J; supplemental Figure 7G-H).
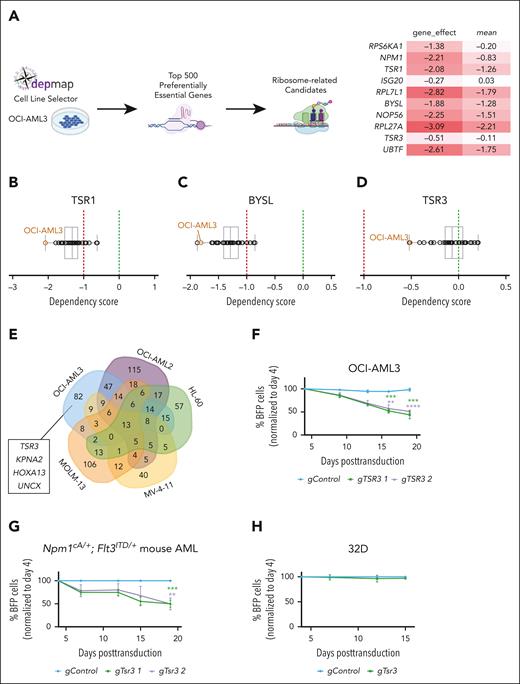
NPM1c+ AML cells are preferentially sensitive to inhibition of the 40S maturation pathway
Ribosome biogenesis is a highly orchestrated process involving more than 200 different factors.7 ActD targets the first step in the process, namely transcription of the polycistronic 47S pre-rRNA precursor, but in higher concentrations can have pleiotropic effects and associated toxicities.38 Thus, we next wanted to explore whether ribosome biosynthesis could be targeted in NPM1c+ AML cells in alternative ways, by inhibiting specific ribosome-related factors.
To investigate this, we started by evaluating the DepMap database, which contains CRIPSR knockout data for hundreds of different cancer cell lines.24,25 We isolated the top 500 preferentially essential genes for OCI-AML3 (the only NPM1c+ cell line in the database), whose knockout is more detrimental to this cell line vs other cancer cell lines. From this list, we selected all genes with a known role in ribosome biogenesis, including ribosomal components and factors involved in rRNA transcription, modification, assembly, and maturation (Figure 6A). Candidates recovered in the list included NPM1 itself, the rDNA transcription factor UBTF, NOP56, which is a core component of box C/D small nucleolar ribonucleoprotein particles, and RPS6KA1, a member of the RSK family of kinases that is involved in the phosphorylation of RPS6 and is druggable43,44 (supplemental Figure 8A-F).
NPM1c+ AML cells are preferentially sensitive to the inhibition of TSR3 and related ribosome biogenesis factors. (A) Analysis of DepMap data was used to identify the top 500 preferentially essential genes for the OCI-AML3 cell line. Of these, candidates with an established role in ribosome biogenesis are highlighted (right). The “gene_effect” represents the dependency score of each candidate for OCI-AML3 cells, where a score of 0 indicates that the gene is not essential, whereas a score of −1 corresponds to the median of all common essential genes in the database. The mean represents the mean gene effect across all cancer cell lines in the database. (B-D) Differential dependency of 37 AML cell lines to CRISPR knockout of TSR1 (B), BYSL (C), or TSR3 (D) according to DepMap. (E) Venn diagram of gene dropouts in a CRISPR screen with 5 AML cell lines (OCI-AML3, OCI-AML2, MOLM-13, HL-60, and MV-4-11).23 (F-H) Relative proliferation of OCI-AML3 (F), murine Npm1cA/+; Flt3ITD/+AML (G), or nonleukemic 32D (H) cells on knockout of TSR3 as measured by a competitive proliferation assay of gRNA-BFP positive (BFP+) transduced cells vs untransduced BFP negative (BFP−) cells in 15 to 18 days. BFP percentage was normalized to the day 4 reading and for panel G to the “gControl” (mean ± SD, n = 3 independent experiments). (F-G) t test was used to calculate P values between groups. ∗∗P ≤ .01; ∗∗∗P ≤ .001; ∗∗∗∗P ≤ .0001. (A-D) CRISPR scores originate from the DepMap Public 23Q2+Score, Chronos data set.24,25 Panel A was created with BioRender.com. Vassiliou, G. (2025) https://BioRender.com/9tt8kww.
NPM1c+ AML cells are preferentially sensitive to the inhibition of TSR3 and related ribosome biogenesis factors. (A) Analysis of DepMap data was used to identify the top 500 preferentially essential genes for the OCI-AML3 cell line. Of these, candidates with an established role in ribosome biogenesis are highlighted (right). The “gene_effect” represents the dependency score of each candidate for OCI-AML3 cells, where a score of 0 indicates that the gene is not essential, whereas a score of −1 corresponds to the median of all common essential genes in the database. The mean represents the mean gene effect across all cancer cell lines in the database. (B-D) Differential dependency of 37 AML cell lines to CRISPR knockout of TSR1 (B), BYSL (C), or TSR3 (D) according to DepMap. (E) Venn diagram of gene dropouts in a CRISPR screen with 5 AML cell lines (OCI-AML3, OCI-AML2, MOLM-13, HL-60, and MV-4-11).23 (F-H) Relative proliferation of OCI-AML3 (F), murine Npm1cA/+; Flt3ITD/+AML (G), or nonleukemic 32D (H) cells on knockout of TSR3 as measured by a competitive proliferation assay of gRNA-BFP positive (BFP+) transduced cells vs untransduced BFP negative (BFP−) cells in 15 to 18 days. BFP percentage was normalized to the day 4 reading and for panel G to the “gControl” (mean ± SD, n = 3 independent experiments). (F-G) t test was used to calculate P values between groups. ∗∗P ≤ .01; ∗∗∗P ≤ .001; ∗∗∗∗P ≤ .0001. (A-D) CRISPR scores originate from the DepMap Public 23Q2+Score, Chronos data set.24,25 Panel A was created with BioRender.com. Vassiliou, G. (2025) https://BioRender.com/9tt8kww.
Notably, among the list of preferentially essential genes for OCI-AML3 cells, we found an enrichment for factors that are involved in related steps of the 40S ribosome maturation pathway, including the ribosome assembly factors BYSL, TSR1, and TSR3. BYSL and TSR1 bind early 40S precursors in the nucleoplasm and are exported bound to the pre-40S subunit to the cytoplasm, where TSR3 is then recruited for the final stages of maturation.7 Loss of any of these 3 factors had a strong, preferential antileukemic effect against OCI-AML3 cells, compared with a panel of 37 different AML cell lines (Figure 6B-D; supplemental Figure 9A).
TSR3 is an aminocarboxypropyl (acp) transferase responsible for the hypermodification of the 18S rRNA during late 40S maturation steps in the eukaryotic cytoplasm. Unlike BYSL and TSR1 which are common essential genes,24,25,TSR3 is not broadly essential for cell survival,45,46 making it an attractive therapeutic target. Notably, an independent CRISPR knockout screen also identified TSR3 as a specific vulnerability of OCI-AML3 cells, but not of the other 4 different AML cell lines tested (OCI-AML2, MOLM-13, HL-60, and MV-4-11)23 (Figure 6E). We decided to focus on TSR3 for validation studies for the following reasons: (1) specificity against NPM1c+ AML; (2) cross-validation in 2 independent CRISPR screens23-25; (3) participation in the 40S subunit maturation, disruption of which is particularly detrimental for NPM1c+ cells; (4) nonessential function in normal cells45,46; and (5) potentially druggable profile.46 We confirmed that TSR3 knockout inhibits the growth of OCI-AML3 cells (Figure 6F) but has little or no effect on the proliferation of NPM1wt cell lines (supplemental Figure 9B). In addition, Tsr3 targeting reduced the proliferation of murine Npm1cA/+; Flt3ITD/+ leukemic cells (Figure 6G), but not of murine AML cells driven by Flt3ITD/+ and oncogenic MLL gene fusions (supplemental Figure 9C). Finally, Tsr3 knockout did not affect the growth of the non-leukemic murine hematopoietic progenitor line 32D (Figure 6H).
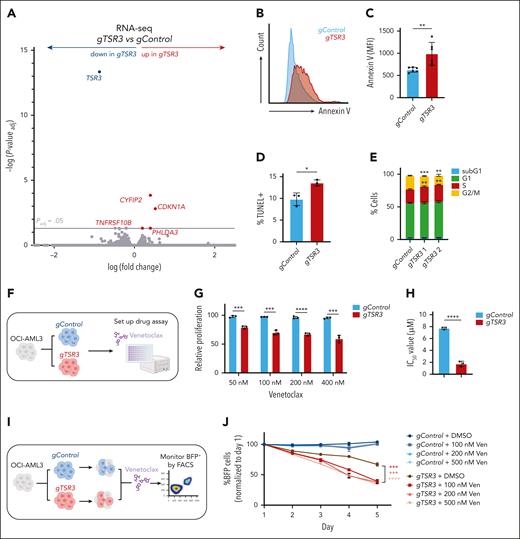
Upregulation of p53 targets and induction of apoptosis in TSR3-depleted NPM1c+ AML cells
Next, we wanted to investigate the effects of TSR3 knockout on NPM1c+ AML cells. RNA-seq analysis revealed the upregulation of many direct p53 target genes, including CYFIP2, CDKN1A, and PHLDA3, shortly after TSR3 depletion (Figure 7A; supplemental Table 7). We performed gene set enrichment analysis of RNA-seq from TSR3-depleted OCI-AML3 cells and found an enrichment for genes involved in apoptosis and a depletion of E2F targets and G2M checkpoint genes (supplemental Figure 10A-C). Conversely, no apoptosis or cell cycle-related gene sets were significantly altered in TSR3-depleted OCI-AML2 cells (NPM1wt) at the same time point (supplemental Figure 10D-F).
TSR3 knockout activates a p53-dependent response and resensitizes resistant NPM1c+ AML cells to Ven. (A) Volcano plot revealing differentially expressed genes in OCI-AML3 cells transduced with a TSR3 gRNA (gTSR3) vs a nontargeting control gRNA (gControl). (B) Representative histogram plot of Annexin V staining in gTSR3 vs gControl-transduced OCI-AML3 cells. (C) Annexin V staining, (D) TUNEL staining, and (E) cell cycle distribution in gTSR3 vs gControl-transduced OCI-AML3 cells. (F) Schematic representation of the OCI-AML3 presensitization experiment. OCI-AML3 cells were transduced with gTSR3 or gControl and their response to venetoclax (Ven) was assessed using the MTS assay. (G) Proliferation of gTSR3 vs gControl-transduced OCI-AML3 cells after 4 days of incubation with Ven at the indicated doses (MTS assay). (H) IC50 values of gTSR3 vs gControl-transduced OCI-AML3 cells after 4 days of incubation with Ven (MTS assay). (I) Schematic representation of resensitization experiment using OCI-AML3 cells. Cells were transduced with BFP-expressing gTSR3 or gControl and were mixed with BFP-negative untransduced cells. Next, they were exposed to Ven and the BFP+ percentage was monitored for a course of 5 days by flow cytometry. (J) Percentage of BFP+gTSR3 or gControl-transduced cells on treatment with Ven at the indicated doses. BFP percentage was normalized to the reading of day 1. (A) Unpaired t test with multiple-hypothesis–testing correction (FDR, Benjamini-Hochberg method). (C-E,G-H,J) t test was used to calculate P values between groups. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001; ∗∗∗∗P ≤ .0001. Data represent 3 to 6 independent experiments (mean ± SD). (G-H) Absorbance values were normalized to DMSO-treated cells. Data represent 3 independent experiments (mean ± SD). MFI, mean fluorescence intensity. Panels F and I were created with BioRender.com. Vassiliou, G. (2025) https://BioRender.com/9tt8kww.
TSR3 knockout activates a p53-dependent response and resensitizes resistant NPM1c+ AML cells to Ven. (A) Volcano plot revealing differentially expressed genes in OCI-AML3 cells transduced with a TSR3 gRNA (gTSR3) vs a nontargeting control gRNA (gControl). (B) Representative histogram plot of Annexin V staining in gTSR3 vs gControl-transduced OCI-AML3 cells. (C) Annexin V staining, (D) TUNEL staining, and (E) cell cycle distribution in gTSR3 vs gControl-transduced OCI-AML3 cells. (F) Schematic representation of the OCI-AML3 presensitization experiment. OCI-AML3 cells were transduced with gTSR3 or gControl and their response to venetoclax (Ven) was assessed using the MTS assay. (G) Proliferation of gTSR3 vs gControl-transduced OCI-AML3 cells after 4 days of incubation with Ven at the indicated doses (MTS assay). (H) IC50 values of gTSR3 vs gControl-transduced OCI-AML3 cells after 4 days of incubation with Ven (MTS assay). (I) Schematic representation of resensitization experiment using OCI-AML3 cells. Cells were transduced with BFP-expressing gTSR3 or gControl and were mixed with BFP-negative untransduced cells. Next, they were exposed to Ven and the BFP+ percentage was monitored for a course of 5 days by flow cytometry. (J) Percentage of BFP+gTSR3 or gControl-transduced cells on treatment with Ven at the indicated doses. BFP percentage was normalized to the reading of day 1. (A) Unpaired t test with multiple-hypothesis–testing correction (FDR, Benjamini-Hochberg method). (C-E,G-H,J) t test was used to calculate P values between groups. ∗P ≤ .05; ∗∗P ≤ .01; ∗∗∗P ≤ .001; ∗∗∗∗P ≤ .0001. Data represent 3 to 6 independent experiments (mean ± SD). (G-H) Absorbance values were normalized to DMSO-treated cells. Data represent 3 independent experiments (mean ± SD). MFI, mean fluorescence intensity. Panels F and I were created with BioRender.com. Vassiliou, G. (2025) https://BioRender.com/9tt8kww.
Reflecting these findings, TSR3 knockout increased apoptosis in OCI-AML3 cells and murine Npm1cA/+; Flt3ITD/+ leukemia cells (Figure 7B-7D; supplemental Figure 10G-I), while also affecting cell cycle distribution (Figure 7E; supplemental Figure 10J). In contrast, we did not observe any significant alterations in apoptosis or the cell cycle distribution on TSR3 knockout in OCI-AML2 cells (supplemental Figure 10K-M).
TSR3 depletion resensitizes resistant NPM1c+ AML cells to venetoclax
Finally, we wanted to investigate whether TSR3 depletion could restore venetoclax sensitivity of resistant NPM1c+ leukemia cells, as found with low-dose ActD treatment. For this, we knocked out TSR3 in OCI-AML3 cells and reassessed response to venetoclax (Figure 7F). Strikingly, we observed that on TSR3 depletion, OCI-AML3 cells responded to concentrations as low as 50 nM, whereas control guide RNA (gRNA)-transduced cells required 100 to 200 times higher concentrations to display a similar effect (Figure 7G; supplemental Figure 11A-B). Overall, IC50 was four and a half-fold lower in TSR3-depleted compared with control OCI-AML3 cells (Figure 7H; supplemental Figure 11C).
Next, we mixed BFP-expressing control gRNA and TSR3 gRNA-transduced OCI-AML3 cells with nontransduced cells and treated with increasing venetoclax concentrations for a course of 5 days, while also monitoring the gRNA abundance using flow cytometry (Figure 7I). As expected, control gRNA-transduced cells demonstrated no decreased survival in the evaluated concentrations. In contrast, gTSR3-transduced cells demonstrated decreased survival that was also reduced on venetoclax treatment, further revealing the selective sensitivity of the TSR3-depleted cell population (Figure 7J; supplemental Figure 11D-G).
Discussion
NPM1c mutations are found in 30% of all AML cases.13 Yet, despite some recent advances in understanding their molecular consequences,47,48 current therapeutic outcomes remain suboptimal, highlighting the need for new effective treatments.18 In this study, we identified NPM1c-induced proteomic changes and investigated their therapeutic potential. The use of a faithful conditional knockin mouse model allowed us to study the proteomic effects of NPM1c mutations in isolation, by overcoming the confounding effects of different comutations19 or other sources of biological variation, including potential in vitro adaptive changes in transformed cell lines. This enabled us to establish a direct connection between the mutated NPM1c protein and observed phenotypes.
Our findings revealed that levels of many proteins involved in ribosome biogenesis are significantly reduced in NPM1c+ cells, without significant changes in expression of their cognate messenger RNAs (RNA-seq), in keeping with altered posttranscriptional regulation. This downregulation could be either directly or indirectly triggered by the reduced levels of NPM1wt in mutant cells, although the precise mechanisms involved require further investigation. Potential scenarios include the following: (1) the interaction of NPM1wt with rRNA and ribosome biogenesis factors may stabilize them, with loss of this interaction in the mutant cells leading to protein degradation and (2) NPM1 haploinsufficiency may alter the availability of its chaperone partners or disrupt the structural organization of the nucleolus,9,10 affecting interactions, levels, and stability of ribosome biogenesis factors. Interestingly, the altered nucleolar structure was recently found to be detectable by holotomography.49 On the basis of the observed depletion of ribosome biogenesis factors, we hypothesized that targeting nucleolar homeostasis may selectively affect NPM1c+ cells. Indeed, NPM1-mutant cells were selectively more sensitive to multiple RNA pol I inhibitors (ActD, CX-5461, and BMH-21) but displayed no differential response to translation inhibitors such as cycloheximide.
Importantly, reduced ribosomal factor levels are also evident after leukemia development, providing a rationale for reevaluating the concept of targeting ribosome biogenesis in NPM1c+ AML, as first suggested and tested by Falini et al.31,32 More recently, Gionfriddo et al33 revealed that ActD could induce complete remission in patients with relapsed/refractory NPM1-mutant AML, with evidence that the treatment initiated a nucleolar stress response. Interestingly, Wu et al50 discovered an additional effect of ActD in the mitochondria which contributed to its antileukemic effects.
In the second part of this study, we explored whether specific ribosome biogenesis factors can be targeted in NPM1-mutant leukemic cells. Using DepMap24,25 and another independent genome-wide AML CRISPR screen,23 we identified TSR3, a gene whose knockout preferentially reduced the proliferation of NPM1c+ AML cells. TSR3 acts at the late cytoplasmic stages of the 40S ribosome maturation and is responsible for introducing the N1-methyl-N3-aminocarboxypropylpseudouridine (m1acp3Ψ) mark on 18S rRNA.46 Interestingly, loss of the m1acp3Ψ is a common event in cancer,45 though it is unclear whether this loss confers a growth advantage or is simply a byproduct of the high ribosome production rates in cancer cells.
Currently, the mechanism underlying the dependency of NPM1c+ AML on TSR3 is poorly understood, and it remains uncertain whether it relates to its enzymatic activity. The significantly reduced levels of the ribosome factors KRI1 and its interactor KRR1, both of which are acting upstream of TSR3 in 40S maturation,7 could provide a potential link to explain this vulnerability. In this model, TSR3 knockout, similarly to RNA pol I inhibition, could be exacerbating an already impaired ribosome biogenesis process in NPM1c+ AML, lowering the threshold for p53 activation and leading to apoptosis.
Notably, both low-dose ActD treatments and TSR3 knockout synergized with venetoclax and displayed the ability to induce at least partial resensitization in NPM1-mutant, venetoclax-resistant AML models. Previous studies have revealed that nucleolar stress, such as that caused by RNA pol I inhibition, leads to the diffusion of ribosomal proteins into the nucleoplasm, where they bind and sequester MDM2, resulting in p53 stabilization and upregulation of key apoptotic genes.35,51-53 In addition, induction of p53-driven apoptosis was demonstrated in ActD-treated NPM1-mutant AML cells.33 In line with these findings, our RNA-seq analysis of TSR3-depleted NPM1c+ cells revealed the upregulation of direct p53 targets, including CDKN1A (p21), and an enrichment for genes involved in apoptosis. Importantly, p53 and the apoptotic response network have a central role in controlling sensitivity to venetoclax.54,55 Taken together, these data suggest that activation of p53 and its targets by ActD or TSR3 knockout works synergistically with BCL-2 inhibition (venetoclax) to promote apoptosis in NPM1-mutant AML cells.
Overall, our findings lead us to propose that acquisition of NPM1c and the accompanying loss of 1 NPM1wt allele result in the posttranscriptional depletion of multiple players involved in ribosome biogenesis, a consequence that can be exploited therapeutically to target NPM1-mutant AML cells (supplemental Figure 12).
Acknowledgments
The authors thank the Cancer Research UK Cambridge Institute Proteomics team, the Cambridge Biomedical Research Centre Phenotyping Hub, the Anne McLaren Biomedical Research Facility (especially Charlotte Townend), and the Cambridge Stem Cell Institute Genomics facility and Imaging facility (especially James Boyd) for providing expert technical assistance and support for our research project.
This work was funded by grants from Cancer Research UK (C22324/A24482) and AstraZeneca (G113590). G.S.V. is supported by a Cancer Research UK Senior Cancer Fellowship (C22324/A23015), and work in his laboratory is also funded by the European Research Council, Leukemia and Lymphoma Society, Rising Tide Foundation for Clinical Cancer Research, Kay Kendall Leukaemia Fund, Blood Cancer UK, and Wellcome Trust. A.D. is supported by Cancer Research UK (C22324/A24482) and AstraZeneca (G113590). K.T., E.Y., and M.E. are supported by Wellcome Trust (grants RG94424, RG83195, RG106133, and RG127005) and UKRI Medical Research Council (grant RG83195). K.T. and M.E. are supported by Leukaemia UK (grants 2020/JGF/004 and 2022/FuF/001). E.Y. is supported by a Leukaemia UK John Goldman Fellowship (2024/JGF/009). Work in the laboratory of A.J.W. is supported by Cancer Research UK (DRCNPG-Jun24/100002), UK Medical Research Council (MR/T012412/1), European Cooperation in Science and Technology, Action Translacore (CA21154), Blood Cancer UK (21002), Rosetrees Trust (PGL22/100032), Isaac Newton Trust (G125517), and Addenbrookes Charitable Trust (900426). J.-E.S. was supported by the Programme Investissement d’Avenir Projets de Recherche et Développement Structurants Pour la Compétitivité (Initiative de Modèles Innovants) and the Laboratoire d’Excellence Toulouse Cancer (TOUCAN; contract ANR11-LABEX). A. Sahal was a fellow from the European Regional Development Fund through the Interreg V-A Spain–France–Andorra (Programa Interreg España-Francia-Andorra) program, project PROTEOblood (EFA360/19; J.-E.S.).
Authorship
Contribution: G.S.V. conceived and supervised the study; G.S.V. and A.D. designed the experimental plan; A.D. carried out most of the experiments with help from R.W.; M. Gozdecka contributed to experimental design and provided scientific expertise; the in vivo patient-derived xenograft (PDX) experiment was performed with the help from G.G. (study design, animal procedures, imaging), R.A. (animal procedures, animal collection), and G.S.V. (intravenous injections); M.E. contributed to immunofluorescence imaging and animal collection and processing; M. Gu performed bioinformatic analysis and data deposition for the RNA sequencing experiments; primary AML characterization, sample collection, and PDX generation were carried out by C.R. (clinician), V.M.-D.M. (biobanking), F.V. (diagnosis), A. Sahal (PDX generation), B.V. (passaging and introduction of luciferase reporter), and I.J. and J.-E.S. (supervision); E.K.P., A. Sawle, and C.D'S. performed, analyzed, and provided scientific expertise for the preleukemic tandem mass tag proteomics experiment; E.Y., M.D., X.L., J. Russell, and J. Rak contributed to animal collection and processing; C.H. performed polysome profiling; I.J., J.-E.S., K.T., B.J.P.H., A.J.W., and O.T. provided scientific expertise; and G.S.V. and A.D. prepared the manuscript with help from all authors.
Conflict-of-interest disclosure: G.S.V. is a consultant to STRM.BIO and holds a research grant from AstraZeneca (G113590) that funded part of the research presented here. K.T. has received stock options and research funding from Storm Therapeutics Ltd. O.T. is an AstraZeneca employee and shareholder. The remaining authors declare no competing financial interests.
Correspondence: George S. Vassiliou, Department of Haematology, Cambridge Stem Cell Institute, University of Cambridge, Puddicombe Way, Cambridge, CB2 0AW, United Kingdom; email: gsv20@cam.ac.uk; and Aristi Damaskou, Department of Haematology, Cambridge Stem Cell Institute, University of Cambridge, Puddicombe Way, Cambridge, CB2 0AW, United Kingdom; email: ad971@cam.ac.uk.
References
Author notes
Mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium (data set identifier PXD053249).
Data are available on request from the corresponding authors, George S. Vassiliou (gsv20@cam.ac.uk) and Aristi Damaskou (ad971@cam.ac.uk).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.