Abstract
The granulocyte-macrophage colony-stimulating factor (GM-CSF ) receptor is expressed on normal and malignant hematopoietic cells as well as on cells from other organs in which it transduces a variety of functions. Despite the widespread expression and pleiotropic nature of the GM-CSF receptor, little is known about its assembly and activation mechanism. Using a combination of biochemical and functional approaches, we have found that the human GM-CSF receptor exists as an inducible complex, analogous to the interleukin-3 (IL-3) receptor, and also as a preformed complex, unlike the IL-3 receptor or indeed other members of the cytokine receptor superfamily. We found that monoclonal antibodies to the GM-CSF receptor α chain (GMRα) and to the common β chain of the GM-CSF, IL-3, and IL-5 receptors (βc ) immunoprecipitated both GMRα and βc from the surface of primary myeloid cells, myeloid cell lines, and transfected cells in the absence of GM-CSF. Further association of the two chains could be induced by the addition of GM-CSF. The preformed complex required only the extracellular regions of GMRα and βc , as shown by the ability of soluble βc to associate with membrane-anchored GMRα or soluble GMRα. Kinetic experiments on eosinophils and monocytes with radiolabeled GM-CSF, IL-3, and IL-5 showed association characteristics unique to GM-CSF. Significantly, receptor phosphorylation experiments showed that not only GM-CSF but also IL-3 and IL-5 stimulated the phosphorylation of GMRα-associated βc . These results indicate a pattern of assembly of the heterodimeric GM-CSF receptor that is unique among receptors of the cytokine receptor superfamily. These results also suggest that the preformed GM-CSF receptor complex mediates the instantaneous binding of GM-CSF and is a target of phosphorylation by IL-3 and IL-5, raising the possibility that some of the biologic activities of IL-3 and IL-5 are mediated through the GM-CSF receptor complex.
GRANULOCYTE-MACROPHAGE colony-stimulating factor (GM-CSF ) is a pleiotropic cytokine that exhibits effects on most cell types in the hematopoietic compartment.1,2 GM-CSF exhibits overlapping biologic activities with interleukin-3 (IL-3) on several hematopoietic cells owing to a similar pattern of receptor expression and to the sharing of a communal signal transducing receptor subunit that is also shared with IL-5.3 Indeed, on eosinophils that express GM-CSF, IL-3, and IL-5 receptors, these three cytokines stimulate the same functions with very similar potency.4 The functional receptors for GM-CSF, IL-3, and IL-5 are closely related and are composed of two subunits: a ligand-specific α chain and the communal β chain (βc ).5-7 The receptor α chains bind their cognate cytokine ligands with low affinity but are largely unable to mediate signalling alone, although some reports have suggested a role for GM-CSF receptor α chain (GMRα) in glucose transport.8 The communal β chain, βc , is unable to bind any cytokine alone, but confers high-affinity binding on a ligand: α chain complex (kd ∼100 pmol/L) and is required for receptor signalling.5-7 Functional high-affinity receptors for GM-CSF, IL-3, or IL-5 can be reconstituted on cells that do not normally express these receptors by coexpressing cytokine-specific α chains and β9-11c ; however, the relationship and assembly of these subunits on the cell surface are unknown.
The mechanism of activation of the GM-CSF receptor is likely to involve receptor dimerization, although the molecular basis of this phenomenon is poorly understood. Ligand-induced receptor dimerization is a common theme among the cytokine receptor superfamily and is usually a prerequisite for receptor activation. For example, IL-6 induces IL-6Rα and gp130 dimerization12 with homodimerization of gp130 causing receptor phosphorylation.13 Similarly, ciliary neurotrophic factor induces receptor dimerization and subsequent receptor activation.14 In the case of the IL-3 receptor that is closely related to the GM-CSF receptor,15 IL-3 induces receptor α:βc heterodimerization followed by covalent disulphide bridging between receptor α chain and βc .16 The structural similarities and functional overlap between the GM-CSF and IL-3 receptor systems have suggested that activation of the GM-CSF receptor may follow a similar pattern of events. Indeed, GM-CSF has been shown to induce coassociation of GMRα with βc ,17 and a general mechanism has been noted that involves disulphide bridging between receptor α chain and a cysteine motif in βc that is essential for activation of GM-CSF, IL-3, and IL-5 receptors (Bagley et al18 and unpublished observation). Despite exhibiting some common features of activation with other receptors, the GM-CSF receptors also appear to exhibit some unusual features. For example, mutant forms of GMRα that are deficient in GM-CSF binding when expressed alone on cells are able to support binding when coexpressed with βc ,19,20 suggesting that βc can compensate for losses in binding affinity. Conversely, a mutation in GM-CSF abolishes the ability of the molecule to compete for low-affinity binding but retains the ability to compete for high-affinity binding.21 Lastly, a recent report showed that a naturally occurring soluble form of GMRα is retained at the cell surface when coexpressed with βc , although coimmunoprecipitation of the two subunits could only be demonstrated in the presence of GM-CSF.22
We report here that the human GM-CSF receptor exists as both an inducible complex and, unlike other cytokine receptors, as a preformed receptor complex. Using monoclonal antibodies (MoAbs) specific for the GM-CSF receptor α and βc , we found that both subunits could be coimmunoprecipitated in the absence of GM-CSF regardless of whether they were surface expressed or expressed as soluble forms by the same cells. Consistent with there being two types of GM-CSF receptor complex, we show in kinetic experiments on eosinophils that GM-CSF exhibits unique association kinetics with two types of binding site; one type exhibits association kinetics very similar to those of IL-3 and IL-5, whereas the other type shows virtually instantaneous association. Significantly, stimulation of cells not only with GM-CSF but also with IL-3 and IL-5 induces the phosphorylation of βc associated with GMRα. A model is proposed in which IL-3 and IL-5 recruit the performed GM-CSF receptor into a high order complex, raising the possibility that some of the biologic activities of IL-3 and IL-5 are mediated indirectly through activation of the preformed GM-CSF receptor complex.
MATERIALS AND METHODS
Cell lines. Chronic myelogenous leukemia (CML) cells were obtained as described previously16 and cultured in RPMI supplemented with 10% fetal calf serum (FCS). Mo7e cells and Ba/F-3 cells expressing GMRα/βc were maintained in DMEM (GIBCO, Melbourne, Australia) supplemented with 20 mmol/L HEPES supplemented with 10% FCS and 5 or 2 ng/mL GM-CSF, respectively. TF-1.8 cells were maintained in RPMI supplemented with 10% FCS and 2 ng/mL GM-CSF. Factor dependent cells were routinely starved of growth factor overnight before cytokine treatment. Chinese hamster ovary (CHO) cells were maintained in F12 medium supplemented with 10% FCS and transfected as described previously.21
Plasmid construction. The cDNA for the human βc was cloned by polymerase chain reaction (PCR) from cDNA prepared from the KMT-2 cell line.23 A soluble form of the βc (sβc ) was created by PCR using the following synthetic oligonucleotides: (1) 5′-TGAATTCGCCTGTCCAGAGCTGACCAGGG-3′ that starts 25 nucleotides 5′ of the ATG and contains an engineered HindIII site and (2) 5′- ATACACTCTATATCACGACTCGGTGTCCCAGGAGCG -3′ that contains an inframe termination codon immediately 5′ of the transmembrane region followed by an engineered Xba I site. The PCR product obtained from these primers was subcloned into the Neomycin resistance conferring expression vector pRc/CMV (Invitrogen Corp, San Diego, CA) giving rise to sβcpRc/CMV. A soluble form of the human GMRα (sGMRα) was made in a similar fashion using the following synthetic oligonucleotides: (1) 5′-ATACACAAGCTTAGCACCATGCTTCTCCTGGTG-3′ that starts 18 nucleotides 5′ of the ATG and contains an engineered HindIII site and (2) 5′-ATACACTCTAGATCACCCGTCGTCAGAACCAAATTC-3′ that contains an inframe termination codon immediately 5′ of the transmembrane region followed by an engineered Xba I site. The PCR product obtained from this set of primers was subcloned into pRc/CMV to produce sGMRαpRc/CMV.
To allow for dual stable transfection of two receptors, pRc/CMV was engineered such that the neomycin resistance gene (NeoR) was replaced with the puromycin resistance gene (pac ) from pRuf puro.24 Briefly, the 1.5-kb Kpn I-BamHI fragment from pRc/CMV containing NeoR and its flanking SV40 early promoter and poly-adenylation region was subcloned into pUC19. The NeoR gene was removed by EcoRV-Nae I digestion and pac introduced as a Sal I-Cla I fragment from pRuf puro. The puromycin resistance gene plus flanking SV40 early promoter and poly-adenylation region was excised from pUC19 as a Kpn I-BamHI fragment and subcloned into Kpn I-partial BamHI digested sβcpRc/CMV, resulting in sβcpRc/CMVpuro. Subsequently, full-length βc cDNA was introduced in on an EcoRI-Xba I fragment thereby generating βcpRc/CMVpuro.
Construction of stable CHO cell lines. The CHO cell lines, sβcCHO and sGMRα CHO, were developed as described previously for the GMRα CHO cell line, A9/C7.21 CHO lines expressing sGMRα or GMRα were subsequently cotransfected with either βcpRc/CMVpuro or sβcpRc/CMVpuro by the same method and selected in 2.5 μg/mL Puromycin (Calbiochem, La Jolla, CA). Cell surface expression of transfected receptors was confirmed by flow cytometry as described previously16 and analyzed on an EPICS Profile II Flow Cytometer (Coulter Electronics, Hialeah, FL).
Purification of human eosinophils and monocytes. Eosinophils were purified from the peripheral blood of eosinophilic individuals by centrifugation on a hypertonic gradient of metrizamide as described previously.25 Monocytes were purified from the peripheral blood of normal donors obtained from the Adelaide Red Cross Transfusion Service as described previously.26
Antibodies. MoAbs directed against GMRα, IL-3Rα, or βc were generated as previously described27 and purified and characterized as detailed elsewhere.16,27,28 The MoAbs 8E4 and 4F3 were selected for their ability to specifically immunoprecipitate βc , 8G6 for GMRα, and 9F5 for IL-3Rα. The MoAb 1C1 and an antipeptide polyclonal rabbit antibody (against residues 131-241 of βc ) were used for immunoblotting βc and an MoAb 8D10 for immunoblotting GMRα. Phosphotyrosine residues were detected by immunoblot using directly horseradish peroxidase-conjugated PY20 antibody (Sapphire Bioscience, Alexandria, New South Wales, Australia). MoAbs 4F3, 8G6, and 6H6 were used for cell surface expression staining for βc , GMRα, and IL-3Rα, respectively. The anti-βc antibody, 3D7,28 was used for affinity purification of sβc protein. The MoAbs were purified from ascites as described.27 A rabbit polyclonal anti–GM-CSF antibody was used for immunoprecipitating GM-CSF.21
Purification of recombinant soluble human βc receptor. Soluble βc protein was purified from conditioned medium from CHO cells stably expressing the protein using a 3D7 anti-βc MoAb affinity column. Bound soluble βc was eluted with a linear gradient from 3 to 1 mol/L KSCN in 10 mmol/L Tris-HCl, pH 8.0, and subsequently buffer exchanged into phosphate-buffered saline (PBS) containing 0.02% (vol/vol) Tween 20 [polyoxyethylene (20)-sorbitan monolaurate].
125I-surface labeling and immunoprecipitation conditions. Cells were cell-surface labeled with 125I by the lactoperoxidase method as described previously.29 Approximately 108 cells were labeled with 1 mCi 125I (NEN, Boston, MA) in PBS. Cells were lysed in lysis buffer consisting of 137 mmol/L NaCl, 10 mmol/L Tris-HCl (pH 7.4), 10% glycerol, and 1% nonidet P-40 (NP40) with protease and phosphatase inhibitors (10 μg/mL leupeptin, 2 mmol/L phenylmethlsulphonyl fluoride, 10 μg/mL aprotonin, and 2 mmol/L sodium vanadate) for 30 minutes at 4°C followed by centrifugation of the lysate for 15 minutes at 4°C. After 1 hour of preclearance with protein A-sepharose (Pierce, Rockford, IL) at 4°C, the supernatant was incubated for 18 hours with 10 μg/mL antibody. Protein-Ig complexes were captured by incubation for 1 hour with protein A-sepharose followed by 6 subsequent washes in lysis buffer and then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Immunoprecipitation of proteins from conditioned medium was performed similarly.
Deglycosylation conditions. Deglycosylation of proteins was performed after immunoprecipitation with the protein still attached to the protein A-sepharose beads. The immunoprecipitated protein was first incubated in 200 mmol/L sodium cacodylate, pH 7.0, 0.1% SDS and then in 0.75% NP40 with neuraminidase, O-glycanase (Genzyme, Castle Hill, New South Wales, Australia), and N-glycanase (New England Biolabs, Arundel, Australia) for 18 hours at 37°C before separation by SDS-PAGE.
SDS-PAGE and silver staining. Immunoprecipitated proteins were analyzed by SDS-PAGE on 7.5% or 10% polyacrylamide gels as stated. Samples were boiled for 10 minutes in either the presence or absence of 2-mercaptoethanol (ie, reducing or nonreducing) before separating immunoprecipitated proteins by SDS-PAGE. Molecular weights (MW) were estimated using SeeBlue Pre-Stained Standards (Novex, San Diego, CA). Radiolabeled proteins were visualized using an ImageQuant Phosphorimager (Molecular Dynamics, Sunnyvale, CA). Silver staining of gels was performed as described previously.30
Immunoblotting and enhanced chemiluminescence (ECL) detection. Immunoprecipitated proteins separated by SDS-PAGE were transferred to nitrocellulose membrane by electroblotting. Nitrocellulose membranes were routinely blocked in a solution of PBS, 0.05% Tween 20 (vol/vol) (PBT) containing 5% skim milk (wt/vol) or in 10 mmol/L Tris (pH 8.0), 150 mmol/L NaCl, 0.05% Tween 20 (vol/vol) (TNT) containing 5% bovine serum albumin (wt/vol) and probed with antibody, followed where appropriate by either rabbit antimouse horseradish peroxidase (Dako, Carpenteria, CA) or goat antirabbit horseradish peroxidase (Dako). Immunoreactive proteins were detected by chemiluminescence using the ECL kit (Amersham, Little Chalfont, UK) following the manufacturer's instructions. Stripping of membranes was performed by incubating nitrocellulose membrane for 30 minutes at 50°C in 100 mmol/L 2-mercaptoethanol, 2% SDS, 62.5 mmol/L Tris-HCl, pH 6.7, followed by two sequential washes in PBT or TNT. Membranes were reblocked for 1 hour before reprobing.
Production and radio-iodination of GM-CSF, IL-3, and IL-5. Recombinant GM-CSF was produced in Escherichia coli as described previously.21 For the kinetic experiments, recombinant GM-CSF, IL-3, and IL-5 were produced in yeast as described previously.31 Radio-iodination of cytokines was performed by the iodine monochloride method32 and the iodinated proteins separated from iodide ions on a Sephadex G-25 PD-10 column (Pharmacia, Uppsala, Sweden) and eluted with PBS containing 0.02% Tween 20 and stored at 4°C for up to 4 weeks. The yeast derived radio-iodinated cytokines were purified before use as described previously.31
Saturation binding assays. Binding assays were performed on CHO cells grown to confluency in 96-well plates over a concentration range of 10 pmol/L to 10 nmol/L 125I-labeled GM-CSF in binding medium (RPMI containing 0.5% [wt/vol] bovine serum albumin and 0.1% [wt/vol] sodium azide) with nonspecific binding determined at each concentration with excess unlabeled GM-CSF. After incubation at room temperature for 2 hours, radioligand was removed and the wells were washed briefly twice in binding medium. Where stated, low-affinity binding was then removed with five sequential 15-minute washes in binding medium. Specific counts were determined after lysis of the cell monolayer with subsequent transfer and counting on a γ-counter (Cobra Auto Gamma; Packard Instruments Co, Meridien, CT). Dissociation constants were calculated using the EBDA and LIGAND programs33 (Elsevier Biosoft, Cambridge, UK).
Binding assays were performed on soluble receptors in solution in a similar fashion to soluble receptor assays described previously.34 Aliquots of soluble receptor (100 μL) were incubated with 125I-labeled GM-CSF (10 μL) over a concentration range of 0.5 to 20 nmol/L. An excess of unlabeled GM-CSF was added to assays to determine nonspecific binding. Assays were incubated at room temperature for 1 hour, and then Con A-sepharose (10 μL of 50% slurry in PBS) was added to each tube and allowed to bind over 1 hour. Sepharose (100 μL of 50% slurry in PBS) was then added to each assay to increase the amount of precipitable material, and the tubes were centrifuged to pellet the beads. Pelleted beads were washed once in PBS and then the radioactivity was determined by counting on a γ-counter.
Kinetic binding assays. Association kinetics were determined at 4°C with eosinophils and monocytes using radio-iodinated cytokines at 200 pmol/L. Cells (2 to 4 × 106 per tube) were incubated in 0.15 mL of binding medium containing radioligand with or without 100-fold excess unlabeled cytokine in borosilicate tubes on a rotating table. Assays were harvested at time points after addition of radio-iodinated cytokine by overlaying onto 0.2 mL FCS and spinning for 30 seconds at maximum speed in a Beckman microfuge (Beckman, Gladesville, New South Wales, Australia). The visible cell pellet was removed by cutting and the radioactivity in the pellet determined on the γ-counter. The apparent association rate (Kobs ) was calculated using the KINETIC program (Elsevier Biosoft) from the specific binding data. Kobs is a composite function encompassing both on and off rates (Kon and Koff , respectively) from the receptor: Kobs = Kon [L] + Koff .
RESULTS
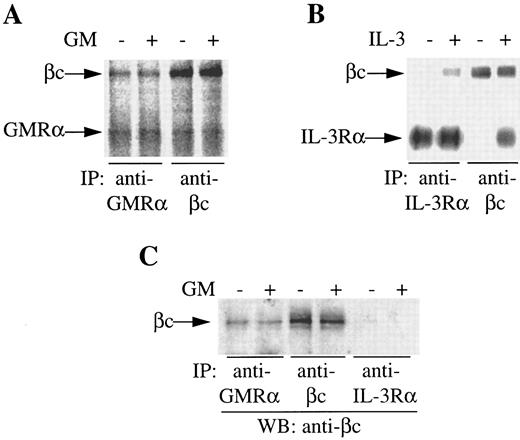
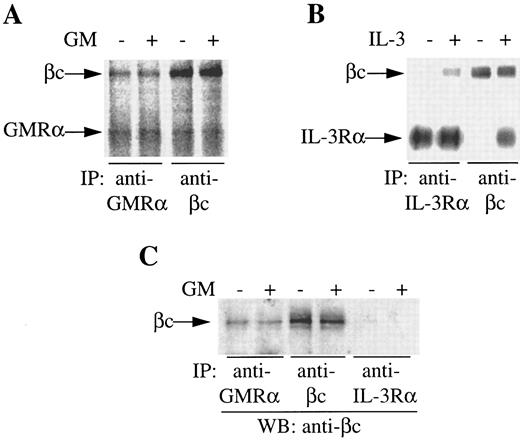
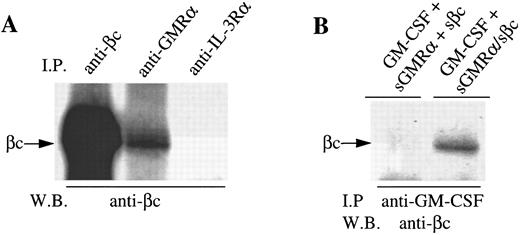
GMRα and βc are preassociated on the cell surface. During the course of our studies on IL-3 receptor complex formation, we previously observed coimmunoprecipitation of an 80,000 MW protein with βc from 125I-surface–labeled primary CML cells in the absence of exogenous stimuli.16 The size of this protein suggested it could be the GMRα. To examine this possibility, we conducted immunoprecipitation of 125I-surface–labeled CML cells either in the presence or absence of GM-CSF or IL-3 with anti-GMRα, anti–IL-3Rα, or anti-βc antibodies. Immunoprecipitation of unstimulated cells with anti-GMRα antibody 8G6 immunoprecipitated a protein of 80,000 MW, consistent with the size of GMRα (Fig 1A). A second protein of 120,000 MW, corresponding in size to βc , coimmunoprecipitated with GMRα in the absence of GM-CSF and its level did not increase with the addition of GM-CSF (Fig 1A). Reciprocally, immunoprecipitation with anti-βc antibody 4F3 immunoprecipitated both the 120,000 MW βc protein and the 80,000 MW GMRα protein in the presence or absence of GM-CSF (Fig 1A). Coimmunoprecipitation of GMRα with βc with either anti-GMRα or anti-βc antibodies could be the result of these antibodies recognizing similar epitopes on both receptor chains. However, in previous studies, we have shown that these antibodies are absolutely specific for their respective receptor chains and show no cross-reactivity.16, 28
Coimmunoprecipitation of GMRα and βc from primary CML cells. (A and B) CML cells were 125I–surface-labeled, treated with (+) or without (−) GM-CSF or IL-3 (6 nmol/L) for 5 minutes at 4°C and immunoprecipitation was performed either with anti-GMRα (8G6), anti–IL-3Rα (9F5), or anti-βc (4F3) MoAb. Immunoprecipitated proteins were separated on 7.5% SDS-PAGE and visualized by phosphorimager and are presented at exposure levels appropriate for the specific signal obtained. (C) Proteins immunoprecipitated from CML cells with different antireceptor antibodies either in the presence (+) or absence (−) of GM-CSF were subjected to Western transfer and immunoblotted using a polyclonal anti-βc antibody.
Coimmunoprecipitation of GMRα and βc from primary CML cells. (A and B) CML cells were 125I–surface-labeled, treated with (+) or without (−) GM-CSF or IL-3 (6 nmol/L) for 5 minutes at 4°C and immunoprecipitation was performed either with anti-GMRα (8G6), anti–IL-3Rα (9F5), or anti-βc (4F3) MoAb. Immunoprecipitated proteins were separated on 7.5% SDS-PAGE and visualized by phosphorimager and are presented at exposure levels appropriate for the specific signal obtained. (C) Proteins immunoprecipitated from CML cells with different antireceptor antibodies either in the presence (+) or absence (−) of GM-CSF were subjected to Western transfer and immunoblotted using a polyclonal anti-βc antibody.
In contrast to the coimmunoprecipitation seen with GMRα and βc , coimmunoprecipitation of IL-3Rα and βc by either anti–IL-3Rα or anti-βc antibodies was only seen in the presence of IL-3 (Fig 1B), as shown previously.16 The phosphorimage signal for the IL-3 receptor (Fig 1B) is strong relative to the signal obtained for GM-CSF receptor (Fig 1A) owing to the high level of IL-3 receptor expression relative to GM-CSF receptor on these cells.35 As stated previously, a protein of 80,000 MW, consistent in size with GMRα, coimmunoprecipitated with βc in either the presence or absence of IL-3, although at much weaker intensity than either βc or IL-3Rα16 and is hence not visible at the exposure shown (Fig 1B).
To confirm the identity of the 120,000 MW protein coimmunoprecipitated by anti-GMRα antibody 8G6, we performed immunoprecipitations with unlabeled cells before and after treatment with GM-CSF using anti-GMRα (8G6), anti-βc (4F3), and anti–IL-3Rα (9F5) antibodies. After Western transfer, an immunoblot with anti-βc antibody was performed. An 120,000 MW protein was clearly detected in the presence or absence of GM-CSF in both GMRα and βc immunoprecipitates but not in the IL-3Rα immunoprecipitate (Fig 1C). This indicates that βc is associated with GMRα but not with IL-3Rα in the absence of added cytokine on these primary CML cells.
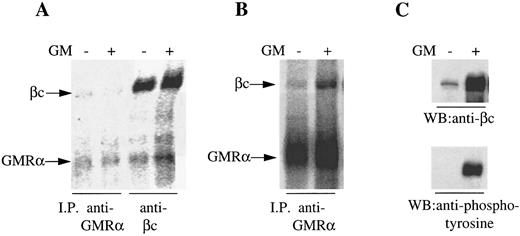
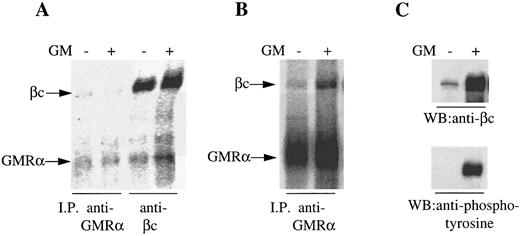
One possible explanation for the preassociation of GMRα with βc was the autocrine production of GM-CSF by the CML cells. However, we were unable to detect either GM-CSF protein by enzyme-linked immunosorbent assay or GM-CSF mRNA by Northern analysis or reverse transcription-PCR (data not shown). Nevertheless, to confirm the GM-CSF–independent association between GMRα and βc and to determine the generality of this observation, we performed immunoprecipitation experiments on a human GM-CSF–dependent cell line (Mo7e) and on a mouse cell line (Ba/F-3) transfected with the human GM-CSF receptor. Mo7e cells maintained in IL-3 and murine Ba/F-3 cells expressing human GMRα and βc maintained in GM-CSF were starved overnight before GM-CSF stimulation. Cells were 125I-surface labeled and proteins were immunoprecipitated with anti-GMRα (8G6) or anti-βc (8E4) before and after treatment with GM-CSF. We observed coimmunoprecipitation of the 120,000 MW βc protein and the 80,000 MW GMRα protein with either antibody in the presence or absence of GM-CSF (Fig 2A and B), although the signal observed on Mo7e cells was weak relative to the CML and Ba/F-3 cells, presumably due to low receptor expression. However, the relative intensity of the two proteins immunoprecipitated from Mo7e cells was similar regardless of whether GM-CSF was present or not, whereas with the GM-CSF receptor expressing Ba/F-3 cells, GM-CSF stimulation enhanced the association of βc with GMRα (Fig 2A and B), indicating that only a proportion of GM-CSF receptors are preformed in these cells. To confirm that the 120,000 MW protein coimmunoprecipitated from Ba/F-3 cells with GMRα was human βc and not a mouse β chain protein, an immunoblot was performed on the immunoprecipitates with anti-βc antibody. The anti-βc antibody was highly immunoreactive against the 120,000 MW protein immunoprecipitated by anti-GMRα antibody in either the presence or absence of GM-CSF, confirming the 120,000 MW protein as human βc (Fig 2C). Reprobing the immunoblot with antiphosphotyrosine antibody PY20 showed that the βc was phosphorylated only after treatment of the Ba/F-3 cells with GM-CSF (Fig 2C), indicating that the preformed GMRα:βc complex is not activated and that this complex was not the result of residual cytokine on the cells after overnight factor depletion. These findings strongly suggest that GMRα and βc are associated at the cell surface in the absence of GM-CSF as a preformed complex.
Coimmunoprecipitation of GMRα and βc from Mo7e and hGMRα/hβc -expressing Ba/F-3 cells. (A) Mo7e cells were starved overnight and 125I–surface-labeled and treated with (+) or without (−) GM-CSF (6 nmol/L) for 5 minutes and immunoprecipitation was performed either with anti-GMRα MoAb (8G6) or anti-βc MoAb (4F3). Immunoprecipitated proteins were separated on 7.5% SDS-PAGE under reducing conditions and the gel was exposed to phosphorimager. (B) hGMRα/hβc -expressing Ba/F-3 cells were starved overnight and 125I–surface-labeled and treated with (+) or without (−) GM-CSF (6 nmol/L) for 5 minutes and immunoprecipitation was performed with anti-GMRα MoAb (8G6). Immunoprecipitated proteins were separated on 7.5% SDS-PAGE under reducing conditions and the gel was exposed to phosphorimager. (C) Proteins immunoprecipitated from hGMRα/hβc -expressing Ba/F-3 cells with anti-GMRα MoAb (8G6) either in the presence (+) or absence (−) of GM-CSF were subjected to Western transfer and immunoblotted using anti-βc antibody 1C1 (upper panel) or antiphosphotyrosine antibody PY20 (lower panel).
Coimmunoprecipitation of GMRα and βc from Mo7e and hGMRα/hβc -expressing Ba/F-3 cells. (A) Mo7e cells were starved overnight and 125I–surface-labeled and treated with (+) or without (−) GM-CSF (6 nmol/L) for 5 minutes and immunoprecipitation was performed either with anti-GMRα MoAb (8G6) or anti-βc MoAb (4F3). Immunoprecipitated proteins were separated on 7.5% SDS-PAGE under reducing conditions and the gel was exposed to phosphorimager. (B) hGMRα/hβc -expressing Ba/F-3 cells were starved overnight and 125I–surface-labeled and treated with (+) or without (−) GM-CSF (6 nmol/L) for 5 minutes and immunoprecipitation was performed with anti-GMRα MoAb (8G6). Immunoprecipitated proteins were separated on 7.5% SDS-PAGE under reducing conditions and the gel was exposed to phosphorimager. (C) Proteins immunoprecipitated from hGMRα/hβc -expressing Ba/F-3 cells with anti-GMRα MoAb (8G6) either in the presence (+) or absence (−) of GM-CSF were subjected to Western transfer and immunoblotted using anti-βc antibody 1C1 (upper panel) or antiphosphotyrosine antibody PY20 (lower panel).
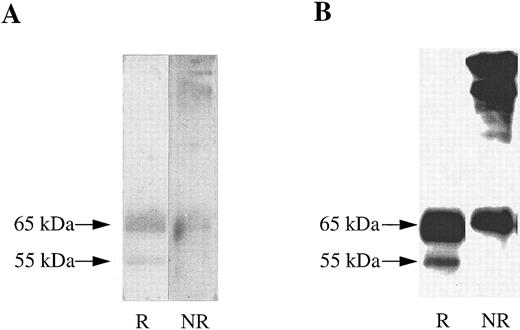
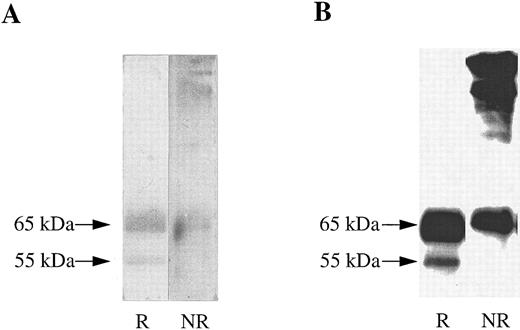
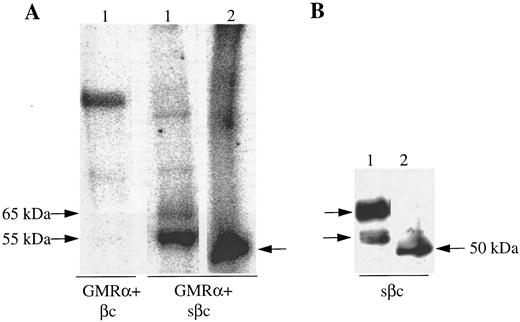
A soluble form of βc interacts with cell surface expressed GMRα. To determine whether the extracellular portions of GMRα and βc are sufficient for ligand-independent GMRα:βc interaction, we made a construct encoding a soluble form of βc (sβc ) comprising the entire extracellular domain but lacking the transmembrane and cytoplasmic regions and examined its ability to associate with GMRα. Initial characterization of sβc was performed by transfection into CHO cells and affinity purification of conditioned medium on an anti-βc antibody 3D7 coupled to CNBr-activated sepharose column. Two proteins of 55,000 and 65,000 MW were specifically eluted from the affinity column and visualized on a reducing SDS-PAGE gel by silver staining (Fig 3A). These two proteins were also detected after Western transfer by immunoblotting with anti-βc antibody (1C1; Fig 3B), implying that they represent two forms of sβc protein. Intriguingly, when the eluted sβc fractions were run on SDS-PAGE under nonreducing conditions, proteins of 120,000 MW and higher were seen by silver staining (Fig 3A) and also by anti-βc immunoblotting (Fig 3B), suggesting that the sβc forms disulphide-linked dimers and higher order complexes. A similar phenomenon was observed with a soluble form of the mouse IL-3–specific β chain, sAIC2A,36 and may relate to the ability of βc to spontaneously form dimers, as previously noted.16,18 37
Soluble βc protein was purified from conditioned medium from CHO transfectants. Soluble purified βc protein was run under reducing (R) or nonreducing (NR) conditions on 10% SDS-PAGE and either (A) silver stained or (B) subjected to Western transfer and immunoblotted with anti-βc antibody (1C1).
Soluble βc protein was purified from conditioned medium from CHO transfectants. Soluble purified βc protein was run under reducing (R) or nonreducing (NR) conditions on 10% SDS-PAGE and either (A) silver stained or (B) subjected to Western transfer and immunoblotted with anti-βc antibody (1C1).
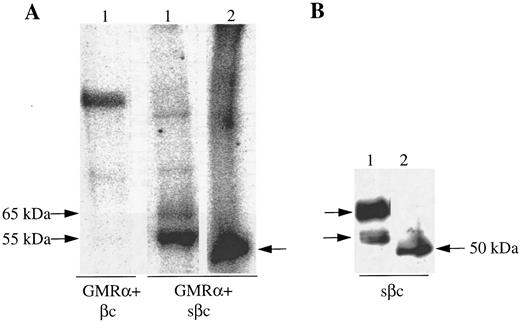
The association of sβc with GMRα was studied by transfecting the sβc construct into CHO cells expressing GMRα and monitoring sβc retention at the cell surface with anti-βc MoAb. Initial flow cytometric analysis showed specific binding of anti-βc MoAb on the surface of CHO cells coexpressing sβc and GMRα but not on CHO cells expressing sβc alone (data not shown). Importantly, the specific association of sβc with GMRα on the surface of CHO cells could be also demonstrated by coimmunoprecipitation experiments. In these experiments we also sought to establish that the retained βc reactivity detected on the GMRα-expressing CHO cells was indeed sβc and not another protein with a common epitope or a fusion protein produced by an anomalous transfection event. To examine surface expressed βc specifically and avoid involvement of βc from intracellular compartments, CHO cells expressing either full-length or soluble βc with or without GMRα were surface labeled with 125I and βc protein immunoprecipitated using an anti-βc antibody (8E4). A single 125I-labeled protein of 120,000 MW was immunoprecipitated from CHO cells expressing full-length βc (Fig 4A). Two 125I-labeled proteins of 55,000 and 65,000 MW were immunoprecipitated from CHO cells expressing GMRα and sβc (Fig 4A) that corresponded in size to the sβc proteins detected in cell supernatants (Fig 3). No labeled protein was immunoprecipitated from CHO cells expressing sβc alone (data not shown), indicating that the sβc retained on the surface of GMRα-expressing cells does not represent sβc protein in the process of secretion but is specifically retained by GMRα.
Soluble βc is retained on the surface of GMRα-expressing CHO cells. (A) CHO cells expressing GMRα and either full-length βc (βc ) or soluble βc (sβc ) were 125I–surface-labeled and immunoprecipitation was performed with anti-βc MoAb (8E4). The immunoprecipitated proteins were then either incubated with (2) or without (1) deglycosylating enzymes and subsequently separated on 7.5% SDS-PAGE under reducing conditions and visualized by phosphorimager. (B) Soluble βc was immunoprecipitated from the medium of CHO cells coexpressing GMRα and soluble βc (sβc ) and the immunoprecipitated proteins were either subjected to enzymatic deglycosylation (2) or not (1) and subsequently separated on 7.5% SDS-PAGE. Western transfer was then performed and immunoblotting with anti-βc antibody 1C1.
Soluble βc is retained on the surface of GMRα-expressing CHO cells. (A) CHO cells expressing GMRα and either full-length βc (βc ) or soluble βc (sβc ) were 125I–surface-labeled and immunoprecipitation was performed with anti-βc MoAb (8E4). The immunoprecipitated proteins were then either incubated with (2) or without (1) deglycosylating enzymes and subsequently separated on 7.5% SDS-PAGE under reducing conditions and visualized by phosphorimager. (B) Soluble βc was immunoprecipitated from the medium of CHO cells coexpressing GMRα and soluble βc (sβc ) and the immunoprecipitated proteins were either subjected to enzymatic deglycosylation (2) or not (1) and subsequently separated on 7.5% SDS-PAGE. Western transfer was then performed and immunoblotting with anti-βc antibody 1C1.
To investigate the nature of the sβc protein doublet detected on GMRα-expressing CHO cells, in vitro deglycosylation was performed on the immunoprecipitated protein before SDS-PAGE. The two 125I-labeled sβc proteins were both rendered to a 50,000 MW protein (Fig 4A). Similarly, the two 55,000 and 65,000 MW forms of sβc immunoprecipitated from conditioned medium were converted to a 50,000 MW protein after in vitro deglycosylation as seen by immunoblot using anti-βc antibody (1C1; Fig 4B). This shows that the 55,000 and 65,000 MW proteins represent differentially glycosylated forms of sβc , as has previously been observed with the full-length βc ,36 and that both forms are retained on GMRα-expressing cells.
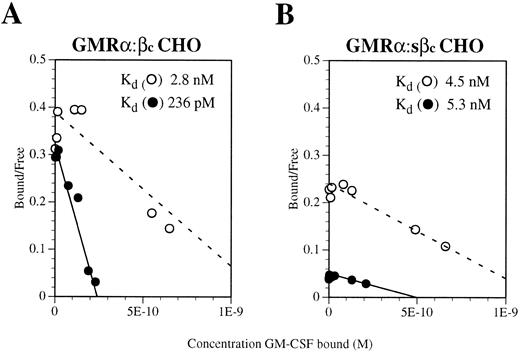
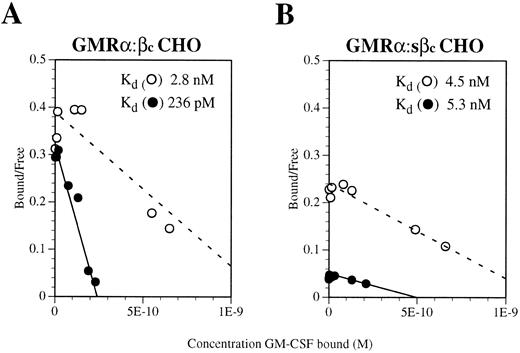
To determine whether the GMRα:sβc complex is able to bind GM-CSF with high affinity, saturation binding assays were performed on GMRα CHO cells coexpressing a similar amount of either sβc or full-length βc . Because of the very high level of GMRα chain expression on these transfectants (5 × 105 sites per cell, as determined by Scatchard analysis) no high-affinity sites could be detected directly from either transfectant (Fig 5A and B). To reduce this interference, dissociation of weakly bound radioligand was performed after binding, thereby removing ligand interacting with low-affinity receptors. Using this approach, high-affinity binding sites (kd 236 pmol/L) were detectable on GMRα cells coexpressing full-length βc (Fig 5A) but not on those coexpressing sβc (kd 5.3 nmol/L; Fig 5B). This finding implies that the sβc protein is unable to confer full high-affinity binding on the GM-CSF:GMRα complex, a function that may require a conformational change facilitated by the transmembrane and cytoplasmic regions of βc .
Scatchard transformation of saturation binding studies performed on CHO cells coexpressing (A) GMRα and full-length βc or (B) GMRα and soluble βc (sβc ). Binding assays were performed with 125I-labeled GM-CSF over a concentration range of 10 pmol/L to 10 nmol/L. After binding had reached equilibrium, the cells were washed briefly and either lysed directly (○) or subjected to five 15-minute washes to remove ligand bound with low affinity (•). The dashed line indicates the best fit for nondissociated binding and the solid line indicates the best fit for dissociated binding.
Scatchard transformation of saturation binding studies performed on CHO cells coexpressing (A) GMRα and full-length βc or (B) GMRα and soluble βc (sβc ). Binding assays were performed with 125I-labeled GM-CSF over a concentration range of 10 pmol/L to 10 nmol/L. After binding had reached equilibrium, the cells were washed briefly and either lysed directly (○) or subjected to five 15-minute washes to remove ligand bound with low affinity (•). The dashed line indicates the best fit for nondissociated binding and the solid line indicates the best fit for dissociated binding.
Soluble GMRα and βc can exist as a complex and bind GM-CSF. Based on our demonstration of a preformed complex between GMRα and βc on the cell surface and also the retention of sβc by cells expressing GMRα chain, we suspected that it may be possible to observe coassociation of a soluble form of GMRα and sβc in solution. To test this idea, we constructed a soluble carboxy-truncated form of GMRα that comprised only the extracellular portion of the receptor, termed sGMRα. By immunoprecipitation and immunoblotting using a GMRα chain specific antibody (8G6), we detected a 65,000 MW protein in the medium of CHO cells transfected with this construct, indicating that the soluble GMRα protein was expressed and was able to bind GM-CSF specifically with low affinity (kd 13.7 nmol/L; data not shown).
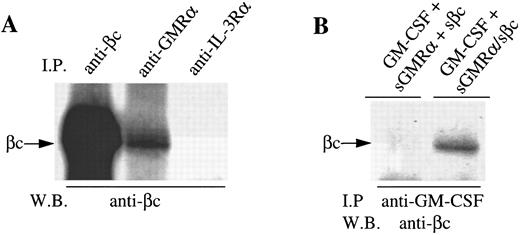
We then cotransfected the sGMRα construct together with the sβc encoding cDNA into CHO cells. Both soluble proteins were detectable by immunoprecipitation and Western blotting with appropriate antibodies in the cell medium of cotransfected cells (data not shown). Significantly, sβc protein was detected by immunoblot when immunoprecipitated not only with anti-βc (4F3) but also anti-GMRα antibody (8G6) but not an irrelevant antibody (9F5) (Fig 6A). This suggests that some but not all sGMRα is associated with sβc in solution. Immunoprecipitation of a mixture of conditioned medium from cells expressing sGMRα and sβc separately did not result in coimmunoprecipitation of the two chains (data not shown). This is consistent with the retention of sβc on GMRα-expressing CHO cells in that it appears that coexpression of the two soluble receptor chains is required for the association to occur.
Soluble forms of GMRα and βc spontaneously associate when coexpressed and bind GM-CSF. (A) Conditioned medium from CHO cells coexpressing soluble GMRα and soluble βc was immunoprecipitated using either anti-βc (4F3), anti-GMRα (8G6), or a control antibody (9F5); and the proteins were separated on 10% SDS-PAGE, Western transferred, and immunoblotted with anti-βc antibody 1C1. (B) Conditioned medium from CHO cells either coexpressing soluble GMRα and soluble βc (sGMRα/sβc ) or a mixture of conditioned medium from CHO cells expressing the soluble proteins separately (sGMRα + βc ) were incubated with GM-CSF and immunoprecipitation was performed with anti–GM-CSF antibody. Proteins were separated on 10% SDS-PAGE, Western transferred, and then immunoblotted with anti-βc antibody 1C1.
Soluble forms of GMRα and βc spontaneously associate when coexpressed and bind GM-CSF. (A) Conditioned medium from CHO cells coexpressing soluble GMRα and soluble βc was immunoprecipitated using either anti-βc (4F3), anti-GMRα (8G6), or a control antibody (9F5); and the proteins were separated on 10% SDS-PAGE, Western transferred, and immunoblotted with anti-βc antibody 1C1. (B) Conditioned medium from CHO cells either coexpressing soluble GMRα and soluble βc (sGMRα/sβc ) or a mixture of conditioned medium from CHO cells expressing the soluble proteins separately (sGMRα + βc ) were incubated with GM-CSF and immunoprecipitation was performed with anti–GM-CSF antibody. Proteins were separated on 10% SDS-PAGE, Western transferred, and then immunoblotted with anti-βc antibody 1C1.
To determine whether the sGMRα:sβc complex is capable of binding ligand, conditioned medium from cells expressing sGMRα and sβc was incubated with GM-CSF and subsequently immunoprecipitation was performed with anti–GM-CSF antibody. Immunoblotting of the precipitated material showed that sβc was associated with the anti–GM-CSF immunoprecipitated complex when conditioned medium from cells coexpressing the two receptor proteins was used, but not when conditioned medium from cells expressing the two chains separately was mixed (Fig 6B). This implies that the association of sβc with GM-CSF is dependent on its interaction with sGMRα in conditioned medium from cells coexpressing sGMRα and sβc .
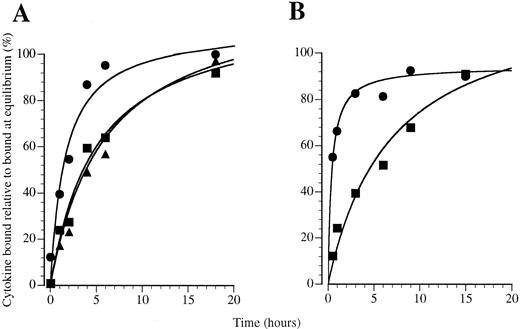
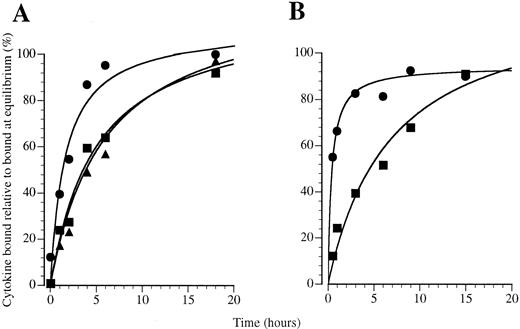
GM-CSF exhibits rapid receptor association compared with IL-3 and IL-5. To examine whether a preformed GM-CSF receptor complex may influence the kinetics of GM-CSF binding, we examined the kinetics of association of 125I–GM-CSF to primary human eosinophils and monocytes. We used these cells because they express IL-3 receptors and, in the case of eosinophils, IL-5 receptors as well as GM-CSF receptors, thus allowing a comparison between different receptor systems. The association of GM-CSF was compared with IL-3 and IL-5 on human eosinophils in binding studies performed at 4°C with 200 pmol/L 125I-labeled cytokine in which specific binding was determined over a 24-hour time course (Fig 7A). We found that GM-CSF binding approached equilibrium faster than IL-3 and IL-5 and that binding was detected at very early time points. Curve fitting analysis showed that a significantly improved fit was obtained for GM-CSF association when binding was resolved into two classes of binding site (Table 1): one site exhibiting a rapid approach to equilibrium about 1,000-fold faster than IL-3 or IL-5 and the other exhibiting similar apparent association kinetics to IL-3 and IL-5 (Table 1). Only a small proportion of the GM-CSF binding sites exhibit rapid binding kinetics, with the majority behaving like IL-3 and IL-5 receptors with a slower apparent association (Table 1). In previous studies, we have shown that eosinophils exhibit only high-affinity binding sites for GM-CSF, IL-3,38 and IL-5.31 From these studies it appears that the GM-CSF receptors exists in two pools that exhibit different kinetic properties.
Association kinetics of 125I-labeled cytokines binding to (A) eosinophils and (B and C) monocytes at 4°C with 200 pmol/L 125I-CSF: (•) GM-CSF, (▪) IL-3, and (▴) IL-5.
Association kinetics of 125I-labeled cytokines binding to (A) eosinophils and (B and C) monocytes at 4°C with 200 pmol/L 125I-CSF: (•) GM-CSF, (▪) IL-3, and (▴) IL-5.
On monocytes, as on eosinophils, the kinetics of GM-CSF binding were rapid and approached equilibrium faster than IL-3 binding (Fig 7B). We have previously shown that the approach to equilibrium by GM-CSF is approximately 10 times faster than IL-3.39 The rate of approach to equilibrium of IL-3 on monocytes is comparable to that seen for IL-3 and IL-5 and the slower binding site for GM-CSF on eosinophils, suggesting that association at these sites may involve similar mechanisms, whereas GM-CSF binding to the rapidly associating sites on eosinophils and monocytes is different.
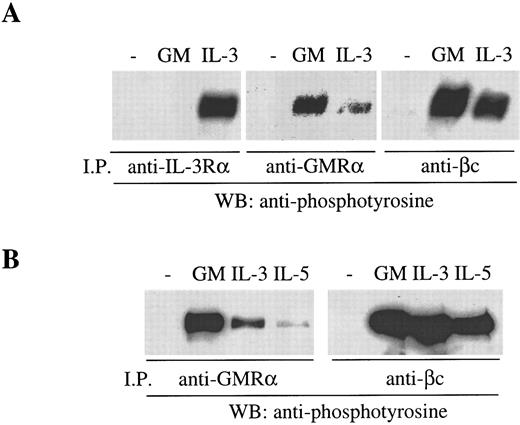
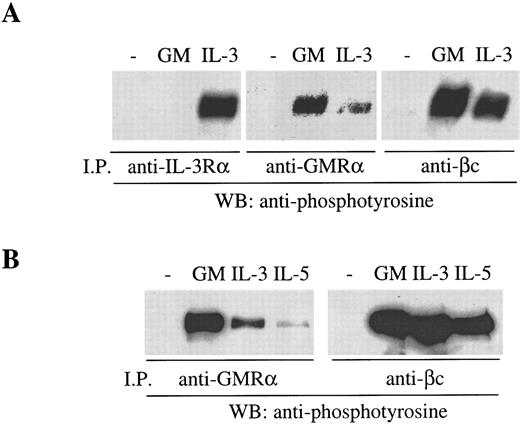
The preformed GMRα:βc can be phosphorylated in response to IL-3 and IL-5. The functional significance of the preformed GMRα:βc complex was examined by means of receptor activation studies. It is known that, in the course of activation of the GM-CSF, IL-3, and IL-5 receptors, βc becomes phosphorylated in response to ligand binding,10 40 a process that requires the ligand-specific α chain. We have examined the phosphorylation of βc induced by cytokines in Mo7e and TF-1.8 cells and have found that phosphorylated βc can be detected by antiphosphotyrosine (PY20) immunoblot after treatment with GM-CSF and immunoprecipitation with either anti-βc (8E4) or anti-GMRα (8G6) antibody (Fig 8A and B). Similarly, treating Mo7e cells with IL-3 also resulted in βc phosphorylation that was immunoprecipitable by either anti-βc (8E4) or anti–IL-3Rα antibody (9F5) (Fig 8A). However, strikingly, we found that anti-GMRα antibody also immunoprecipitated phosphorylated βc in cells treated with IL-3, indicating that GMRα is associated with the IL-3–induced receptor complex (Fig 8A). Similar results were obtained in TF-1.8 cells, with the addition that anti-GMRα antibody also immunoprecipitated βc phosphorylated in response to IL-5 (Fig 8B). However, treatment of TF-1.8 cells with erythropoietin did not result in βc phosphorylation (data not shown), indicating that βc phosphorylation is specific to GM-CSF, IL-3, and IL-5 and not a general activation event. The involvement of GMRα in the IL-3– and IL-5–induced receptor complexes is specific to GMRα and may be mediated by the preformed GMRα:βc complex. Thus, these findings raise the possibility that the preformed GMRα:βc complex can be recruited into an active receptor complex induced not only by GM-CSF but also by IL-3 or IL-5.
βc phosphorylated in response to GM-CSF, IL-3, or IL-5 is immunoprecipitable by anti-GMRα antibody. (A) Mo7e cells were starved overnight and then treated with either 6 nmol/L GM-CSF, IL-3, or medium for 5 minutes at 4°C and then immunoprecipitated with either anti–IL-3Rα (9F5), anti-GMRα (8G6), or anti-βc (8E4) antibody. (B) TF-1.8 cells were starved overnight and then treated with either 6 nmol/L GM-CSF, IL-3, or IL-5 or medium for 5 minutes at 4°C and then immunoprecipitated with either anti-GMRα (8G6) or anti-βc (8E4) antibody. Immunoprecipitated proteins were separated on 7.5% SDS-PAGE, Western transferred, and then immunoblotted with antiphosphotyrosine antibody (PY20).
βc phosphorylated in response to GM-CSF, IL-3, or IL-5 is immunoprecipitable by anti-GMRα antibody. (A) Mo7e cells were starved overnight and then treated with either 6 nmol/L GM-CSF, IL-3, or medium for 5 minutes at 4°C and then immunoprecipitated with either anti–IL-3Rα (9F5), anti-GMRα (8G6), or anti-βc (8E4) antibody. (B) TF-1.8 cells were starved overnight and then treated with either 6 nmol/L GM-CSF, IL-3, or IL-5 or medium for 5 minutes at 4°C and then immunoprecipitated with either anti-GMRα (8G6) or anti-βc (8E4) antibody. Immunoprecipitated proteins were separated on 7.5% SDS-PAGE, Western transferred, and then immunoblotted with antiphosphotyrosine antibody (PY20).
DISCUSSION
We show here the existence of a GMRα:βc complex that is formed in the absence of GM-CSF. We have observed this ligand-independent association between GMRα and βc with both cell surface expressed receptors in several cell lines and also with carboxy-truncated soluble forms of the receptor subunits. The number of preformed GMRα:βc complexes observed on cells varied from cell to cell. In some cases, all of the GMRα and βc chains were apparently coassociated and no further association was induced by GM-CSF treatment, whereas on other cells only a component of GMRαs and βcs were preassociated and further association was induced by GM-CSF treatment. This suggests that two pools of GM-CSF receptors exist: preformed complexes and ligand induced complexes.
The notion of two GM-CSF receptor pools is consistent with previous experiments showing that GM-CSF induces GMRα and βc association17 and reconciles this observation with that of Ronco et al,19 who suggested that the GM-CSF receptor may exist as a preformed complex. This possibility was raised by the inability of a mutant GMRα to bind GM-CSF unless it was coexpressed with βc . This was interpreted as βc preassociated with GMRα compensating for the loss of GM-CSF binding on the mutant GMRα. In an analogous manner, a GM-CSF helix D mutant showed no detectable binding to GMRα alone, yet could bind to cells expressing both GMRα and βc ,21 possibly reflecting the effect of a GMRα:βc preformed complex.
By using soluble receptor constructs, we were able to demonstrate the formation of sGMRα:sβc complexes in solution, indicating that the extracellular domains of the two proteins are sufficient to mediate the interaction. This in turn is dependent on the two soluble receptor chains being expressed by the same cell, because neither the addition of sβc to GMRα expressing cells nor combining separately expressed sGMRα and sβc resulted in complex formation. This suggests that the association between the two proteins occurs as the proteins reach the cell surface, possibly before or during transport to the cell surface. However, interestingly, the retention of sβc by cells expressing GMRα did not result in a detectable increase in affinity for GM-CSF, in contrast to full-length βc that confers high-affinity binding on the GM-CSF:GMRα complex. Under the dissociation conditions used it is possible that binding of intermediate affinity was lost and so we can only conclude that sβc is unable to confer full high-affinity binding on GMRα-expressing cells. This deficiency in binding with sβc may be due to the βc lacking transmembrane and extracellular portions. Our findings are consistent with recent studies in which a naturally occurring soluble form of GMRα was found to be retained on the cell surface when coexpressed with full-length βc on BHK cells.22 The soluble GMRα conferred GM-CSF binding on the cells albeit with intermediate affinity, indicating some deficit in the interaction with βc . These observations suggest that the transmembrane and cytoplasmic regions of these receptor subunits may be required for conformational changes and optimal high-affinity binding. Alternatively, these associations observed with soluble forms of the receptor may not represent normal receptor interactions.
The precise regions in the extracellular domains of GMRα and βc that mediate their spontaneous association in the cell membrane and in solution are not known. From modelling studies and comparison with the growth hormone crystal structure,41 the A-B loop and the E strand in the fourth domain of βc appear to be good candidates for interaction with the second domain of the cytokine receptor module of GMRα. It is worth noting that insertions, deletions, and point mutations in this domain of βc lead to factor-independent activation.42 It is possible that various perturbations of an already preformed complex may result in receptor activation. In our hands we did not observe receptor activation, as measured by antiphosphotyrosine reactivity of the performed complex (Fig 2C). However, it would be interesting to examine this possibility with βc mutants and indeed in human leukemias.
In seeking to determine the functional significance of the preformed GMRα:βc complex, we performed kinetic analysis for GM-CSF association. Using normal cells expressing GM-CSF receptor, we found that the association of GM-CSF to both eosinophils and monocytes is more rapid relative to IL-3 and IL-5 and, in the case of eosinophils, is bimodal. In previous studies we have shown that eosinophils exhibit only high-affinity binding sites for GM-CSF, IL-3,38 and IL-5.31 This suggests that there are sufficient βcs to support full-affinity conversion of GM-CSF receptors and that the receptors exist in two forms: one form approaches equilibrium very rapidly and a second form binds with similar kinetics to IL-3 and IL-5. This is consistent with the presence of two pools of receptor for GM-CSF: a small number of receptors that bind GM-CSF rapidly, possibly representing preformed complexes as described here, and a larger pool, possibly composed of free GMRαs and βcs that exhibit slower association on GM-CSF binding akin to IL-3 and IL-5 binding. We have previously reported that GM-CSF binds more rapidly to monocytes39 and induces their adhesion faster than IL-3.43 The presence of preformed GMRα:βc complexes may also account for these kinetic differences on monocytes by providing a pool of preformed receptors that rapidly associate with GM-CSF.
The binding cross-competition exhibited between GM-CSF, IL-3, and IL-5 has previously been described on normal26,38,44 and leukemia cells.45,46 The molecular basis of this phenomenon is the competition between GM-CSF:GMRα, IL-3:IL-3Rα, and IL-5:IL-5Rα for βc interaction. The proposed preformed GMRα:βc complex might be expected to have an effect on this phenomenon, sequestering βc for the exclusive binding of GM-CSF. However, cross-competition experiments performed previously on eosinophils38 and CML cells35 show that IL-3 is able to compete for 125I–GM-CSF binding effectively, with up to 90% competition. This suggests that the βc associated with GMRα in the preformed complex is in equilibrium with free βc and is therefore competable by IL-3. This may also explain the relative numbers of preformed complexes observed on cells in that the level of preformed complex would be dependent on the relative level of expression of βc . Thus, cells that express excess βc and thus exhibit high-affinity binding sites only may have relatively more preformed sites compared with cells that express limiting amounts of βc .
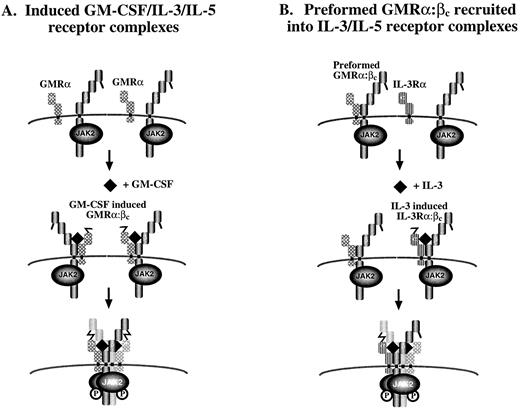
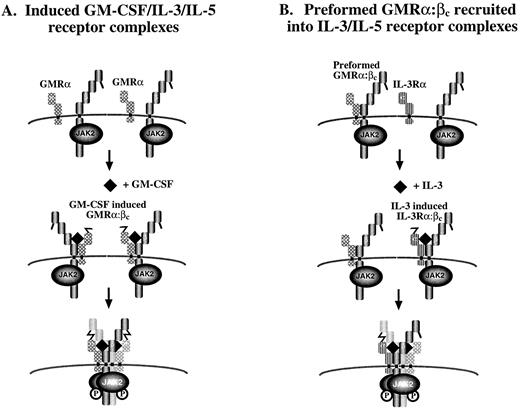
The stoichiometry of the active GM-CSF receptor is not known and may involve a GMRα:βc ratio of 1:1 or a 2:2 complex. Because of the disulphide-linked GMRα:βc heterodimer and molecular modelling of the extracellular region of βc , we favor the second possibility.18 This is also consistent with the observations that at least two molecules of GMRα are required for receptor activation47 and that phosphorylation of βc dimers36 and disulphide-linked βc containing complexes18 occurs in response to GM-CSF. In this model, binding of ligand may render cysteine residues in GMRα and βc reactive, leading to disulphide bond formation across two receptors in a hexameric configuration, thus bringing together two βc -associated JAK-2 molecules, thereby inducing receptor activation (Fig 9A). Given the observation of the preformed GMRα:βc , we speculate that it could be recruited into an IL-3 or IL-5 receptor complex (Fig 9B). Consistent with this possibility, we found that anti-GMRα antibodies immunoprecipitated phosphorylated βc when cells were stimulated not only with GM-CSF but also with IL-3 and IL-5. In contrast, GM-CSF did not induce phosphorylation of βc associated with IL-3Rα (Fig 8A), consistent with IL-3 receptor existing only as a fully inducible receptor.
Proposed models for assembly of (A) GM-CSF–, IL-3–, and IL-5–induced receptor complexes and (B) preformed GM-CSF receptor complexes into activated receptors. In (A), GMRα, IL-3Rα, or IL-5Rα are in close proximity to (although not associated with) βc on the cell surface. Ligand binding to the appropriate a chain induces α:βc heterodimerization and a conformational change in a chain that allows its disulphide linkage to βc . Modelling of βc suggests that this bridging would only be possible if the unpaired cysteines in the α chain of receptor 1 formed a disulphide bridging with cysteine of βc in receptor 2.18 The bringing together of two βc with their associated JAK-2 molecules would then lead to receptor activation. In (B), it is postulated that the binding of IL-3 or IL-5 to their specific α chain and βc triggers a conformational change in the α subunit analogous to model (A). However, in this case, a disulphide bridge can be formed between the free cysteine in the IL-3Rα or IL-5Rα and a cysteine in βc that is already noncovalently associated with GMRα chain in a preformed complex. This interaction may be sufficient to bring together two βc and two JAK-2 kinases leading to receptor activation. This model raises the possibility that some of the functions mediated by IL-3 and IL-5 are mediated inducibly through the activation of a preformed GMRα:βc complex.
Proposed models for assembly of (A) GM-CSF–, IL-3–, and IL-5–induced receptor complexes and (B) preformed GM-CSF receptor complexes into activated receptors. In (A), GMRα, IL-3Rα, or IL-5Rα are in close proximity to (although not associated with) βc on the cell surface. Ligand binding to the appropriate a chain induces α:βc heterodimerization and a conformational change in a chain that allows its disulphide linkage to βc . Modelling of βc suggests that this bridging would only be possible if the unpaired cysteines in the α chain of receptor 1 formed a disulphide bridging with cysteine of βc in receptor 2.18 The bringing together of two βc with their associated JAK-2 molecules would then lead to receptor activation. In (B), it is postulated that the binding of IL-3 or IL-5 to their specific α chain and βc triggers a conformational change in the α subunit analogous to model (A). However, in this case, a disulphide bridge can be formed between the free cysteine in the IL-3Rα or IL-5Rα and a cysteine in βc that is already noncovalently associated with GMRα chain in a preformed complex. This interaction may be sufficient to bring together two βc and two JAK-2 kinases leading to receptor activation. This model raises the possibility that some of the functions mediated by IL-3 and IL-5 are mediated inducibly through the activation of a preformed GMRα:βc complex.
The unidirectional activation of the GM-CSF receptor by IL-3 is reminiscent of trans-downmodulation experiments in the mouse in which IL-3 was found to trans-downmodulate GM-CSF, macrophage colony-stimulating factor (M-CSF ), and granulocyte colony-stimulating factor (G-CSF ) receptors, but GM-CSF or G-CSF were unable to trans-downmodulate the mouse IL-3 receptor.48 49 The transphosphorylation of GMRα-associated βc we observe appears to be limited to the GM-CSF/IL-3/IL-5 receptor system in that erythropoietin is ineffective and is probably a reflection of the unique mode of assembly of this heterodimeric receptor family. GM-CSF receptors are expressed by many cells of the hematopoietic lineage and, intriguingly, most cells that express either IL-3 or IL-5 receptors also express GM-CSF receptors. The data presented here suggest that IL-3 and IL-5 are able to activate preformed GM-CSF receptors, thus raising the possibility that the biologic functions of IL-3 and IL-5 are mediated in part by signalling through the GM-CSF receptor. A further possibility is that the GM-CSF preformed complex may act to potentiate the effects of IL-3, IL-5, and GM-CSF by reducing the need for multiple ligand-induced heterodimerization events. A single receptor oligomerization event (ie, hexameric complex formation) in the absence of preformed complexes would require the formation of two ligand-induced receptor heterodimers. However, the presence of preformed complexes may theoretically reduce the number of ligand-induced receptor heterodimers required to produce a functional signal. These possibilities are currently being investigated.
ACKNOWLEDGMENT
The authors acknowledge Mara Dottore for her excellent technical assistance and Dr Tom Gonda for critically reading the manuscript.
Supported by grants from the National Health and Research Council of Australia. C.J.B. is a Rotary Peter Nelson Leukaemia Research Fellow of the Anti-Cancer Foundation of the Universities of South Australia.
Address reprint requests to Angel F. Lopez, MD, PhD, Division of Human Immunology, Hanson Centre for Cancer Research, Institute of Medical and Veterinary Science, Box 14 Rundle Mall Post Office, Adelaide, South Australia 5000.