Abstract
Fanconi anemia (FA) is an autosomal recessive cancer susceptibility syndrome. The phenotype includes developmental defects, bone marrow failure, and cell cycle abnormalities. At least eight complementation groups (A-H) exist, and although three of the corresponding complementation group genes have been cloned, they lack recognizable motifs, and their functions are unknown. We have isolated a binding partner for the Fanconi anemia group C protein (FANCC) by yeast two-hybrid screening. We show that the novel gene, FAZF, encodes a 486 amino acid protein containing a conserved amino terminal BTB/POZ protein interaction domain and three C-terminal Krüppel-like zinc fingers. FAZF is homologous to the promyelocytic leukemia zinc finger (PLZF) protein, which has been shown to act as a transcriptional repressor by recruitment of nuclear corepressors (N-CoR, Sin3, and HDAC1 complex). Consistent with a role in FA, BTB/POZ-containing proteins have been implicated in oncogenesis, limb morphogenesis, hematopoiesis, and proliferation. We show that FAZF is a transcriptional repressor that is able to bind to the same DNA target sequences as PLZF. Our data suggest that the FAZF/FANCC interaction maps to a region of FANCC deleted in FA patients with a severe disease phenotype. We also show that FAZF and wild-type FANCC can colocalize in nuclear foci, whereas a patient-derived mutant FANCC that is compromised for nuclear localization cannot. These results suggest that the function of FANCC may be linked to a transcriptional repression pathway involved in chromatin remodeling.
FANCONI ANEMIA (FA) is an autosomal recessive disorder characterized by progressive pancytopenia, diverse congenital anomalies, and predisposition to cancer, particularly acute myeloid leukemia (AML) and squamous cell carcinoma.1-5 The diagnostic hallmark of FA cells is a unique hypersensitivity to DNA crosslinking agents such as mitomycin C (MMC) and diepoxybutane (DEB). In addition, FA cells have an abnormal cell cycle profile, described as an elongation or arrest at the G2 phase, and this abnormality is exacerbated by treatment with MMC.6,7 Because of associated genomic instability and cancer predisposition, FA has been classified along with xeroderma pigmentosum, Cockayne’s syndrome, trichothiodystrophy, ataxia telangiectasia, Bloom syndrome, Werner’s syndrome, and hereditary nonpolyposis colorectal cancer as one of the “caretaker gene diseases.”8 However, the basic defect in FA is unknown.
FA is genetically heterogeneous, comprising at least eight different complementation groups (A-H) identified by somatic cell hybrid analysis.9,10 Genes mutated in three of the FA complementation groups have been identified (FANCA,FANCC, and FANCG).11-13 In addition,FANCD has been mapped to chromosome 3p22-26,14 andFANCE has recently been mapped to chromosome 6p21-22.15 The proteins predicted to be encoded by the known FA genes lack recognizable motifs, and their functions are unknown. Because FA patients from different complementation groups have similar clinical and cellular phenotypes, a common molecular pathway in which the FA gene products participate has been hypothesized.
Although FA appears to have the characteristics of a DNA repair disorder, early work with the first known gene product, Fanconi anemia group C protein (FANCC), indicated that FANCC was located primarily in the cytoplasm, suggesting an indirect role in DNA repair.16,17 However, FANCC was subsequently also detected in the nuclear compartment, confirming that at least some FANCC is available in the nucleus for DNA-related activities.18,19Attempts to investigate even these most basic issues regarding subcellular localization of FANCC have been confounded by low endogenous expression levels, changes in expression during the cell cycle, and presence in both nuclear and cytoplasmic compartments.16-20
It is likely that the activities of FA proteins are regulated or mediated by other cellular proteins. In support of this notion, evidence based on coimmunoprecipitation and in vitro binding experiments suggest that FANCC binds to several cytosolic proteins, including the molecular chaperone GRP94, nicotinamide adenine dinucleotide phosphate (NADPH) cytochrome P450 reductase, the cyclin dependent kinase cdc-2, and Fanconi anemia complementation group A (FANCA).18,20-22 Indeed, there is growing awareness that the FA proteins may interact with each other within a large protein complex.18,23 24
We report here that FANCC interacts with a new human BTB/POZ (for Broad Complex, tramtrack, and Bric á Brac/pox virus and zinc finger) domain protein (for reviews see Albagli et al25 and Bardwell et al26), which we have named FAZF. The FAZF protein (for Fanconi anemia zincfinger) is similar to the promyelocytic zinc finger protein, PLZF, a Krüppel-like transcription factor involved chromosomal translocations with the retinoic acid receptor alpha (RARα) leading to acute promyelocytic leukemia (APL; for recent review, see Melnick et al27). In hematopoetic cells, PLZF is localized in nuclear speckles, which become delocalized in APL. PLZF represses transcription of specific targets by recruitment of histone deacetylase through the SMRT-mSin3-HDAC corepressor complex.28-32 In mammalian cells, FAZF is also located in nuclear speckles, which colocalize with wild-type FANCC, but not an inactivated FANCC protein. In addition, we show that FAZF is a transcriptional repressor and it readily forms heterodimers with PLZF. BTB/POZ proteins are implicated in oncogenesis,29,33-37hematopoiesis,38-40 and limb development,41-43suggesting that FANCC/FAZF interaction and the implied connection to transcriptional repression may be involved in the FA pathway.
MATERIALS AND METHODS
Plasmids, cell lines, antibodies, and chemicals.
The bacterial expression plasmids for GST-FAN2 and GST-FAC1 have been described.19 To make the two-hybrid bait expressing FANCC amino acid #1-168 (pAS-FAN), polymerase chain reaction (PCR) primers (159-192 [5′-TTCGCTTTTTCCACCATGGCTCAAGATTCAGTAG-3′] and 660-704 [5′-GGGAGCCATTCGCCTTGGATCCTTCTATCCATTAAGATGATTCT-3′]) were used to amplify the FANCC sequence from a full-length cDNA contained in pLFACXN.19 The primers incorporatedNcoI and BamHI restriction sites in the amplified fragment for use in ligation. For p2HA-FANCC, full-length FANCC was obtained by PCR from pLFACXN with primers FAC5′ BamHI (5′-CGGGATCCGATGGCTCAAGATTCAGTAGATCTTTCT-3′) and FACW3′ BamHI (5′-CCGGATCCTAGACTTGAGTTCGCAGCTCTTTAAGGA-3′) and ligated into BamHI-digested/calf intestinal phosphatase (CIP)-treated pCEPHA2.RIG.G, a plasmid that encodes two tandem hemagglutinin (HA) epitopes (kindly provided by Dr Hans Joenje, Free University, Amsterdam, The Netherlands). 2HA-L554P was constructed similarly except that primer FACM3′ BamHI (5′-CCGGATCCTAGACTTGAGTTCGCGGCTCTTTAAGGA-3′) was used instead of FACW3 BamHI. For construction of the expression vector encoding the epitope-tagged pFlag-FAZF, total RNA was extracted from normal human peripheral blood, and primers FAZF5 BGL2ATC (5′-TACCCAAGCCAAGGCAAGATCTCAATGTCCCTGCCCCCCAT-3′) and FAZF3 BGL2 (5′-GCTACCGACACCCCGTAGATCTCAGGTGGTGGAGGAAGAA-3′) were used to obtain the cDNA by reverse transcription (RT)-PCR. The amplified fragment was digested withBglII, and ligated into BamHI-digested/CIP-treated vector pCEP4-Flag (kindly provided by Dr Naumovski, Stanford University, Stanford, CA). All PCR-derived sequences and ligation joints were verified by DNA dideoxy sequencing using an ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin Elmer, Foster City, CA). Anti-HA was obtained from Roche Molecular Biochemicals (Indianapolis, IN). The monoclonal antibody (MoAb) specific for FANCC was produced by standard methods against the keyhole limpet hemacyanin-conjugated peptide C-ARELLKELRTQV, corresponding to FANCC aa #547-558.44 A detailed description of the antibody will be published elsewhere (M.E.H., in preparation). All chemicals were obtained from Sigma (St Louis, MO), unless otherwise indicated.
Two-hybrid library screen.
The two-hybrid screen was performed essentially as described previously.45 Briefly, Saccharomyces cerevisiae Y190 was transformed with the pAS-FAN bait (binding domain [BD] fusion) plasmid and selected for Trp prototrophy. After ensuring that the resulting strain expressed the FANCC bait and did not autoactivate the GAL4 reporter, it was transformed with a human B lymphocyte library encoding the activation domain (AD)–library hybrids. Approximately 300,000 His+ primary clones were obtained, and 59 of these were positive for β-galactosidase (gal) activity. Plasmid DNA prepared from several of the positive strains was transformed into Escherichia coli strain HB101, and transformants were selected. Candidate plasmids that did not repeat with the original bait, that autoactivated the reporter, or that interacted with unrelated baits were eliminated. The remaining interactors were further analyzed. A second screen was also performed with the same library using a BD expression vector encoding FANCC aa #116-558. For confirmation of two-hybrid interactions of PLZF and FAZF, the PJ69-4A strain of S cerevisiae46 was transformed with BD fusion constructs encoding full-length FAZF, PLZF, or the PLZF POZ domain. The transformed strains were selected on media lacking leucine, tryptophan, and adenine. Yeast colonies were counted and then selected in duplicate for liquid β-galactosidase assays as directed (Clontech, Palo Alto, CA). Results were normalized relative to the dimerization of PLZF. A full-length GAL4 plasmid was used as a positive control (Clontech). Other two-hybrid positive and negative control AD and BD domain plasmids were purchased from Clontech.
In vitro protein binding assays.
Expression and purification of Glutathione S-transferase (GST)-fusion proteins used in these assays have been described previously.19 Radiolabeled protein encoded by the library plasmid pAct8-1 was made by amplifying the insert using T7-promoter containing primers specific for flanking sequences in the pACT vector (Clontech). In vitro translations were performed using a commercial transcription-translation system (TnT, Promega, Madison, WI). Equal amounts of radiolabeled protein were added to GST fusion proteins immobilized on glutathione-Sepharose beads (GST, GST-FAC2, and GST-FAC1) and allowed to bind for 4 hours at 4°C in binding buffer (10 mmol/L Tris HCl/150 mmol/L NaCl/1 mmol/L EDTA/1% Nonidet P-40/1% deoxycholic acid). After extensive washing with the buffer, the beads were boiled in sample buffer and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
Northern blotting.
A multiple tissue blot of human poly (A+) RNA (Clontech) was probed with a radiolabeled 525-bp (BamHI/XhoI) restriction fragment from the library plasmid pAct8-1.
Immunoprecipitation and immunoblotting.
Cells were washed twice with phosphate-buffered saline (PBS), and cell lysates were solubilized in lysis buffer 1 (20 mmol/L Tris-Cl [pH 7.5], 150 mmol/L NaCl, 1% Triton X-100, 10 mmol/L EDTA, 1% deoxycholate, 1.5% aprotinin, 1 mmol/L phenylmethylsulfonyl fluoride [PMSF]). Anti-Flag M2-affinity gel (Sigma) or anti-HA (Roche Molecular Biochemicals) followed by Protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) was added as indicated in the text, and the sample was rocked overnight at 4°C. The immune complexes were sedimented by centrifugation, washed in lysis buffer, mixed with Laemmli sample buffer, boiled, and separated by SDS-PAGE. Proteins were electroblotted onto Bio-Blot nitrocellulose (Costar, Cambridge, MA) and probed with anti-HA, anti-Flag M2 (Sigma), or anti-FANCC MoAb 3A11 as previously described.19 For the PLZF-FAZF interaction, cells were lysed in lysis buffer 2 (1% NP40, 50 mmol/L NaCl, 50 mmol/L Tris pH 8.0, 1 mmol/L MgCl2, 10 mmol/L ZnCl2, 4% glycerol and Complet protease inhibitors [Roche Molecular Biochemicals]). Immune complexes were obtained essentially as described above using anti-Flag MoAb (Kodak, Rochester, NY) and Protein A agarose (Roche Molecular Biochemicals). The electrophoresed proteins were transferred to Immobilon PVDF membrane (Millipore, Bedford, MA) and were processed as described above using anti-PLZF polyclonal antibody.47
Transient transfection and immunofluorescence microscopy.
293 Epstein-Barr nuclear antigen (EBNA) cells (Invitrogen, Carlsbad, CA) were grown on chamber slides and transfected using lipofectamine as directed by the manufacturer (GIBCO, Grand Island, NY). Plasmids encoding epitope-tagged FAZF, FANCC, L554P, or parental vectors as negative controls for antibody specificity were used as indicated in text. Cells were processed as described previously19 using monoclonal anti-HA 12CA5 (Roche Molecular Biochemicals) and/or rabbit polyclonal anti-Flag (Zymed, South San Francisco, CA) and Oregon Green–conjugated goat antimouse and/or Texas Red–conjugated goat antirabbit (Molecular Probes, Eugene, OR) as secondary antibodies. Double-labeled images were acquired with a Bio-Rad MRC 1024 ES laser scanning confocal imaging system (Bio-Rad Laboratories, Richmond, CA) attached to an inverted Nikon Eclipse TE300 microscope (Nikon, Melville, NY). The acquisition system (LaserSharp, Hercules, CA) uses a krypton/argon laser with excitation lines at 488 and 564 nm, and sequential detection using three 8-bit photomultiplier tubes (PMTs). Single-labeled images were acquired with a Leica 900 confocal laser-scanning microscope (Leica Inc, Deerfield, IL) equipped with a krypton/argon detector. Collected images were imported into Adobe Photoshop 4.0 (Adobe, San Jose, CA) and overlapped to produce merged images. Localization study data (see Table 2) was gathered with a Leitz Orthoplan 2 fluorescence microscope (Ernst Leitz GMBH, Wetzlar, Germany) using green (488 nm) or red (568 nm) filters and a ×100 oil lens. One hundred cells expressing the protein of interest were counted for each cell line. Cells were scored as positive if they contained two or more nuclear foci. Standard error was computed using StatView 5.0 (SAS Institute, Cary, NC).48
Electrophorectic mobility shift assay (EMSA).
293T (ATCC No. CRL1573) cells were transfected with 20 μg of DNA using the Ca PO4 method.49 Nuclear extracts were prepared 48 hours after transfection, frozen in liquid nitrogen, and stored at −70°C.50 Synthetic duplexes (Table 1) were end-labeled using the large Klenow fragment of E coli DNA polymerase and [α-P32] dCTP (3,000 Ci/mmol), and purified by spin column or PAGE. Each binding reaction contained approximately 2 mg nuclear extract protein in 20 mmol/L HEPES pH 7.5, 1 mmol/L MgCl2, 10 mmol/L ZnCl2, 4% glycerol, 100 mg/mL bovine serum albumin (BSA), and 1 mg dIdC, and was incubated on ice for 30 minutes. Unlabeled oligonucleotide competitors or anti-Flag antibody (Kodak) was added at least 20 minutes before the addition of 10 femtomoles of labeled duplex as indicated in the text. After a further incubation of 20 minutes, protein-DNA complexes were separated by electrophoresis on a 4% nondenaturing polyacrylamide (30:1, acrylamide:bis-acrylamide) gel.
Transcriptional repression assays.
Plasmids encoding PLZF in the pSG5 expression vector were previously described.51 The Flag-epitope tagged version of FAZF was constructed in the pSG5 vector by standard methods. Four copies of a PLZF binding site found within the interleukin (IL)-3 receptor promoter (5′-TCGAGGATGACTGCGAGTACAGTTGCAACGG-3′)51a were cloned 5′ of the thymidine kinase promoter driving transcription of a luciferase reporter gene (IL-3R-Luc). For transcription assays, 2 × 105 293T cells per well of a 12-well dish were transfected with 1 μg of DNA, or as indicated in text, and 5 μL of Superfect (Qiagen, Valencia, CA). Luciferase levels were measured 48 hours after transfection using a Dual Luciferase kit (Promega).
RESULTS
Identification of a FANCC-interacting protein by yeast two-hybrid screening.
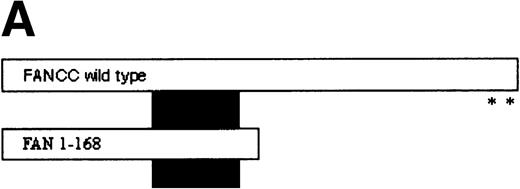
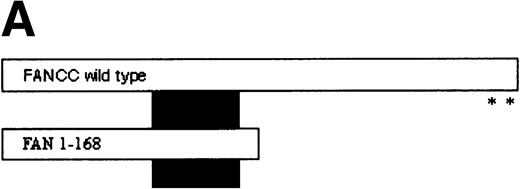
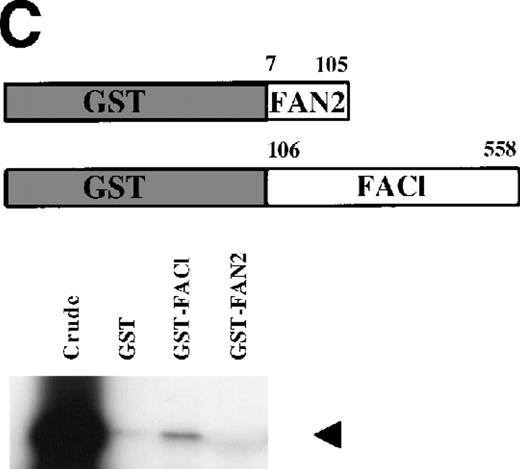
FANCC has been shown to bind to other cellular proteins and may be part of a large intracellular complex.18,20-22,24,52 We used a yeast two-hybrid assay to screen for proteins that bind to FANCC in vivo in an effort to identify binding partners that might shed light on the function of the FA pathway.45 The bait was designed (Fig 1A) to contain a region of FANCC deleted in patients with a severe phenotype (IVS4-4A-T), reasoning that this may be an area for functionally important protein-protein contact.14,53 Related strategies to compare strains expressing wild-type and mutant proteins (eg, full-length FANCC baits, and the inactive L554P protein) were unsuccessful because the strains were unstable and could not be used for screening (data not shown). The bait strain containing N-terminal sequences (FAN, aa #1-168) was used to screen a human B-cell library.45 Transformants were selected that were HIS+, and β-gal+, that did not autoactivate the reporter, interacted with the FAN bait on retesting, and did not have reporter activity with unrelated baits. Figure 1B shows the two-hybrid interaction of a representative interacting clone encoded by library plasmid pACT8-1. Southern blot analysis of candidates from the screen using an internal fragment from clone 8-1 indicated that at least three clones of different sizes contained hybridizing sequences, suggesting that the same candidate had been selected in the screen several times. Additional screening performed with a FANCC bait containing aa #116-558 also resulted in capturing sequences contained in clone 8-1, although this bait proved to be too unstable for further analysis (data not shown). DNA sequence analysis of the hybridizing clones indicated that the clones contained overlapping regions of the same sequence (data not shown).
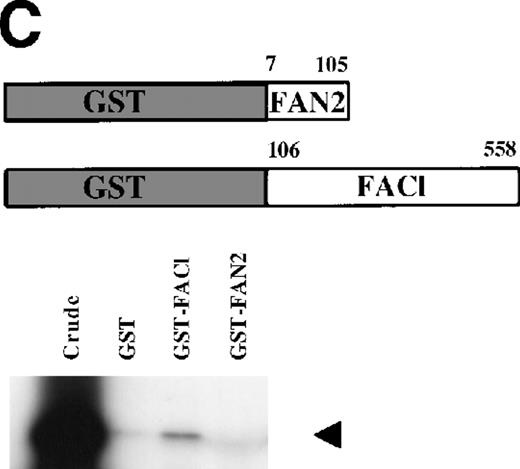
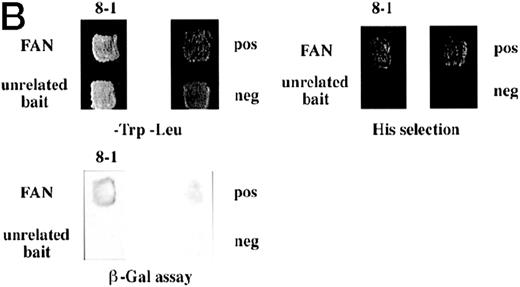
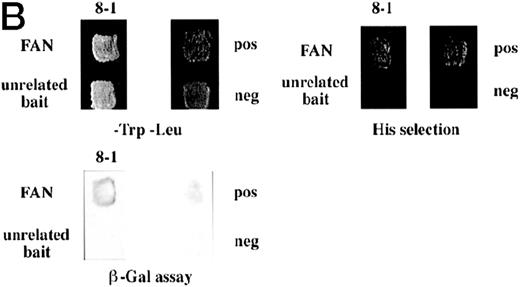
Identification of a FANCC-interacting protein by two-hybrid screening. (A) FANCC two-hybrid bait. The upper open rectangle represents wild-type FANCC. Asterisks indicate location of the R548X and L554P inactivating mutations.11,53,82 The lower open rectangle represents the region of FANCC expressed as a two-hybrid bait. The numbers indicate the corresponding amino acid residue of FANCC (the binding domain of the two-hybrid fusion is not shown for simplicity). The overlap with wild-type is indicated by the arrangement of the boxes in which the shaded area corresponds to the deletion in the IVS4 4A-T inactivating mutation that leads to a severe disease phenotype.14 53 (B) (see page 3740) Two-hybrid interaction. The Y190 yeast strain was cotransformed with the FAN bait plasmid and a representative prey plasmid (8-1) isolated by two-hybrid screening. Controls are shown for strains containing the prey plasmid 8-1 and an unrelated bait, and strains known to be positive and negative for interaction. The cotransformed strains are shown growing on media selective for both bait and prey plasmids (-Trp, -Leu), and under HIS selection. The lower panel shows results of the β-gal assay. (C) The protein encoded by clone 8-1 interacts with an amino terminal region of FANCC in vitro. Radiolabeled protein made by coupled in vitro transcription and translation from 8-1 cDNA bound specifically to immobilized GST-FANC1, but not to immobilized GST or GST-FAN2.
Identification of a FANCC-interacting protein by two-hybrid screening. (A) FANCC two-hybrid bait. The upper open rectangle represents wild-type FANCC. Asterisks indicate location of the R548X and L554P inactivating mutations.11,53,82 The lower open rectangle represents the region of FANCC expressed as a two-hybrid bait. The numbers indicate the corresponding amino acid residue of FANCC (the binding domain of the two-hybrid fusion is not shown for simplicity). The overlap with wild-type is indicated by the arrangement of the boxes in which the shaded area corresponds to the deletion in the IVS4 4A-T inactivating mutation that leads to a severe disease phenotype.14 53 (B) (see page 3740) Two-hybrid interaction. The Y190 yeast strain was cotransformed with the FAN bait plasmid and a representative prey plasmid (8-1) isolated by two-hybrid screening. Controls are shown for strains containing the prey plasmid 8-1 and an unrelated bait, and strains known to be positive and negative for interaction. The cotransformed strains are shown growing on media selective for both bait and prey plasmids (-Trp, -Leu), and under HIS selection. The lower panel shows results of the β-gal assay. (C) The protein encoded by clone 8-1 interacts with an amino terminal region of FANCC in vitro. Radiolabeled protein made by coupled in vitro transcription and translation from 8-1 cDNA bound specifically to immobilized GST-FANC1, but not to immobilized GST or GST-FAN2.
To confirm the in vivo interactions we found in yeast, we tested a radiolabeled in vitro translated protein encoded by clone 8-1 for binding to bacterially expressed GST-FANCC fusion proteins immobilized on glutathione Sepharose. In these protein affinity experiments, radiolabeled 8-1 protein bound to immobilized GST-FAC1 (aa #106-558), but not to GST-FAN2 (aa #7-106) or to GST alone (Fig 1C), indicating that the binding between the 8-1 protein and FANCC is direct and specific for the amino terminal region of FANCC. Because the FAN yeast bait contains aa #1-168, the in vitro and in vivo binding data suggest that the region of FANCC participating in the interaction is approximately between aa #106-168. Thus, binding between protein encoded by clone 8-1 and FANCC corresponds roughly to the deletion beginning at aa #116, resulting from the IVS4-4A-T mutation.54
Characterization of FANCC interacting clone 8-1.
PCR primers were used to amplify the inserts from the library plasmid pAct8-1. DNA sequence analysis revealed an open reading frame of 300 amino acids in-frame with two-hybrid activation domain sequences. A BLASTX database search (http://www.ncbi.nlm.nih.gov/BLAST/)55 56 with the 8-1 sequence indicated similarities to multiple zinc finger–containing proteins, and near identity with sequences contained within exons of a genomic clone from human chromosome 19q13.1 (cosmid F24109, GenBank accession No. AD000671). Thus, FAZF does not map to the location of a mapped FA gene. This cosmid clone contained additional 5′ sequences, suggesting that clone 8-1 was not complete. Therefore, the 1,461-bp full-length FAZF was cloned by RT-PCR using total RNA from normal human peripheral blood. Recently, another group has also deposited sequences in agreement with our data (accession no. AF130255, unpublished).
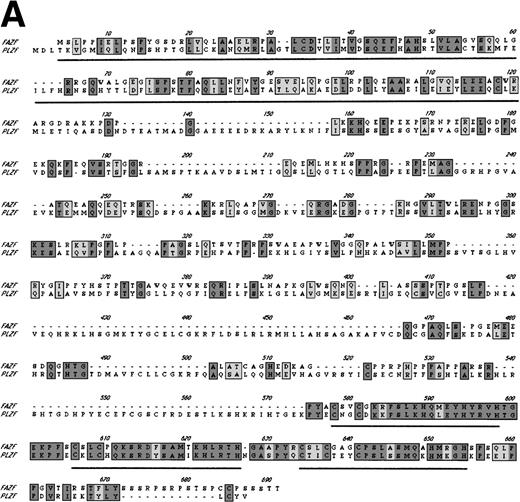
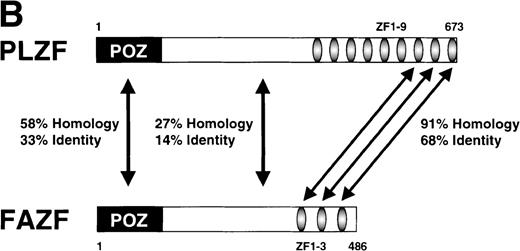
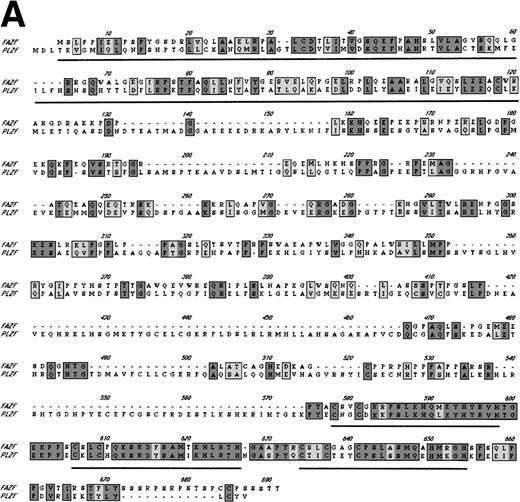
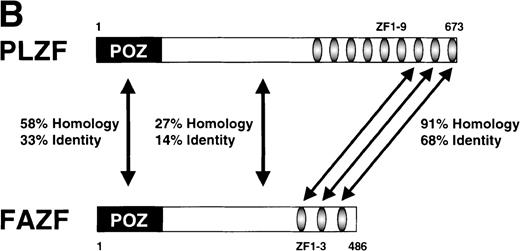
Examination of the predicted FAZF protein sequence indicated a significant degree of homology with the PLZF protein of t(11;17) (q21;q23)–associated APL (reviewed in Melnick et al27; Fig 2A). FAZF contains an amino terminal POZ/BTB domain25,57 sharing highly conserved residues appearing in this subfamily that includes LRF,41LAZ-3/BCL-6,33,36,37 and the recently cloned Kaiso.58 The POZ domain can mediate protein dimerization59,60 and transcriptional repression by PLZF.61 PLZF binds to specific DNA sequences and can do so using only the last five to seven of nine zinc finger motifs.61,62 FAZF’s three zinc fingers are closest in homology (91% similar) to the last in the series of nine at the C-terminus of PLZF (Fig 2B), and the POZ/BTB domain is nearly 60% similar. The degree of similarity between PLZF and FAZF suggested that these proteins might have common transcriptional functions. Examination of the genomic database also indicated a possible evolutionary relationship between PLZF and FAZF. A search of the EST database (expressed sequence tags; NCBI/NLM, Bethesda, MD) showed that the FAZF sequence (UniGene Hs. 99430;http://www.ncbi.nlm.nih.gov/UniGene/) is contained in a clone that also includes the gene HRX2, a gene very similar to theMLL/HRX (mixed lymphoid or mixed lineage leukemia/homologue of trithorax) gene,63-66 located adjacent to the PLZF gene on chromosome 11. This finding suggests that the simpler FAZF gene with its three zinc fingers might be an evolutionary antecedent to PLZF. The central portion of FAZF has no significant homology to PLZF or any other sequences in the database. The lack of homology in sequences between the BTB/POZ domain and the zinc fingers is a common feature in other BTB/POZ and Krüppel-like proteins. The FAZF sequences between aa #1-98, including part of the BTB/POZ domain (aa #1-109), are apparently not required for FANCC interaction, because we obtained only clones missing the amino terminal portion of the protein in the two-hybrid screens.
FAZF is similar to PLZF. (A) Comparison of the amino acid sequences of FAZF and PLZF showing BTB/POZ domains (underlined at the amino termini) and zinc fingers (underlined at the C-termini) in CLUSTAL W format. The FAZF sequence has been submitted to GenBank (accession no. AF165097). (B) Schematic representation of the PLZF and FAZF proteins indicating regions of homology and similarity.
FAZF is similar to PLZF. (A) Comparison of the amino acid sequences of FAZF and PLZF showing BTB/POZ domains (underlined at the amino termini) and zinc fingers (underlined at the C-termini) in CLUSTAL W format. The FAZF sequence has been submitted to GenBank (accession no. AF165097). (B) Schematic representation of the PLZF and FAZF proteins indicating regions of homology and similarity.
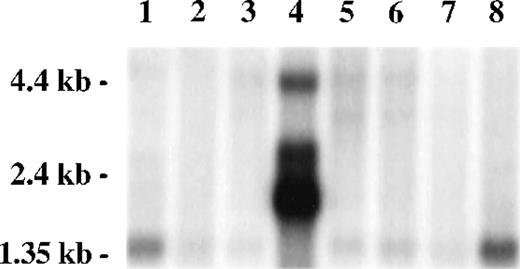
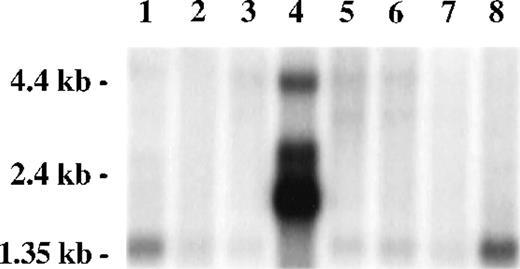
A Northern blot survey of RNA from multiple tissues probed with a radiolabeled restriction fragment from clone 8-1 revealed a predominant weak transcript of 1.4 kb in most tissues, except for testis, in which multiple strong transcripts were observed (2.0, 2.7, and 4.4 kb) and the 1.4-kb transcript was absent. Transcripts present in testis, particularly the 4.4-kb band, were also present in other tissues as minor transcripts. A strong signal was observed in peripheral blood leukocytes, consistent with the presence of FAZF hybridizing sequences in the mRNA from cells similar to that used for construction of the two-hybrid expression library (Fig 3).
FAZF is ubiquitously expressed. Multiple human tissue northern blot (Clontech) was probed using radiolabeled restriction fragment from clone 8-1. Lane 1, spleen; lane 2, thymus; lane 3, prostate; lane 4, testis; lane 5, ovary; lane 6, small intestine; lane 7, colon; lane 8, peripheral blood mononuclear leukocyte. Longer exposure of this blot showed a 1.4-kb FAZF band in all tissues except testis.
FAZF is ubiquitously expressed. Multiple human tissue northern blot (Clontech) was probed using radiolabeled restriction fragment from clone 8-1. Lane 1, spleen; lane 2, thymus; lane 3, prostate; lane 4, testis; lane 5, ovary; lane 6, small intestine; lane 7, colon; lane 8, peripheral blood mononuclear leukocyte. Longer exposure of this blot showed a 1.4-kb FAZF band in all tissues except testis.
FAZF coimmunoprecipitates with FANCC.
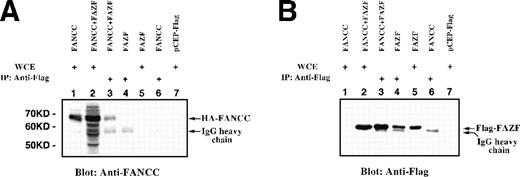
To determine whether FANCC associates with FAZF in vivo, expression vectors for epitope-tagged FANCC and full-length FAZF were cotransfected into 293 EBNA cells. Immobilized anti-Flag MoAb was added to lysates prepared from an asynchronous population of transfected cells, and the resulting complexes were separated by SDS-PAGE and analyzed by immunoblotting with anti-Flag. Whole cell extracts (WCE) were analyzed as controls for protein expression. As shown in Fig 4B, Flag-FAZF is detected as an Mr = 56,000 protein in whole cell extracts (lanes 2 and 5) and in lysates first immunoprecipitated with anti-Flag (lanes 3 and 4). The Flag-FAZF signal is absent in lysates from cells transfected with the parental vector (lane 7), or in lysates from cells expressing HA-FANCC (lane 1). We conclude that FAZF is expressed, is approximately the size expected, and is specifically detected by immunoprecipitation and by simple blotting in the whole cell lysates of cells transfected with the Flag-FAZF expression vector.
FANCC and full-length FAZF interact in vivo. 293 EBNA cells were transiently transfected with epitope-tagged expression vectors for HA-FANCC and/or Flag-FAZF. Lysates were electrophoresed directly as WCE or after immunoprecipitation with anti-Flag epitope tag. Proteins were detected by immunoblotting with anti-Flag or anti-FANCC. (A) Lane 1, FANCC WCE; lane 2, FANCC + FAZF WCE; lane 3, FANCC + FAZF IP; lane 4, FAZF IP; lane 5, FAZF WCE; lane 6, FANCC IP; lane 7, pCEP-Flag WCE. (B) The same loading order as in (A), probed with anti-Flag.
FANCC and full-length FAZF interact in vivo. 293 EBNA cells were transiently transfected with epitope-tagged expression vectors for HA-FANCC and/or Flag-FAZF. Lysates were electrophoresed directly as WCE or after immunoprecipitation with anti-Flag epitope tag. Proteins were detected by immunoblotting with anti-Flag or anti-FANCC. (A) Lane 1, FANCC WCE; lane 2, FANCC + FAZF WCE; lane 3, FANCC + FAZF IP; lane 4, FAZF IP; lane 5, FAZF WCE; lane 6, FANCC IP; lane 7, pCEP-Flag WCE. (B) The same loading order as in (A), probed with anti-Flag.
To determine if FANCC was coimmunoprecipitated with Flag-FAZF, the blot was stripped and reprobed with an antibody specific for FANCC (Fig 4A). In cell lysates containing Flag-FAZF, FANCC was detected (lane 3), and this band comigrated with the HA-FANCC in lanes 1 and 2 (whole cell lysates containing HA-FANCC and both HA-FANCC and Flag-FAZF, respectively). We conclude that HA-FANCC coimmunoprecipitates with Flag-FAZF. The reverse experiment was performed on the same lysates by first immunoprecipitating HA-FANCC with anti-HA serum followed by immunoblotting with either anti-FANCC or anti-Flag. Although the background was much higher using the HA antibody, we found that Flag-FAZF was specifically coimmunoprecipitated with HA-FANCC (data not shown).
FAZF is a nuclear protein with a punctate pattern.
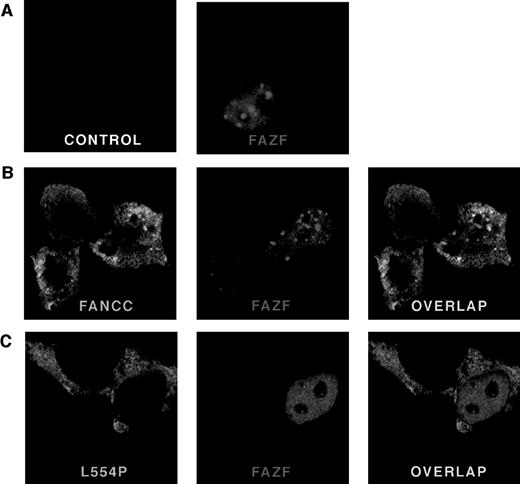
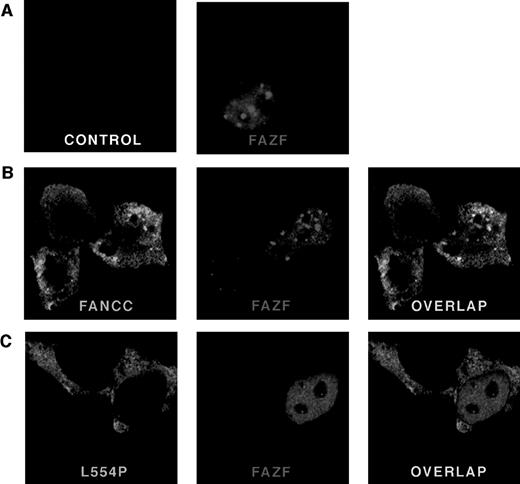
To determine the subcellular localization of FAZF, a Flag epitope-tagged protein was expressed in 293 EBNA cells and analyzed by indirect immunofluorescence. Cells were transfected with pFlag-FAZF or pFlag parental vector (as a negative control for specificity of the Flag antisera) and processed 20 hours later for confocal microscopy. Figure 5 (see page 3740) shows representative views of FAZF expression patterns. Consistent with observations reported for PLZF and other BTB/POZ-containing proteins, FAZF is almost entirely nuclear, with most cells containing foci superimposed on a microspeckled nuclear pattern.26,40,47,60 67 Like PLZF foci, FAZF nuclear foci varied in size and number, but were present in nearly all cells observed (Table 2). Staining was absent in cells transfected with pFlag.
FANCC and FAZF colocalize in nuclear bodies. 293 EBNA cells were transfected with pFlag-FAZF, pHA-FANCC, or pHA-L554P, or the pFlag parental vector (control panel), as indicated. The fixed and permeabilized cells were stained with antibodies specific for the epitope tags, followed by staining with fluorescent-conjugated secondary antibodies (Red = FAZF and Green = FANCC or L554P). Immunofluoresence was examined by confocal laser microscopy. For double labeling, the green and red channels were recorded independently and then overlaid. (A) Subcellular localization of FAZF. Original magnification ×63. (B) Colocalization of FANCC and FAZF in nuclear foci. Original magnification ×60. (C) The mutant L554P protein does not colocalize with FAZF.
FANCC and FAZF colocalize in nuclear bodies. 293 EBNA cells were transfected with pFlag-FAZF, pHA-FANCC, or pHA-L554P, or the pFlag parental vector (control panel), as indicated. The fixed and permeabilized cells were stained with antibodies specific for the epitope tags, followed by staining with fluorescent-conjugated secondary antibodies (Red = FAZF and Green = FANCC or L554P). Immunofluoresence was examined by confocal laser microscopy. For double labeling, the green and red channels were recorded independently and then overlaid. (A) Subcellular localization of FAZF. Original magnification ×63. (B) Colocalization of FANCC and FAZF in nuclear foci. Original magnification ×60. (C) The mutant L554P protein does not colocalize with FAZF.
Wild-type but not mutant FANCC colocalizes with FAZF in nuclear bodies.
To determine if FAZF and FANCC proteins colocalize, we performed experiments using indirect immunofluorescence and confocal microscopy to examine cells that were doubly transfected with constructs encoding tagged FAZF and FANCC. A survey of cells expressing FANCC (either singly or coexpressed with FAZF) showed that FANCC is distributed in both cytoplasmic and nuclear compartments, as reported previously.16-19 In addition, we observed that the distribution of FANCC between cytoplasm and nucleus varied, and that among cells expressing FANCC in an asynchronous population, 24% of cells contained two or more nuclear foci (Table 2 and Fig 5B, FANCC panel). The amount of FANCC appearing in the nucleus, or in foci, did not seem to be correlated with higher expression levels. Indeed, in many cells where FANCC expression was highest, no FANCC was observed in the nuclear compartment at all (data not shown). In cells that contained foci, the typical number ranged from approximately 5 to 20. The foci were typically superimposed on a diffuse nuclear stain, which was excluded from the nucleoli. Table 2 summarizes the percentage of cells containing FANCC foci observed in this experiment. Similar results were obtained from a 293 cell line stably expressing FANCC without an epitope tag using a FANCC-specific MoAb (data not shown). Interestingly, a similarly complex and variable pattern was recently described for another FA protein, FANCA, by D’Andrea et al,68 suggesting that foci formation may be an important and newly appreciated feature of FANCC and FANCA subnuclear localization. Not all cells expressing FANCC contain foci, as described above, but when FAZF and FANCC foci did appear in the same cell, we observed extensive overlap. Figure 5C shows a representative cell in which there is overlap of nuclear foci. No colocalization was observed between FANCC and PLZF, or between promyelocytic leukemia gene product (PML) and FANCC in colocalization experiments performed similarly (data not shown).
Although the FAZF interaction site on FANCC appears to be between aa #106 and 168, well away from the C-terminus of the FANCC, we wondered whether the L554P mutant protein could colocalize with the FAZF nuclear foci, because other features of the defective protein besides this interaction might influence colocalization. To answer this question, we expressed an HA-epitope–tagged L554P protein in 293 EBNA cells and observed the staining pattern of L554P with and without FAZF coexpression. In contrast with the pattern observed for wild-type FANCC, L554P staining shows intense, diffuse cytoplasmic staining with minimal nuclear staining (Fig 5C, L554P panel). In addition, although the expression of L554P was comparable with that of wild-type FANCC, nuclear foci were very rare; of 100 cells expressing L554P, only one foci in each of two cells was observed (Table 2), suggesting that the ability of L554P to enter the nucleus is compromised under these conditions. A recent report by Savoia and others described similar observations for L554P overexpressed in Hela cells.69 When L554P and FAZF are expressed in the same cells, the proteins are localized to different parts of the cells (Fig 5C [overlap]). In the cell shown, the FAZF staining shows a micropunctate nuclear staining pattern, and the L554P staining is mainly cytoplasmic. When these two photos were overlaid, no colocalization was observed. No FAZF/L554P colocalization was observed in other cells examined, although other cells coexpressing L554P and FAZF contained larger FAZF foci (data not shown) consistent with FAZF’s overall variable nuclear staining pattern.
FAZF binds to PLZF target sequences.
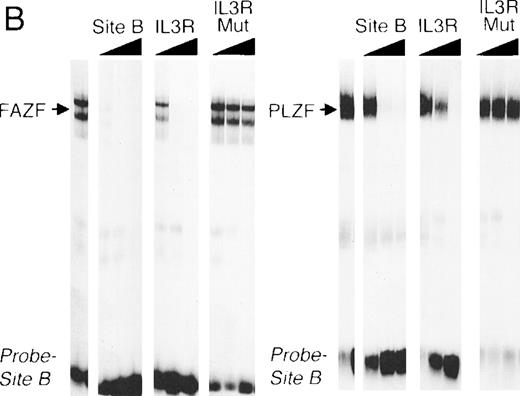
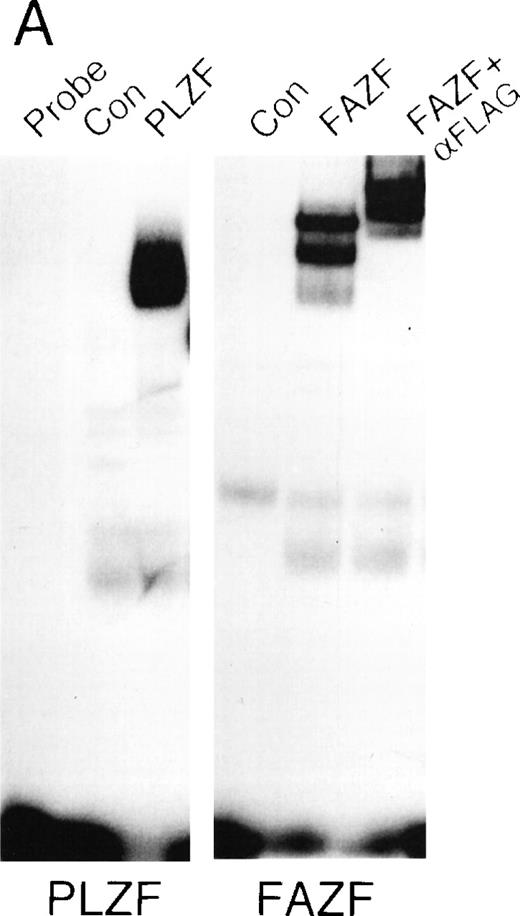
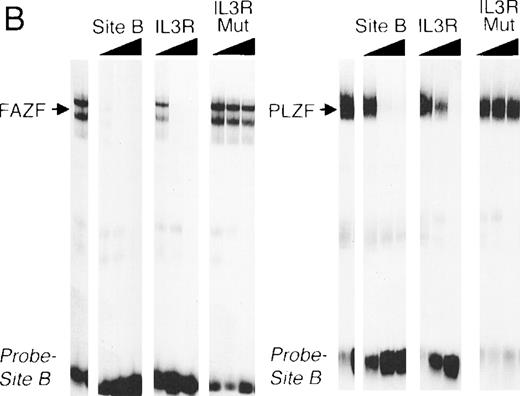
The striking homology between PLZF and FAZF suggested that the two proteins may have similar functions. PLZF has been shown to bind to specific DNA target sequences.61 To determine if FAZF could bind to the same sequences as PLZF, we performed an EMSA with nuclear extracts of 293T cell transfected with the PLZF or FAZF expression constructs, using a radiolabeled high-affinity PLZF binding site (Table1, Site B) derived from a CpG island library.51aFigure 6A shows that both PLZF and FAZF formed DNA-protein complexes with this site, indicated by the appearance of shifted bands in lanes containing FAZF or PLZF extracts. No shifted bands appear in the control lanes where no extracts, or mock transfected extracts, were added. The complexes caused by the epitope-tagged FAZF protein could be supershifted by the addition of an anti-Flag antibody (Fig 6A, right panel), confirming that the shifted band contains tagged FAZF. In a competition assay, the PLZF- and FAZF-DNA complexes were abrogated by addition of a molar excess of unlabeled site B, a high affinity site from the IL-3 receptor promoter (Fig 6B) as well as the Lex operator (data not shown), which is also a target of PLZF.62 A missense mutant within the IL3R site abrogated binding to both PLZF and FAZF. Together, these data suggest that PLZF and FAZF may bind the same or a closely overlapping set of target genes.
FAZF and PLZF bind to the same DNA sequences. (A) Nuclear extracts from untransfected cells (control) or from 293T cells transfected with a PLZF expression vector or a Flag-FAZF expression vector were allowed to interact with a high affinity PLZF binding site (Site B, Table 1), followed by nondenaturing gel electrophoresis. A supershift assay of the Flag-FAZF complexes was performed by the addition of Flag M2 MoAb to the reaction. (B) Competition analysis of FAZF and PLZF DNA binding. Extracts containing PLZF or FAZF were allowed to bind to a high affinity PLZF site (Site B, Table 1) in the absence of competitor or in the presence of a 10-fold, 100-fold, or 1,000-fold molar excess of the indicated unlabeled competitor oligonucleotide. The DNA protein complexes were resolved as described above.
FAZF and PLZF bind to the same DNA sequences. (A) Nuclear extracts from untransfected cells (control) or from 293T cells transfected with a PLZF expression vector or a Flag-FAZF expression vector were allowed to interact with a high affinity PLZF binding site (Site B, Table 1), followed by nondenaturing gel electrophoresis. A supershift assay of the Flag-FAZF complexes was performed by the addition of Flag M2 MoAb to the reaction. (B) Competition analysis of FAZF and PLZF DNA binding. Extracts containing PLZF or FAZF were allowed to bind to a high affinity PLZF site (Site B, Table 1) in the absence of competitor or in the presence of a 10-fold, 100-fold, or 1,000-fold molar excess of the indicated unlabeled competitor oligonucleotide. The DNA protein complexes were resolved as described above.
FAZF interacts with PLZF.
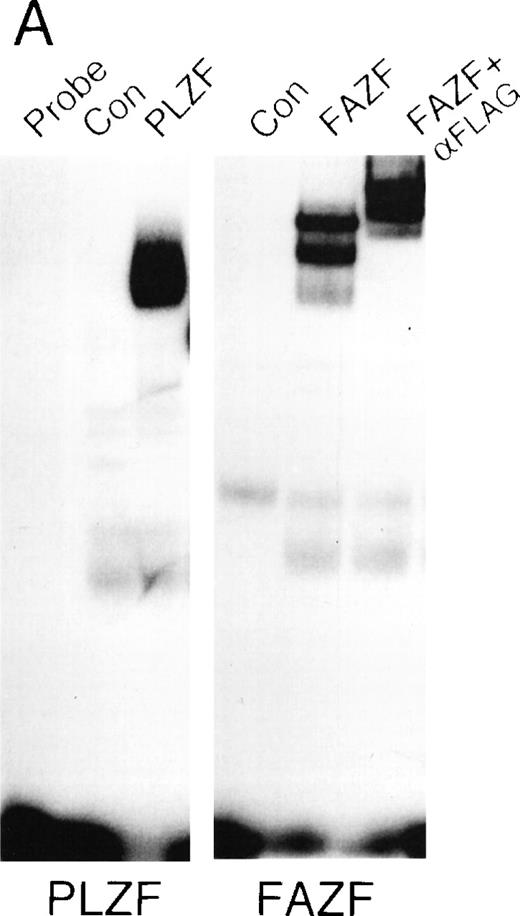
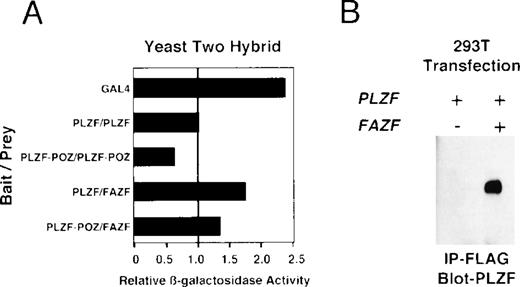
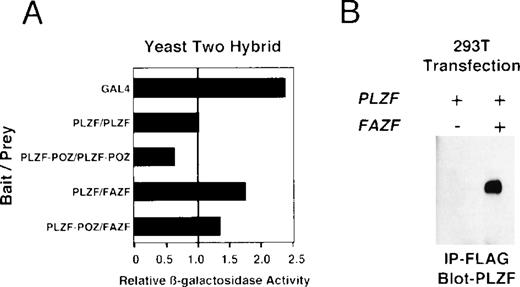
Because BTB/POZ domain-containing proteins have been known to form functional dimers,26 59 we next determined whether PLZF and FAZF might physically interact. PLZF and FAZF combinations were coexpressed as AD/BD fusion proteins in yeast, and assayed for reporter activity. We found that PLZF protein self-associated in this assay, as expected, and the POZ domain of PLZF (POZ-PLZF) was sufficient for this interaction (Fig 7A). FAZF (AD) could similarly interact with a BD (full-length) PLZF or BD-POZ-PLZF. We conclude that the PLZF POZ domain is sufficient for interaction of PLZF and FAZF. To confirm the interaction observed in yeast, we tested for PLZF-FAZF interaction by coimmunoprecipitation in mammalian cells. In 293T cells cotransfected with expression vectors encoding PLZF and a Flag-tagged FAZF expression vector, PLZF could be detected after immunoprecipitation of the epitope-tagged FAZF (Fig 7B).
PLZF and FAZF interact in vivo. (A) Yeast two-hybrid assay. PJ69-4A yeast were transformed with a bait plasmid encoding full-length PLZF or the POZ domain of PLZF linked to the GAL4 DNA binding domain and the indicated prey plasmids containing full-length FAZF or PLZF or the POZ domain of PLZF linked to an acidic activation domain. Yeast colonies grown on leucine and tryptophan deficient media were expanded, and a liquid β-galactosidase assay was performed. Levels of β-galactosidase were normalized, setting the level of β-galactosidase obtained in the presence of a PLZF bait and PLZF prey plasmid to 1.0. The GAL4 protein acted as positive control for transcription. (B) In vivo–coimmunoprecipitation. 293T cells were transfected with PLZF or Flag-FAZF plasmids as indicated. Lysates from the cells were subjected to immunoprecipitation with the Flag M2 MoAb followed by immunoblotting with a PLZF polyclonal antibody.47
PLZF and FAZF interact in vivo. (A) Yeast two-hybrid assay. PJ69-4A yeast were transformed with a bait plasmid encoding full-length PLZF or the POZ domain of PLZF linked to the GAL4 DNA binding domain and the indicated prey plasmids containing full-length FAZF or PLZF or the POZ domain of PLZF linked to an acidic activation domain. Yeast colonies grown on leucine and tryptophan deficient media were expanded, and a liquid β-galactosidase assay was performed. Levels of β-galactosidase were normalized, setting the level of β-galactosidase obtained in the presence of a PLZF bait and PLZF prey plasmid to 1.0. The GAL4 protein acted as positive control for transcription. (B) In vivo–coimmunoprecipitation. 293T cells were transfected with PLZF or Flag-FAZF plasmids as indicated. Lysates from the cells were subjected to immunoprecipitation with the Flag M2 MoAb followed by immunoblotting with a PLZF polyclonal antibody.47
FAZF is a transcriptional repressor.
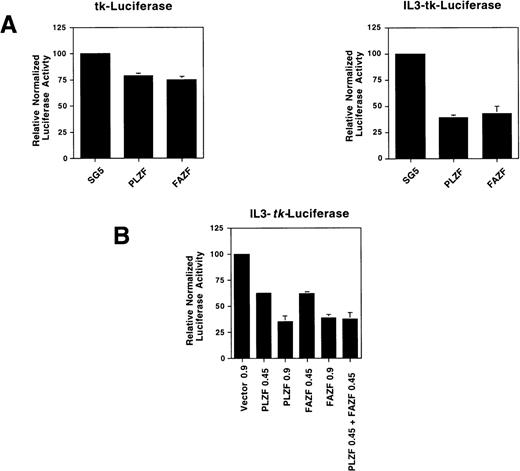
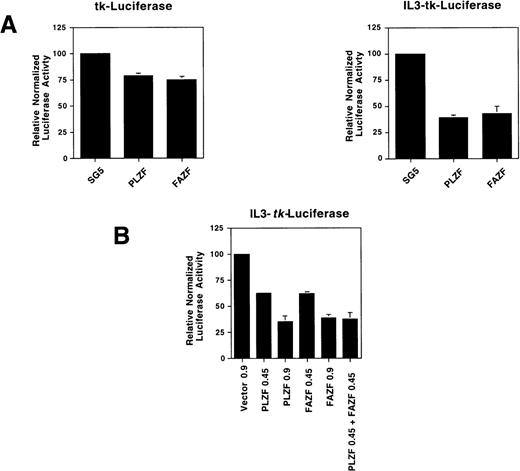
To determine if FAZF is able to act as a transcriptional repressor, we tested FAZF by coexpressing the protein along with a reporter gene linked to a promoter containing multimerized IL3R-PLZF target sites. We found that both PLZF and FAZF specifically repressed expression of this reporter gene, but not the parental tk-luciferase reporter (Fig 8A). The transcriptonal repression was dependent on the amount of plasmid transfected, and when both PLZF and FAZF were expressed, there was an additive effect on repression (Fig8B). Taken together, the binding and repression data suggest that not only do PLZF and FAZF proteins bind to the same DNA sequence, but they repress transcription by a similar mechanism.
FAZF is a transcriptional repressor. (A) 293T cells were cotransfected in quadruplicate with a luciferase reporter gene containing four copies of a high-affinity PLZF binding site (0.15 μg) from the IL-3–receptor promoter linked 5′ to the HSV-tk promoter or the parental tk-luciferase reporter (0.15 μg) along with 0.85 μg of expression vectors for PLZF or FAZF as indicated or the pSG5 expression vector. At 48 hours after transfection, the cells were assayed for luciferase activity. (B) Increasing amounts of the PLZF or FAZF expression vector or a combination of both vectors was cotransfected in quadruplicate along with the IL3-tk-luciferase promoter (0.1 μg). Luciferase activity was assayed as above.
FAZF is a transcriptional repressor. (A) 293T cells were cotransfected in quadruplicate with a luciferase reporter gene containing four copies of a high-affinity PLZF binding site (0.15 μg) from the IL-3–receptor promoter linked 5′ to the HSV-tk promoter or the parental tk-luciferase reporter (0.15 μg) along with 0.85 μg of expression vectors for PLZF or FAZF as indicated or the pSG5 expression vector. At 48 hours after transfection, the cells were assayed for luciferase activity. (B) Increasing amounts of the PLZF or FAZF expression vector or a combination of both vectors was cotransfected in quadruplicate along with the IL3-tk-luciferase promoter (0.1 μg). Luciferase activity was assayed as above.
DISCUSSION
We report here that FANCC interacts with a novel protein, FAZF, which we show functions as a transcriptional repressor. The FAZF/FANCC interaction maps to a central region in FANCC, which is deleted in patients with a severe FA phenotype.14 53 Moreover, we found that wild-type FANCC, but not the L554P mutant protein, can colocalize with FAZF in nuclear foci. Thus, disturbing the FANCC/FAZF interaction may play a role in FA.
The FAZF protein is similar to transcriptional repressors of the BTB/POZ subfamily of Krüppel-like zinc finger proteins (reviewed in Albagli et al25 and Bardwell et al26), which have been shown to be important for development, tissue-specific proliferation and differentiation, and neoplasia. In particular, FAZF is similar to the PLZF gene, which is translocated and fused to the RARα locus t(11;17)(q23;q21) in a subset of patients with APL.27,29,35 Recently, the mechanism of transcriptional repression of PLZF has been shown to occur through binding of specific promoter sequences61,62 followed by recruitment of nuclear receptor Sin-3/histone deactylase corepressor complexes.30-32 70-72 Like PLZF, we found that FAZF is able to repress transcription, suggesting that the function of these two proteins is, at least in part, overlapping.
The BTB/POZ protein-protein interaction motif is a highly conserved domain found in a number of eukaryotic proteins involved in cancer including the PLZF protein,29 BCL-6,37 and HIC (hypermethylated in cancer).73 The BTB/POZ domain mediates self-association26,59,60 and specific heterodimeric association with other BTB/POZ proteins (eg, PLZF,74 LRF, and LACZ-3/BCL-641). Recent evidence suggests that even though BTB/POZ domains may have significant homology and are able to interact with a related set of corepressor components, they may differ in their interactions with the corepressor complex.58 75Thus, crosstalk through heterodimer formation combined with the subtle differences in corepressor interactions suggests that transcriptional regulation by the proteins in this family is highly permuted. In this regard, we found that the FANCC/FAZF interaction is apparently not dependent on the BTB/POZ domain of FAZF, potentially allowing this region to form homodimeric and heterodimeric complexes with other proteins. Indeed, we report here that FAZF forms homodimers and heterodimerizes with PLZF. It will be interesting to determine if unique binding partners exist for the FAZF POZ domain.
The FANCC/FAZF interaction suggests that FANCC may play a role in the repression of gene expression, potentially in response to DNA damage. The nature of the target genes is unknown. The only PLZF target gene reported so far is cyclin A2. Enforced expression of PLZF delayed the transit of cells though the G1 and S phases of the cell cycle, an effect that was reversed by ectopic expression of cyclin A in these cells.76 77 The cyclin A2 promoter contains two PLZF binding sites that are required for the protein to repress this promoter. It is plausible that FAZF could have a similar effect on cyclin A expression and the cell cycle, perhaps reflecting a role in inhibiting cell growth in response to DNA damage. Although the FAZF protein can bind to the same binding site as PLZF, the fact that the FAZF protein only contains three zinc fingers whereas PLZF contains nine motifs suggests that the range of target genes bound by these two proteins may only partially overlap. Future studies will determine the effects of FAZF in cell growth control and DNA damage response.
Immunofluoresence analysis of FANCC in this study indicated that FANCC is expressed in both cytoplasmic and nuclear compartments, as reported previously.16-19 However, L554P was not found in the nuclear compartment at wild-type levels. Failure, or reduced ability, of L554P to enter the nucleus could account for its defect. Confirmation awaits comparison of endogenous wild-type and L554P localization that is currently not possible because of low endogenous expression levels. We also observed that a subset of FANCC-expressing cells contained nuclear foci, and suggest that this is a feature of wild-type FANCC localization. The following evidence suggests that these foci might be functionally significant. First, although expressed at comparable levels, the signal observed for the L554P mutant protein was greatly reduced in the nucleus as suggested by earlier studies,19 and was consequently unable to form nuclear foci. That L554P could be highly expressed in the cytoplasm and not “forced” into the nucleus also argues against the notion that nuclear localization and nuclear foci are simply an artifact of overexpression. Second, FANCA has also been observed in nuclear foci.68 Third, D’Andrea et al18,78 have reported that the nuclear fractions of FANCA and FANCC, but not L554P, form a complex and that this nuclear complex is functional. Taken together, these observations support the notion that foci formation may be biologically significant. Finally, we determined that FAZF, like other members of its family, is located primarily in nuclear foci.26,40,47,60,67 Our coimmunoprecipitation and colocalization experiments independently support the FAZF/FANCC interaction detected by two-hybrid analysis and in vitro binding. We found that in some cells, colocalization of FANCC and FAZF foci was extensive, whereas only partial overlap was observed in other cells, suggesting that the FAZF/FANCC interaction may be transient or conditional. This pattern of partially overlapping foci has been described for a series of colocalized proteins including BRCA1/hRad51,79 Arenavirus proteins/PML,80retinoblastoma protein/PML,81 and PML/PLZF.74Given these complexities, and the fact that the FA pathway is novel, further studies will be required to understand the consequences of the FAZF/FANCC interaction and the potential role of transcriptional repression in the genomic instability observed in FA.
ACKNOWLEDGMENT
The authors are grateful to their colleagues Mike Forte and Beth Blanchly-Dyson for initial assistance with the two-hybrid studies. The authors thank the members of the Fanconi anemia research community for stimulating discussions, in particular Hans Joenje, Mike Heinrich, Johan de Winter, Quinten Waisfisz, and Henri van de Vrugt for their interest and advice.
Supported by NIH grants HL56045 (M.E.H.), CA59938 (J.D.L., A.Z.), and K08 CA73762 (A.M.); the Medical Research Foundation of Oregon and the Fanconi Anemia Research Fund (M.E.H.); and the American Cancer Society DHP160 (J.D.L.). J.D.L. is a Scholar of the Leukemia Society of America.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Maureen E. Hoatlin, PhD, Division of Hematology and Medical Oncology, Oregon Health Sciences University, 3181 SW Sam Jackson Park Rd, Portland, OR 97201; e-mail:hoatlinm@OHSU.edu.