Abstract
Acute promyelocytic leukemia (APL) is typified by the t(15;17), generating the PML-RARα fusion and predicting a beneficial response to retinoids. However, a sizeable minority of APL cases lack the classic t(15;17), prompting the establishment of the European Working Party to further characterize this group. Such cases were referred to a workshop held in Monza, Italy and subjected to morphologic, cytogenetic, and molecular review, yielding 60 evaluable patients. In the majority (42 of 60), molecular analyses revealed underlying PML/RARα rearrangements due to insertions (28 of 42) or more complex mechanisms, including 3-way and simple variant translocations (14 of 42). Metaphase fluorescence in situ hybridization (FISH) demonstrated that insertions most commonly led to formation of the PML-RARα fusion gene on 15q. In 11 of 60 workshop patients, PLZF/RARα rearrangements were identified, including 2 patients lacking the t(11;17)(q23;q21). In one case with a normal karyotype, FISH analysis revealed insertion ofRARα into 11q23, and PLZF-RARα was the sole fusion gene formed. Two patients were found to have t(5;17), one with a diffuse nuclear NPM staining pattern and with NPM-RARαand RARα-NPM transcripts detected. In the other with an unbalanced der(5)t(5;17)(q13;q21) and a nucleolar NPM localization pattern, an NPM/RARα rearrangement was excluded, and FISH revealed deletion of one RARα allele. In the remaining 5 workshop patients, no evidence was found for a rearrangement ofRARα, indicating that in rare instances, alternative mechanisms could mediate the differentiation block that typifies this disease. This study highlights the importance of combining morphologic, cytogenetic, and molecular analyses for optimal management of APL patients and better understanding of the pathogenesis of the disease.
Introduction
Acute promyelocytic leukemia (APL) is defined by particular morphologic features (see the accompanying article in this issue, by Sainty et al1). A number of key clinical features set APL apart from other forms of acute myeloid leukemia (AML), which underlie the need for accurate diagnosis. These include a potentially devastating coagulopathy, which carries a high risk of mortality unless specifically addressed (reviewed by Tallman and Kwaan2; Barbui et al3), and sensitivity to retinoid differentiating agents including all-trans retinoic acid (ATRA) (reviewed by Degos et al4) and to novel agents such as arsenic trioxide (As2O3).5,6 Early studies suggested that retinoids reduce the hemorrhagic complications of APL, whereas use of ATRA in combination with chemotherapy has been shown to confer significant improvements in overall survival compared with treatment with chemotherapy alone.7-11 Hence, combination therapy with ATRA and chemotherapy has now been adopted as the standard treatment approach for this disease. For the majority of APL patients achieving complete remission (CR), the long-term outlook is now favorable because of a relatively low risk of relapse, and routine use of bone marrow transplantation (BMT) in first CR is no longer recommended.
APL is characterized by the reciprocal translocation t(15;17)(q22;q21), disrupting the PML and RARα genes, which are localized to chromosomes 15q and 17q, respectively (reviewed by Melnick and Licht12). The t(15;17) generates 2 chimeric genes: PML-RARα is formed on the derivative 15 [der(15)], whereas the reciprocal RARα-PMLfusion is located on the derivative 17 [der(17)]. PML and RARα have both been implicated in normal hemopoiesis.13-16 PML possesses growth suppressor and proapoptotic activity16-19and is predominantly localized to discrete multiprotein nuclear structures (PML nuclear bodies), which become disrupted in the presence of the PML-RARα fusion protein20,21 (reviewed by Hodges et al22). RARα is a transcription factor that mediates the effect of retinoic acid (RA) at specific response elements; high-affinity binding of the receptor to DNA is achieved through heterodimerization with a member of the distinct family of retinoid X receptors (RXR; reviewed by Chambon23). Previous studies suggested that integrity of the retinoid signaling pathways is necessary for normal myeloid differentiation.13-15 The PML-RARα protein retains key functional domains of both PML and RARα, suggesting that it plays an important role in leukemogenesis. PML-RARα may impair the growth suppressor and proapoptotic functions of PML, contributing to leukemic transformation, and also may induce a block in myeloid differentiation by repression of RA target genes through recruitment of co-repressor molecules and histone deacetylase; the latter phenomenon may also be compounded by sequestration of RXR (reviewed by Melnick and Licht12; Grimwade24). The important role played by PML-RARα in leukemogenesis has been confirmed recently using transgenic mice (reviewed by He et al25; Westervelt and Ley26). However, it should be noted that in these studies, less than a third of the animals expressing PML-RARα ultimately developed APL. Furthermore, a latent period of several months was observed before manifestation of the leukemia, leading to the suggestion that additional oncogenic events are required and arousing interest as to whether the reciprocal RARα-PML fusion product plays a role in this process.27
Previous studies suggested that the PML-RARα fusion protein not only induces the differentiation block that characterizes APL, but paradoxically is also important for mediating the differentiation response to ATRA.12,24 Hence, APL patients with cryptic formation of the PML-RARα fusion gene share the beneficial response to retinoids and the favorable prognosis associated with the group with documented t(15;17).11,28 29 This finding highlights the importance of establishing the presence of thePML/RARα rearrangement in patients with morphologic APL, not only for optimal management such that all patients who could benefit are not denied treatment with ATRA, but also for meaningful analysis of clinical trials involving retinoids.
Over the last few years, considerable reliance has been placed on conventional cytogenetics to confirm a morphologic diagnosis of APL, as a means of determining the treatment approach. In the majority of cases the t(15;17) is detected30; however, more recently a series of alternative chromosomal aberrations have been reported, including t(11;17)(q23;q21),31,32t(5;17)(q35;q12-21),33 t(11;17)(q13;q21),34and der(17), 35 whereby RARα is fused to thePLZF, NPM, NuMA, and STAT5bgenes, respectively. In common withPML-RARα–associated APL, patients with fusion genes involving NPM and NuMA appear to be sensitive to ATRA.34,36 In contrast, APL associated with aPLZF/RARα rearrangement is typified by lack of a differentiation response to retinoids, and patients with this disease treated with ATRA alone have a poor prognosis.37 Recent studies have correlated ATRA sensitivity with ligand-dependent dissociation of the co-repressor complex from the APL-associated chimeric fusion proteins (reviewed12,24). At pharmacologic levels of ATRA, the co-repressor complex is released from the retinoid receptor moiety of PML-RARα, NPM-RARα, PLZF-RARα, and presumably NuMA-RARα fusion proteins; however, the PLZF-RARα fusion additionally binds co-repressors to its PLZF moiety in a retinoid-insensitive fashion. This latter phenomenon has been proposed to account for the lack of response to ATRA that characterizes cases of APL with the t(11;17)(q23;q21). However, it remains a possibility that the reciprocal derived RARα-PLZF fusion could also contribute to retinoid resistance in this subtype of APL because its expression is potentially up-regulated by ATRA, which could induce persistent deregulation of the cell cycle.12 37 Clearly, molecular characterization of cases of APL with alternative translocations has provided insights not only into the pathogenesis of the disease, but also into the mechanisms underlying the response to retinoids. In the present study, the European Working Party performed morphologic, cytogenetic, and molecular review of 60 evaluable APL patients lacking the classic t(15;17) and sought to determine the frequency of such cases.
Patients and methods
Patient characteristics
The European Working Party sought to characterize AML cases classified as APL, but lacking the t(15;17). Overall, 42 institutions from 6 European countries in addition to Memphis, TN participated in this study, as detailed in the accompanying paper.1 Ninety cases of suspected APL were reviewed in a workshop held in Monza, Italy in June 1997. Cases were subjected to central morphologic review.1 The corresponding karyotypes and molecular data were reviewed simultaneously but separately. Morphologic reviewers were ignorant of the cytogenetic and molecular data; in a second step, the reviewed data were combined and considered in the context of the clinical features. Patients were considered eligible for inclusion in the study only if all the following criteria were satisfied: (1) Morphologic features were consistent with or evocative of FAB type M3 or M3v. (2) Karyotype analysis was successful and the t(15;17) had been excluded. For patients with a normal karyotype, at least an overnight culture had to be performed to avoid normal metaphases from erythroblasts. (3) Molecular analysis had been performed by at least one of the following techniques: fluorescence in situ hybridization (FISH), reverse transcriptase-polymerase chain reaction (RT-PCR), or Southern blot analysis. On this basis, 30 patients were excluded from further consideration: 4 lacked features of APL (1 was classified as FAB type M1, 3 as FAB M2); in 15 cases there was no suitable material for further molecular analyses; in 4 cases cytogenetic review revealed a minor clone with t(15;17); and 7 cases presented with ider(17q), which was deemed to be a secondary abnormality to the t(15;17). Morphologic and immunophenotypic features of the evaluable patients are considered in the accompanying manuscript,1 which shares the same case numbers.
FISH analyses
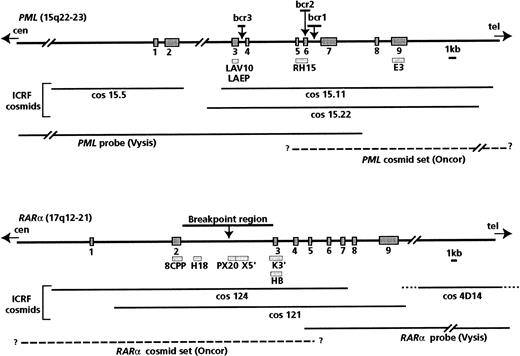
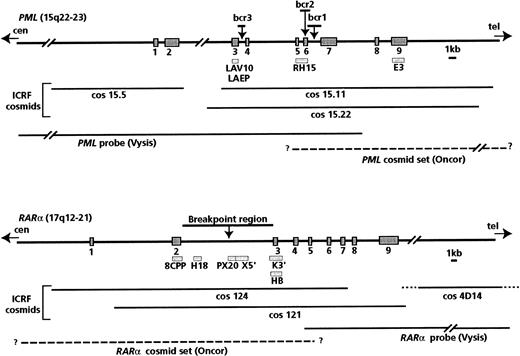
Analyses documenting the occurrence of PML/RARαrearrangements were generally performed using ICRF PML andRARα cosmid probes (from Ellen Solomon, Guy's, King's, and St. Thomas' School of Medicine, London), which were distributed among workshop members (courtesy of K. Howe and F. Birg). Details of the probes used and their positions relative to the PML andRARα breakpoint regions are shown in Figure1; methods used by the participating laboratories have been fully described previously.38,39The genomic map of RARα and exon numbering were according to Hjalt and Murray.40 In some instances, commercially available probes were also used, in accordance with the manufacturer's instructions. The Oncor t(15;17) probe set (Gaithersburg, MD) contains digoxigenin-labeled cosmids specific for the 17q21 region and biotin-labeled cosmids specific for the 15q22 region. These probes are designed to detect the RARα-PML fusion gene on the der(17), but more precise mapping details are not provided by the manufacturer. Thus, we initially evaluated this probe set on a series of 5 patients with t(15;17); secondary RARα andPML signals were detected on the der(15) in 5 of 5 and 4 of 5 cases, respectively, leading to 2 fusion signals in metaphases and nuclei. This result suggests that the RARα probe spans the t(15;17) breakpoint, as does the PML probe in some instances.
Map of the probes used for Southern blot and FISH analyses.
Bars indicate Southern blot probes; lines indicate probes used for FISH analyses. The P63 probe, which includes the entire RARαcDNA, was also used in FISH experiments.
Map of the probes used for Southern blot and FISH analyses.
Bars indicate Southern blot probes; lines indicate probes used for FISH analyses. The P63 probe, which includes the entire RARαcDNA, was also used in FISH experiments.
The Vysis probe set (Downers Grove, IL), which is designed to detect the PML-RARα fusion gene, comprises a mixture of directly labeled probes: a PML probe, which begins in intron 7 and extends toward the centromere for 180 kb, and a RARαprobe, which begins in intron 4 and extends toward the telomere for 400 kb (Figure 1).
In some instances, especially for complex karyotypes, whole chromosome painting (wcp) probes and centromeric probes (Cambio, Cambridge, UK; Oncor; Vysis) were used in single or dual-color FISH experiments.
Twenty-four–color FISH karyotyping41 was carried out on the unique case presenting with RARα-PML as the sole fusion gene using a 24XCyte kit (MetaSystems, GmbH, Altlussheim, Germany). Multicolor banding (mBAND) of chromosome 5 was carried out in the 2 t(5;17) cases as described recently,42 using an XCyte 5 kit (MetaSystems) according to the manufacturer's protocol. A DMRB epifluorescence microscope equipped with a motorized filter wheel and specific filters was used (Leica, Rueil-Malmaison, France). Images were captured and processed using the Isis/M-FISH (Multicolor FISH) imaging system (MetaSystems).
RT-PCR and Southern blot analyses
PML-RARα and RARα-PML fusion genes were detected by nested or semi-nested RT-PCR according to one of the previously described methods.43-45PLZF-RARαand RARα-PLZF fusion transcripts were detected as described previously37,38,46; however, for cases found by PLZF-RARα RT-PCR to have a 3′ breakpoint inPLZF (leading to retention of 3 PLZF zinc fingers in the PLZF-RARα fusion protein),37,47PLZF primersR1 and R2 (Table 1) were used for amplification of reciprocal RARα-PLZFtranscripts. To detect NPM-RARα andRARα-NPM fusion genes, we performed nested RT-PCR usingNPM primers detailed in Table 1 in conjunction with previously described external and internal RARαprimers.44 Identical RARα primers were used with STAT5b primers (Table 1) and previously describedNuMA primers34 (N2a [external],Alt1b, Alt2b, N2b[internal]) for nested RT-PCR to screen forSTAT5b-RARα and NuMA-RARα fusion genes, respectively. Where availability of DNA permitted, workshop cases were also subjected to Southern blot analysis using the probes shown in Figure 1, as described previously.48-50
ATRA in vitro differentiation assays
Assays were performed according to a previously described method51 using ATRA at a final concentration of 10−6 mol/L.
Immunofluorescence
Results
Central morphologic, cytogenetic, and molecular review undertaken at the Monza Workshop yielded 60 evaluable patients with confirmed APL lacking the t(15;17). The review process led to the definition of the following subgroups: (1) PML/RARα rearrangements (n = 42), including insertions (28 of 42) and complex chromosomal changes (14 of 42); (2) PLZF/RARα rearrangements (n = 11); (3) t(5;17) (n = 2); and (4) APL lacking rearrangement ofRARα (n = 5). Central morphologic review revealed no major differences between the appearances of material derived from patients with PML/RARα rearrangements and from 20 control patients with documented t(15;17). Among the remaining patients, only those with PLZF/RARα rearrangements were found to have distinct morphologic features allowing their recognition, as described in the accompanying paper.1
Characterization of APL workshop patients lacking the t(15;17), with underlying PML/RARα rearrangements
Insertion (15;17) or (17;15).
In 28 patients including 16 with a normal karyotype, FISH and molecular findings were consistent with PML/RARα rearrangements being mediated by insertion (ins) events.
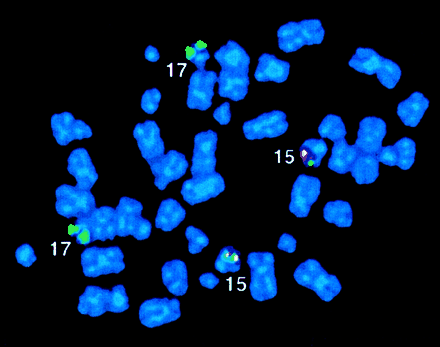
Metaphase FISH was performed in 20 of 28 patients (cases 1-20). In the majority (15 of 20), a fusion or co-localization signal reflecting the formation of PML-RARα was localized to 15q (cases 1-15). In one such patient (case 4), fusion signals were detected on both chromosomes 15, suggesting either loss of the normal 15 and duplication of the der(15) or recombination between the 2 homologs after the insertion event (Figure2). Indeed, a mitotic recombination event leading to BCR-ABL fusion signals on both chromosomes 9 has been described in a Ph-negative case of chronic myeloid leukemia (CML) with submicroscopic ins(9;22).56 In 7 ins(15;17) cases, the Oncor probe set was used in parallel either with ICRFPML 15.5 and RARα 121 cosmids (4 cases) or Vysis probes (3 cases), giving identical results, suggesting that the Oncor RARα probe is not only centromeric, but also spans the 17q breakpoint (see Patients and methods and Figure 1). All of these ins(15;17) patients (n = 15) had apparently normal chromosomes 15 and 17 by conventional cytogenetic analysis and by FISH using wcp probes (7 of 7) and were thus cryptic. Furthermore, diagnostic karyotype was normal in the majority (9 of 15); in such patients, fusion signals were detected in the context of normal metaphases, thereby establishing that the results of cytogenetic analysis reflected sampling of leukemic cells rather than residual normal marrow elements. RT-PCR performed in 10 ins(15;17) patients revealed expression ofPML-RARα (Table 2). Reciprocal RARα-PML transcripts were not detected in the 7 patients investigated, consistent with the occurrence of insertion events in these patients.
FISH analysis of case 4.
The Oncor RARα and PML probes showed fusion signals on both chromosomes 15; similar results were obtained with the Vysis probe set.
FISH analysis of case 4.
The Oncor RARα and PML probes showed fusion signals on both chromosomes 15; similar results were obtained with the Vysis probe set.
In 5 of 20 patients (cases 16-20), metaphase FISH was consistent with insertion of chromosome 15 material into 17q. In 2 of these ins(17;15) patients (cases 18 and 19), this insertion was apparent by conventional cytogenetic analysis and was confirmed by FISH using wcp 15 and 17 probes, whereas in the others, chromosomes 15 and 17 were normal by conventional cytogenetic analysis. In 4 of 5 patients (cases 16-19), formation of the PML-RARα fusion gene was confirmed and localized to 17q in 3 patients studied by metaphase FISH. In the remaining ins(17;15) patient (case 20), molecular analyses suggested that RARα-PML was the sole fusion gene resulting from the insertion event, as reported previously.29 39Twenty-four–multicolor karyotyping performed on the latter patient did not reveal any chromosomal change on the 20 metaphases analyzed (data not shown), confirming the small size of the insertion event and the absence of superimposed chromosomal abnormality at the level of resolution of conventional cytogenetics and M-FISH.
In 8 of 28 patients (cases 21-28), metaphase FISH was not performed. However, these cases were considered as probable insertions because they presented with normal chromosomes 15 and 17 and expressed thePML-RARα transcript. Karyotype was normal in the majority (6 of 8), and RARα-PML transcripts were not detected by RT-PCR in either of the 2 patients tested.
PML immunofluorescence was performed in 6 of 28 insertion cases. In 5 patients with cryptic formation of PML-RARα fusion genes, the classic microparticulate nuclear staining pattern was observed and correlated with a positive in vitro ATRA response in the 2 patients studied (Table 2). In contrast, in case 20, in whichRARα-PML was the sole fusion gene formed, a wild-type nuclear staining pattern was detected, correlating with a negative in vitro ATRA response as reported previously.29 39
Complex rearrangements.
In 14 patients, the PML-RARα fusion gene was formed as a result of complex rearrangements involving at least 3 chromosomes, as detailed in Table 3. Such complex cases can be classified into 3 categories: (1) complex variant t(15;17) due to a 3-way balanced translocation involving 15q22, 17q21, and another chromosome; (2) simple variant t(15;17), apparently involving either 15q22 or 17q21 with another chromosome; and (3) very complex cases.
In 6 patients, a complex variant due to 3-way balanced t(15;17) was defined; all partner chromosomal bands involved were different, as shown in Table 3. In both cases in which metaphase FISH was performed,PML-RARα was found on the der(15).
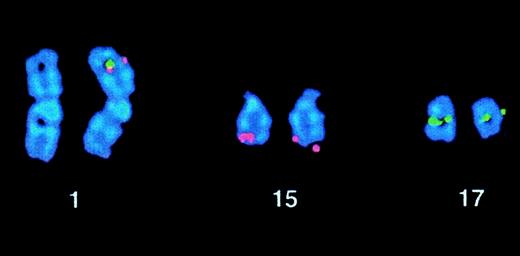
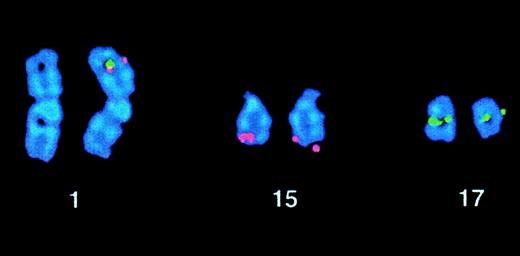
In 2 patients, a simple variant t(15;17) was identified. Case 35 presented with a t(5;15)(q13;q22), but FISH demonstrated aPML-RARα fusion on the der(15). Case 36 was previously reported to have a normal karyotype by R banding, to express aPML-RARα transcript, and was shown to have a t(1;17) by wcp.57 Further analysis was performed by the workshop; DAPI banding permitted visualization of the t(1;17), and FISH demonstrated formation of PML-RARα on 1p34, as shown in Figure 3. These simple variant cases are likely to be due to the combination of a reciprocal translocation and a submicroscopic insertion, leading to the formation of thePML-RARα fusion gene.
Case 36 with t(1;17)(p34;q21).
FISH using ICRF PML 15.5 (red) and RARα 121 (green) cosmid probes demonstrating PML-RARα fusion signals on the der(1).
Case 36 with t(1;17)(p34;q21).
FISH using ICRF PML 15.5 (red) and RARα 121 (green) cosmid probes demonstrating PML-RARα fusion signals on the der(1).
Six patients were classified as very complex cases. In 2 of 6 (cases 37 and 38), formation of the PML-RARα fusion gene was due to a submicroscopic ins(15;17) demonstrated by FISH. In case 39,PML-RARα resulted from a 4-way balanced translocation combined with an insertion of a chromosome 2p segment into the der(17). In case 40, PML-RARα fusion signals were observed in nuclei, but the chromosomal location could not be determined because of the lack of evaluable metaphases. RT-PCR performed in cases 41 and 42 revealed expression of PML-RARα transcripts; however, FISH analysis with Oncor probes did not show any fusion signals, but rather duplication or triplication of RARα signals on the der(17). Because these probes optimally detect theRARα-PML fusion gene on the der(17) in patients with the classic t(15;17), the absence of detectable fusion signals in these patients is consistent with lack of formation of theRARα-PML gene. Unfortunately, insufficient material was available to perform further metaphase FISH documenting the location of the PML-RARα fusion gene.
Cases lacking PML/RARα rearrangements
PLZF-RARα cases.
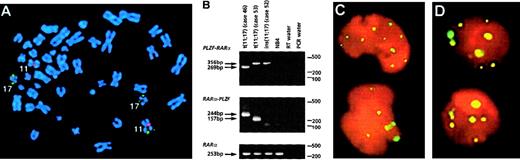
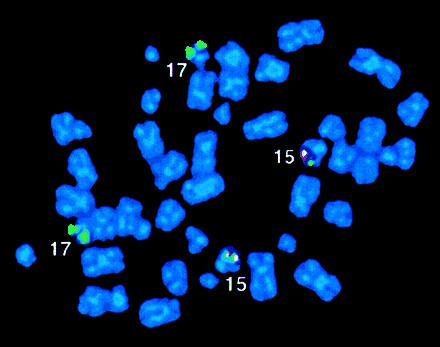
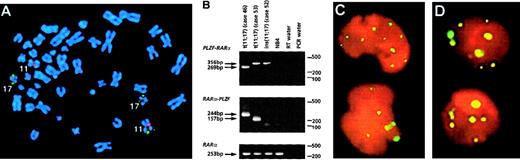
In 11 of 60 workshop patients, APL was associated with aPLZF/RARα rearrangement as determined by RT-PCR, including 5 patients that have not been reported previously (Table4). Nine patients were found to have the reciprocal translocation t(11;17)(q23;q21); in each of the 6 such patients analyzed, reciprocal RARα-PLZF transcripts were detected in addition to PLZF-RARα. RARα-PLZF has been postulated to contribute to leukemogenesis and may play a role in the ATRA resistance associated with this subtype of APL.12Therefore, it was of interest to characterize 2 cases (cases 50 and 52) of PML-RARα– and t(11;17)-negative APL with morphologic features that were typical of patients with the t(11;17).1Case 50 presented with a del(11)(q23), whereas case 52 had a normal karyotype. In both cases, FISH using wcp probes specific for chromosomes 11 and 17 did not show any exchange of material involving these 2 chromosomes. However, FISH using the ICRF RARα 121 probe demonstrated signals on chromosome 11q23 in case 52 (Figure4A). RT-PCR confirmed formation of aPLZF-RARα fusion gene in both patients. In case 52, there was sufficient diagnostic material to evaluate whetherPLZF-RARα was the sole fusion gene formed, and indeedRARα-PLZF transcripts were not detected by RT-PCR, consistent with a submicroscopic insertion event (Figure 4B). Overall,PLZF breakpoints were determined in 10 patients: 7 had a 5′ (intron 2) breakpoint (2 PLZF zinc fingers retained in PLZF-RARα) and 3 had a 3′ (intron 3) breakpoint (3 PLZF zinc fingers retained); introns were numbered according to Zhang et al.47
Molecular analyses of
PLZF-RARα cases. (A) Case 52 with a normal karyotype and formation of PLZF-RARα as the sole fusion gene due to an insertion event. FISH using ICRF RARα 121 (green) cosmid probe and chromosome 11 centromeric probe (red) demonstrated insertion of RARα sequences into band 11q23. (B) RT-PCR revealed expression of PLZF-RARα (3ZFPLZF breakpoint) as the sole fusion transcript in case 52, whereas both fusion transcripts were detected in cases 46 (2ZF) and 53 (3ZF). (C,D) PML immunofluorescence using the PG-M3 antibody on cytospin preparations from case 43 (C) and case 52 (D, courtesy of Francesco Fazi) showed a wild-type pattern, as distinct from the microparticulate PML staining shown in NB4 cells in Figure 8. Images were captured on a Zeiss Axioplan fluorescence microscope.
Molecular analyses of
PLZF-RARα cases. (A) Case 52 with a normal karyotype and formation of PLZF-RARα as the sole fusion gene due to an insertion event. FISH using ICRF RARα 121 (green) cosmid probe and chromosome 11 centromeric probe (red) demonstrated insertion of RARα sequences into band 11q23. (B) RT-PCR revealed expression of PLZF-RARα (3ZFPLZF breakpoint) as the sole fusion transcript in case 52, whereas both fusion transcripts were detected in cases 46 (2ZF) and 53 (3ZF). (C,D) PML immunofluorescence using the PG-M3 antibody on cytospin preparations from case 43 (C) and case 52 (D, courtesy of Francesco Fazi) showed a wild-type pattern, as distinct from the microparticulate PML staining shown in NB4 cells in Figure 8. Images were captured on a Zeiss Axioplan fluorescence microscope.
PML immunofluorescence was performed in 6 patients, revealing in each case discrete nuclear dots in leukemic blasts (Figure 4C,D), indistinguishable from the pattern observed in non-APL controls.38 This contrasted with the characteristic microparticulate distribution detected inPML-RARα–positive patients, as described above and previously.29 38 No terminal granulocytic morphologic differentiation was observed in the presence of 10−6 mol/L ATRA in vitro either in the 5 t(11;17) patients tested or in the ins(11;17) patient (case 52).
Clinical features of workshop patients with PLZF/RARαrearrangements are presented in Table 4. In contrast to a previous study, which highlighted the adverse prognosis of t(11;17) patients treated with ATRA alone,37 each of the 10 patients in the present study treated with combination chemotherapy achieved a CR, in 6 of whom induction chemotherapy was accompanied by ATRA. No cases of ATRA syndrome were observed, consistent with the hypothesis that this phenomenon is associated with modulation of surface adhesion molecules and cytokine release that is correlated with differentiation of the leukemic clone. Five patients are alive in first CR (range, 13-42 months; median, 28 months), including 2 receiving allogeneic BMT; and 5 patients relapsed, of whom 1 remains in remission after allogeneic BMT in second CR.
t(5;17).
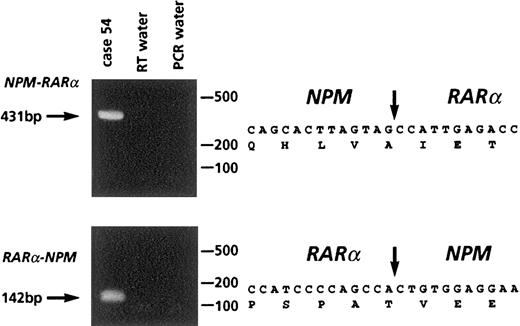
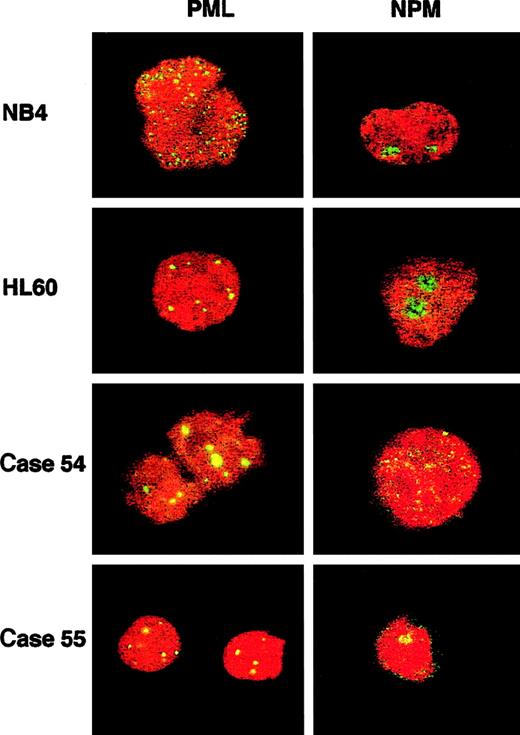
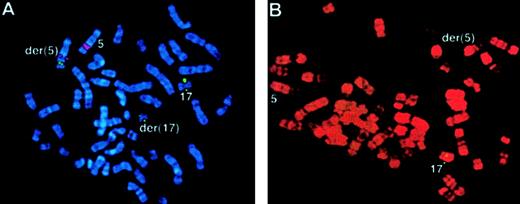
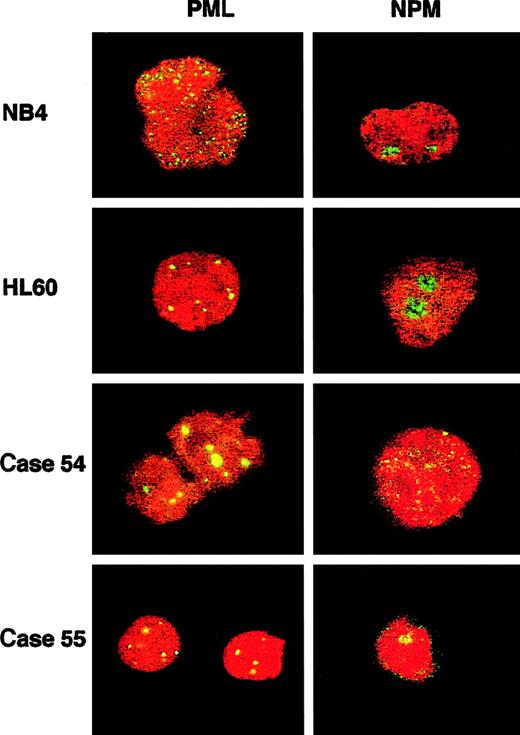
Two workshop patients were found to have a t(5;17); clinical and biologic data are summarized in Table 5. In case 54, conventional cytogenetics revealed t(5;17)(q34;q21) and a deletion of band 5q13 on the der(5). Multicolor banding of chromosome 5 allowed confirmation of the 5q translocation breakpoint and revealed that the del(5)(q13q13) was in fact an insertion of band 5q13 into 3q26 (Figure 5A,C). FISH analysis using Vysis or ICRF RARα 121 probes showed an additional RARαsignal on the der(5) (Figure 6A), and RT-PCR demonstrated expression of NPM-RARα andRARα-NPM fusion transcripts (Figure7). The NPM breakpoint (Figure 7) was identical to that associated with formation of the NPMS-RARα fusion in the 2 previously reported cases of APL with the t(5;17)33,65 and also to that of theNPM-ALK fusion associated with the t(2;5)(p23;q35) in anaplastic large-cell lymphoma.66 In case 55, previously reported,67,68 the molecular review performed in the present study ruled out a RARα rearrangement. Because the translocation was unbalanced, the 5q breakpoint was difficult to assign by conventional cytogenetics; multicolor banding of chromosome 5 allowed this breakpoint to be refined to 5q13 (Figure 5B,C). FISH analyses using all the RARα FISH probes shown in Figure 1revealed signals only on the normal chromosome 17 (Figure 6B), suggesting deletion of the other allele. Work is currently in progress to exclude mutations in the remaining RARα allele. These 2 cases could also be distinguished by NPM immunofluorescence using the NA24 antibody,52 which recognizes NPM-RARα as well as NPM. In the NPM-RARα–positive patient,54diffuse nuclear staining was observed, as distinct from the nucleolar staining52 detected in the patient lacking theNPM-RARα fusion and NB4 and HL60 controls (Figure8). In both t(5;17) patients, a wild-type PML localization pattern was detected (Figure 8).
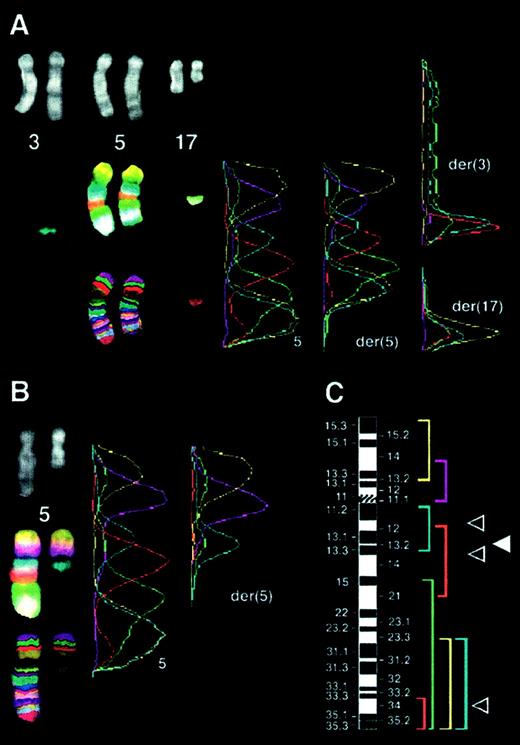
Multicolor banding of chromosome 5 in the t(5;17) cases.
(A) Case 54: insertion of the 5q13 band into band 3q26 and translocation of segment 5q34-qter to band 17q21. Figure shows the translocation of 17q to the der(5q). Left panel: top, DAPI filter; middle, compilation of captures with each filter excluding the DAPI one; bottom, multicolor banding specific for chromosome 5 material obtained after image processing. Right panel: profile of fluorescence intensities along the chromosomal axes. Peaks on der(3) and der(17) are derived from chromosome 5 material. (B) Case 55: localization of the 5q breakpoint to 5q13. Analysis as described in panel A. (C) Location and labeling of chromosome 5 region–specific partial chromosome paints; breakpoints in case 54 (▵) and in case 55 (▴) are shown.
Multicolor banding of chromosome 5 in the t(5;17) cases.
(A) Case 54: insertion of the 5q13 band into band 3q26 and translocation of segment 5q34-qter to band 17q21. Figure shows the translocation of 17q to the der(5q). Left panel: top, DAPI filter; middle, compilation of captures with each filter excluding the DAPI one; bottom, multicolor banding specific for chromosome 5 material obtained after image processing. Right panel: profile of fluorescence intensities along the chromosomal axes. Peaks on der(3) and der(17) are derived from chromosome 5 material. (B) Case 55: localization of the 5q breakpoint to 5q13. Analysis as described in panel A. (C) Location and labeling of chromosome 5 region–specific partial chromosome paints; breakpoints in case 54 (▵) and in case 55 (▴) are shown.
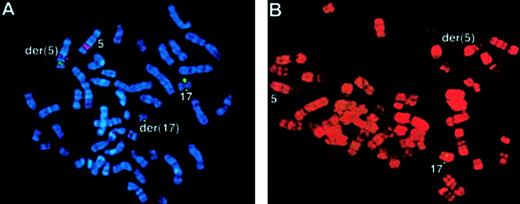
FISH using ICRF
RARα 121 cosmid probe in the t(5;17) cases. (A) Case 54: translocation of RARα sequences (green) to the der(5), identified using a chromosome 5q31 Oncor probe (red) using DAPI banding. (B) Case 55: RARα signals (green) were detected only on the normal chromosome 17. R banding using propidium iodide does not permit capture of double minute chromosomes.
FISH using ICRF
RARα 121 cosmid probe in the t(5;17) cases. (A) Case 54: translocation of RARα sequences (green) to the der(5), identified using a chromosome 5q31 Oncor probe (red) using DAPI banding. (B) Case 55: RARα signals (green) were detected only on the normal chromosome 17. R banding using propidium iodide does not permit capture of double minute chromosomes.
Molecular analysis of case 54 with t(5;17).
Left panel: RT-PCR showing NPM-RARα and RARα-NPMtranscripts. Right panel: sequence analysis of fusion transcripts, with location of cDNA fusion junction.
Molecular analysis of case 54 with t(5;17).
Left panel: RT-PCR showing NPM-RARα and RARα-NPMtranscripts. Right panel: sequence analysis of fusion transcripts, with location of cDNA fusion junction.
PML and NPM immunofluorescence (IF) in t(5;17) cases and cell-line controls.
PML immunofluorescence using the PG-M3 antibody shows a wild-type pattern (discrete nuclear dots) in HL60 and in both patients, and a microparticulate diffuse nuclear pattern in the t(15;17) NB4 cell line. NPM immunofluorescence using the NA24 antibody shows a wild-type nucleolar pattern in both cell lines and in theNPM-RARα–negative patient (case 55), and a diffuse nuclear pattern in the NPM-RARα–positive patient (case 54). Images were captured with a Leica TCS NT confocal microscope.
PML and NPM immunofluorescence (IF) in t(5;17) cases and cell-line controls.
PML immunofluorescence using the PG-M3 antibody shows a wild-type pattern (discrete nuclear dots) in HL60 and in both patients, and a microparticulate diffuse nuclear pattern in the t(15;17) NB4 cell line. NPM immunofluorescence using the NA24 antibody shows a wild-type nucleolar pattern in both cell lines and in theNPM-RARα–negative patient (case 55), and a diffuse nuclear pattern in the NPM-RARα–positive patient (case 54). Images were captured with a Leica TCS NT confocal microscope.
Morphologic APL cases apparently lacking rearrangements of RARα.
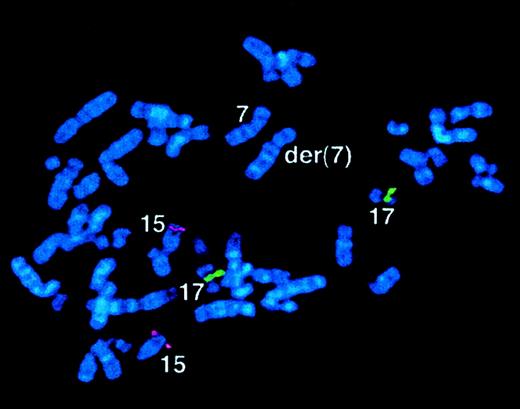
In 5 patients, FISH, Southern blot, and RT-PCR analyses did not reveal rearrangements of RARα (Table6, Figure9); in addition, PML immunofluorescence was performed in case 59, revealing a wild-type pattern. Morphologic review confirmed the diagnosis of APL as described by Sainty et al.1 Investigations are in progress to exclude mutations of RARα in these patients, although to date, no leukemias have been reported in mice expressing mutant RARα.69
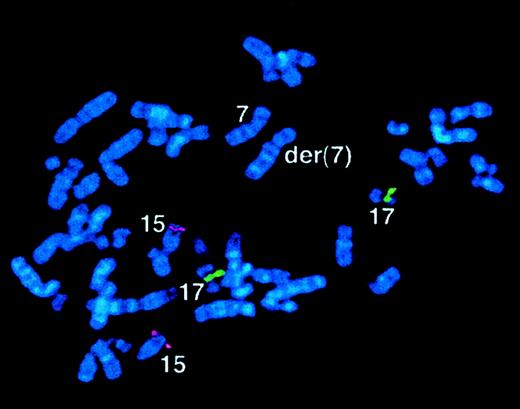
Case 56 with der(7) and lacking
RARα rearrangement. FISH using ICRFPML 15.5 (red) and RARα 121 (green) cosmid probes, demonstrating normal locations of PML andRARα sequences in the malignant clone. See text and Table6.
Case 56 with der(7) and lacking
RARα rearrangement. FISH using ICRFPML 15.5 (red) and RARα 121 (green) cosmid probes, demonstrating normal locations of PML andRARα sequences in the malignant clone. See text and Table6.
Frequency of the classic t(15;17) in patients with APL
To establish the proportion of APL patients lacking the classic t(15;17), we derived epidemiologic data from centers participating in the workshop. For the purposes of this analysis, data collection was restricted to 18 of 42 laboratories that permitted determination of the frequency of specific cytogenetic changes in a completely unselected patient group. Overall, cytogenetic analyses were performed successfully in 611 patients with newly diagnosed APL over an 8-year period, as summarized in Table 7.
Discussion
The t(15;17) is the diagnostic hallmark of APL and initially had been considered to be present in all patients with this condition.30 However, it is now clear from the present study that a sizeable minority actually lack this chromosomal aberration, with epidemiologic data from the Monza workshop indicating that the t(15;17) is not identified in 9% patients with APL after successful diagnostic cytogenetic analysis. Furthermore, this study shows that the majority of cases of morphologic APL lacking the t(15;17) are still associated with formation of thePML-RARα fusion gene, created by insertion events or more complex rearrangements. Such mechanisms occur in approximately 4% and 2% of cases of APL, respectively, and typically lead to the formation of PML-RARα at its usual location on 15q and, less commonly, at the site of the reciprocal fusion gene on 17q or alternative chromosomal locations. These findings are highly analogous to those previously reported in CML. In this condition, 90% of cases are associated with the t(9;22), leading to a rearrangement between theBCR and ABL genes. BCR/ABLrearrangements are also present in approximately half the CML patients lacking the classic t(9;22). In the majority of these patients, chromosomes 9 and 22 are of normal appearance with formation of theBCR-ABL fusion gene at its usual location on chromosome 22; more rarely, the fusion gene is located on chromosome 9 or alternative chromosomal sites, reflecting the occurrence of more complex rearrangements (reviewed by Aurich et al70). The striking similarity between the frequency of the classic translocation and complex and cryptic rearrangements involving the genes disrupted by each respective translocation in CML and APL raises the possibility that similar underlying mechanisms may be involved. This possibility is supported by a recent study documenting proximity of BCR andABL and of PML and RARα genes at specific phases of the cell cycle in hemopoietic progenitors.71
The epidemiologic survey revealed PLZF/RARα as the second most common molecular rearrangement associated with APL, accounting for approximately 0.8% of cases. Identification of this group is extremely important because of the poor response to retinoids as single-agent therapy37 and in view of recent data suggesting that these patients are also resistant to As2O3.58 However, it is clear from the present study that CR is attainable in this group with combination chemotherapy, indicating that cases of PLZF-RARα–positive APL are not necessarily associated with an adverse prognosis, as suggested previously. In addition, the present study has identified cases of APL with cryptic PLZF/RARα rearrangements, including one patient with a normal karyotype in whomPLZF-RARα was the sole fusion gene formed as a result of an insertion event. Rearrangements disrupting STAT5b,NuMA, and NPM appear to be extremely rare, with only isolated case reports in the literature.33-35,65Indeed, we detected no cases involving the former 2 fusion partners and only one case with t(5;17) expressing NPM-RARα. Whereas the 2 previously reported patients with NPM-RARα APL did not receive ATRA before relapse,33 65 our patient was treated with ATRA as part of induction therapy and is alive in first CR at 29 months.
An important aspect of the present study is that it permitted the evaluation of different techniques to establish the presence of thePML/RARα rearrangement as a means of determining the subgroup of APL patients likely to benefit from retinoids and As2O3. It is clear that long-established methods such as conventional cytogenetics are not invariably successful in this regard and must be supplemented by alternative approaches, such as FISH, RT-PCR, Southern blot analysis, or PML immunofluorescence. Nevertheless, cytogenetics should not be abandoned because it detects the t(15;17) in the majority of patients, identifies secondary cytogenetic changes, and has revealed novel translocations in APL, prompting subsequent molecular characterization of their respective breakpoint regions. In many respects, RT-PCR screening of cases of suspected APL affords a number of advantages: providing a rapid diagnostic test, distinguishing PML breakpoint patterns, and defining targets for residual disease monitoring, which has been shown to provide independent prognostic information (reviewed by Grimwade24). Indeed, identification of thePML-RARα fusion by molecular techniques in patients lacking the t(15;17) predicts a beneficial response to ATRA, and such patients share the favorable prognosis of those with the classic t(15;17).11,28 29
PML immunofluorescence techniques are even more rapid than RT-PCR and in some institutions have been incorporated into the standard diagnostic approach to patients with suspected APL.24 It is clear from this study and others29,38 that observation of a microparticulate nuclear staining pattern in leukemic blasts is specific to cases expressing the PML-RARα fusion protein and therefore is predictive of a beneficial response to ATRA and As2O3. This pattern is detected in patients with the classic t(15;17) as well as in those patients in whomPML-RARα is the sole fusion gene formed as a result of insertion events.38 Disruption of PML nuclear bodies has been proposed to play an important role in the pathogenesis ofPML-RARα–associated APL.20 However, this study and others have established that a wild-type PML nuclear staining pattern is associated with APL cases with alternative reciprocal translocations including t(5;17),65t(11;17)(q13;q21),34 and t(11;17)(q23;q21),38,58 as well as in the case in whichRARα-PML was the sole fusion gene formed as a result of an insertion event.29 This finding implies that delocalization of PML does not provide a final common pathway to all molecular subtypes of APL. APL cases associated with NPM orNuMA rearrangements appear to be sensitive to retinoids,34,36,65 whereas cases withPLZF/RARα rearrangements37 or with expression of RARα-PML alone29 fail to differentiate with retinoids as the sole therapeutic agent. This suggests that identification of a normal PML staining pattern in cases of morphologically confirmed APL should not lead to treatment with ATRA alone in the first instance; indeed, whether to use ATRA at any stage (and in any combination) in these patients would await further cytogenetic and molecular characterization or the results of in vitro ATRA differentiation assays. Interestingly, a recent study has suggested that t(11;17)-associated APL may differentiate in response to ATRA in combination with granulocyte colony-stimulating factor.46
There has been considerable interest in the potential contribution of reciprocal fusion gene products to the pathogenesis of APL, and in the course of this study, a single case of morphologically confirmed APL was identified in which RARα-PML (bcr3) appeared to be the sole fusion gene formed. The role of reciprocal fusion genes has been investigated recently using transgenic mice. Whereas expression of bcr3RARα-PML under the control of the human cathepsin G (hCG) promoter did not induce leukemia in its own right,RARα-PML significantly increased the frequency of APL among mice expressing a bcr1 PML-RARα transgene; furthermore, co-expression of both fusion transcripts was suggested to lead to a more aggressive form of the disease.27 However, it remains possible that the phenotype was influenced by the expression of nonreciprocal fusion transcripts with significant overlap of central portions of PML. In man, analysis of large clinical trials has revealed that RARα-PML is not expressed in approximately 30% of cases, including the majority of insertions, and that expression of reciprocal transcripts has no influence on disease characteristics or outcome.11 The RARα-PLZF protein has also been the focus of some attention. This protein contains 6 or 7 zinc fingers, binds DNA, may deregulate the cell cycle, and is up-regulated by ATRA, potentially contributing to leukemogenesis and ATRA resistance.12,37 Interestingly, recent studies have also suggested that RARα-PLZF can modify the leukemic phenotype of PLZF-RARα transgenic mice.72Expression of PLZF-RARα under the hCG promoter induced a myeloproliferative disorder in 100% of mice, whereas co-expression ofPLZF-RARα and RARα-PLZF led to a morphologic picture more reminiscent of APL, implying that both fusion products arising from the t(11;17) are necessary for the leukemic phenotype.72 However, in the present study, a patient with a normal karyotype was identified in whom PLZF-RARα was the sole fusion gene formed because of an insertion event, and in whom the morphologic appearances were indistinguishable from patients with the t(11;17), in which both fusion transcripts were expressed.1 Apparent differences between mouse models of APL and the disease in man could reflect the nature of the hemopoietic progenitor targeted by PML/RARα and PLZF/RARαrearrangements. Results obtained so far from transgenic mouse models imply that more than one step is required to develop APL and that reciprocal fusion genes could influence the rate of development and behavior of leukemias. If this is indeed the case in man, further understanding of these processes may provide insights into the molecular events underlying APL in patients with nonreciprocal rearrangements and also in patients lacking rearrangements ofRARα that were identified by this study. Although it is clear that the latter represent a small subgroup of APL cases, their existence suggests that this disease may arise by mechanisms distinct from the formation of aberrant retinoid receptors. Characterization of the molecular changes underlying such cases may establish whether the APL phenotype is inextricably linked to deregulation of retinoid signaling pathways and could provide further insights into the processes mediating normal myeloid differentiation.
Acknowledgments
This work is dedicated to the memory of Pr Philippe Bernard, Secretary of the Groupe Français de Cytogénétique Hématologique and of the Société Française d'Hématologie. We are very grateful to Daniel Isnardon for technical help in confocal microscopy and to Drs Jackie Cordell, Nick Cross, Pierre Fenaux, Arthur Zelent, and Prof. David Mason for helpful advice.
Other participants are listed in the appendix of the accompanying manuscript by Sainty et al.1
Support: D.G. is supported by the Leukaemia Research Fund of Great Britain; K.H. by the Imperial Cancer Research Fund; E.S. by European Community (BMH4-CT98-3745); A.B. by Fondazione M. Tettamanti, Associazione Italiana Ricerca sul Cancro (AIRC) and MURST; F.L.C. by CNR, Target Project on Biotechnology, AIRC, and MURST; and M.L.P. by the Comité des Bouches-du-Rhône de la Ligue Nationale Française contre le Cancer. This work was supported by the European Community Biomed Concerted Action “CT94-1703,” Molecular Cytogenetic Diagnosis in Haematological Malignancies, by the Flemish Government in the frame of action Kom op tegen Kanker/Vlaamse Kankerliga. This study includes research results of the Belgian program of Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister's Office, Science Policy Programming; scientific responsibility for this work is assumed by the authors.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Marina Lafage-Pochitaloff, Laboratoire de Cytogénétique Hématologique, Institut Paoli-Calmettes et INSERM U119, 232 bd Sainte Marguerite, 13009 Marseille, France; e-mail: cytogen@marseille.fnclcc.fr.