Abstract
The lineage-specific transcription factors GATA-1 and PU.1 can physically interact to inhibit each other's function, but the mechanism of repression of GATA-1 function by PU.1 has not been elucidated. Both the N terminus and the C terminus of PU.1 can physically interact with the C-terminal zinc finger of GATA-1. It is demonstrated that the PU.1 N terminus, but not the C terminus, is required for inhibiting GATA-1 function. Induced overexpression of PU.1 in K562 erythroleukemia cells blocks hemin-induced erythroid differentiation. In this system, PU.1 does not affect the expression of GATA-1 messenger RNA, protein, or nuclear localization. However, GATA-1 DNA binding decreases dramatically. By means of electrophoretic mobility shift assays with purified proteins, it is demonstrated that the N-terminal 70 amino acids of PU.1 can specifically block GATA-1 DNA binding. In addition, PU.1 had a similar effect in the G1ER cell line, in which the GATA-1 null erythroid cell line G1E has been transduced with a GATA-1–estrogen receptor fusion gene, which is directly dependent on induction of the GATA-1 fusion protein to effect erythroid maturation. Consistent with in vitro binding assays, overexpression of PU.1 blocked DNA binding of the GATA-1 fusion protein as well as GATA-1–mediated erythroid differentiation of these G1ER cells. These results demonstrate a novel mechanism by which function of a lineage-specific transcription factor is inhibited by another lineage-restricted factor through direct protein–protein interactions. These findings contribute to understanding how protein–protein interactions participate in hematopoietic differentiation and leukemogenesis.
Introduction
Transcription factors, acting in a combinatorial fashion, play a critical role in the differentiation of hematopoietic lineages.1 As the expression of lineage-specific factors overlaps in early progenitor cells, choice of lineage may be controlled in part through protein-protein interactions that lead either to functional synergy or to inhibition. There are many examples of protein–protein interactions that result in functional synergy, including GATA-1 and Friend of GATA-1 (FOG-1) in erythroid cells,2 PU.1 and PU.1 interaction partner (Pip) in B cells,3,4 and PU.1 and c-Jun in myeloid cells.5 The potential for inhibitory interactions between transcription factors is implied by the observations that overexpression of GATA-1 (or GATA-2) or PU.1 blocks myeloid or erythroid differentiation, respectively.6-12Functional repression could result either from decreased expression of the target transcription factor or from inhibition of that protein's function. The latter possibility is supported by the recent finding that PU.1 and GATA-1 physically interact to inhibit each other's function.10,11 13
GATA-1 regulates the expression of many erythroid genes, is highly expressed in erythroid cells, and is required for terminal differentiation of erythroid precursors.14,15 GATA-1 contains 2 zinc fingers. The carboxyl (C) finger is essential for DNA binding, and the amino-terminal (N) zinc finger stabilizes binding.16 Mutation of one of the cysteines in the C-finger of GATA-1 abolishes DNA binding and results in loss of transactivation function.17 GATA-1 is not appreciably expressed in myeloid cells. Enforced expression of GATA-1 in the progenitor line 416B or multipotential avian precursors blocks myeloid differentiation and induces erythroid, megakaryocytic, and eosinophilic characteristics.6,8 GATA-1 is expressed in some cases of myeloid and megakaryocytic transformation of chronic myelogenous leukemia (CML) into blast crisis but not in lymphoid blast crisis of CML.18 Taken together, these data suggest that expression of GATA-1 may inhibit myeloid differentiation and may be involved in the development of certain types of myeloid leukemia.
The ETS family member PU.1 is a myeloid and B-cell–specific transcription factor that is highly expressed in Friend-virus–induced erythroleukemia.19 Overexpression of PU.1 in long-term culture of bone marrow cells is able to stimulate proliferation but blocks differentiation of proerythroblasts.20 Similarly, overexpression of PU.1 in mouse erythroleukemia cell lines results in a block in chemically induced erythroid differentiation21 and a decrease in the expression of the erythroid marker β-globin,9 which is regulated by GATA-1.22Furthermore, PU.1 transgenic mice developed erythroleukemia.23 Taken together, these data suggest that deregulation of PU.1 increases erythroblast proliferation, blocks erythroid differentiation, and contributes to erythroleukemia.
The mechanism by which PU.1 blocks erythroid differentiation is unclear. We and others have reported that PU.1 physically interacts with GATA-1 and represses GATA-1 function in vitro and in vivo.10,11,13 We previously demonstrated that a small region (β3/β4) no larger than 12 amino acids within the PU.1 C-terminal DNA binding (ETS) domain interacts with GATA-1 (Wara-aswapati et al, unpublished data, July 1999).11,24 As a result of this interaction, GATA-1 inhibits PU.1 function by displacing the PU.1 coactivator c-Jun. It has also been reported that overexpression of PU.1 in murine erythroleukemia (MEL) cells did not affect the expression of GATA-1 messsenger RNA (mRNA) and protein but that GATA-1 DNA-binding activity decreased. Whether inhibition of GATA-1 DNA binding by PU.1 was direct or indirect was not addressed.25 In the present studies, we have further addressed the role of the N-terminal 70 amino acids of PU.1 and determined they are essential to repress GATA-1 function by blocking GATA-1 binding to DNA in vitro and in vivo. These results suggest that the mechanism of PU.1 inhibition of erythroid cell differentiation is mediated by blocking GATA-1 DNA binding.
Materials and methods
Cell culture and development of cell lines expressing PU.1
K562 cells, a multipotential hematopoietic line derived from a patient with CML, were maintained in RPMI medium supplemented with 10% fetal bovine serum (Biowhittaker, Walkersville, MD). The cells were transfected with a linearized human metallothionein promoter26 driving PU.1 expression by electroporation and selected by growth in 400 μg/mL of G418. We isolated 2 K562/PU.1 clones that induced high level of PU.1 protein after treatment with 100 μmol/L of ZnSO4. Erythroid differentiation of K562 and K562/PU.1 cells was induced with 2.5 × 10−5 mol/L hemin.27G1ER cells, a line in which erythroid differentiation of GATA-1 knockout cells can be inducibly restored by a stably transfected GATA-1–estrogen receptor fusion gene (GATA-1–ER), were maintained in Iscove modified Dulbecco medium plus 15% heat-inactivated fetal calf serum in the presence of 2 U/mL erythropoietin, 2.5% stem-cell factor–conditioned medium, and monothioglycerol (all from Biowhittaker).2 Erythroid differentiation was induced with 3.3 × 10−8 mol/L β-estradiol (Biowhittaker), and benzidine staining was performed as described.28
Northern blot analysis
Total RNA was isolated with the use of guanidinium isothiocyanate followed by cesium chloride centrifugation as described29 or guanidinium thiocyanate extraction30; 10 μg of each total RNA was denatured and separated on a 1% agarose-formaldehyde gel and then transferred to a nylon membrane. A full-length human GATA-1 complementary DNA (cDNA) or a murine βmajor-globin cDNA31 was used as a probe. Labeling of the probe with 32P-α-dCTP (deoxycytidine triphosphate) by random primed labeling and hybridization were performed as described.32
Western blot analysis
Whole-cell lysates were made by lysis of cell pellets with a modified RIPA buffer containing 50 mmol/L Tris-HCl (pH 7.4), 1% NP-40, 0.25% sodium deoxycholate, 150 mmol/L NaCL, 1 mmol/L EGTA, 1 mmol/L phenylmethylsulfonyl fluoride, and proteinase inhibitors. Nuclear protein from K562, K562/PU.1, G1ER, and G1ER/PU.1 cells were isolated as described.33 Then, 60 μg of whole-cell lysate or 30 μg of nuclear protein were fractionated on a 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred to nylon membranes. A 1:1000 dilution of rabbit anti-PU.1 polyclonal antibody (sc-352; Santa Cruz Biotechnology, Santa Cruz, CA) or rat anti–GATA-1 monoclonal antibody (sc-265, Santa Cruz Biotechnology) was used to detect protein expression. The amount of β-actin or tubulin in each lane was detected by a mouse antimouse β-actin monoclonal antibody (Sigma, St Louis, MO), or antitubulin monoclonal antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) were used as loading controls for gels with whole-cell lysates. The amount of Sp1 protein detected by goat anti-Sp1 polyclonal antibody (sc-59X7, Santa Cruz Biotechnology) was used as a loading control for gels fractionating nuclear proteins. After incubation with the first antibody, the proteins were detected with an appropriate secondary antibody conjugated with horseradish peroxidase and a chromogen substrate (ECL; Amersham, Uppsala, Sweden).
Purification of glutathione-S-transferase fusion proteins and electrophoretic mobility shift assays
Glutathione-S-transferase (GST) fusion proteins were expressed in BL21 Escherichia coli and purified as described.34 The GST fusion proteins were eluted from the glutathione agarose beads with 20 mmol/L glutathione, 50 mmol/L Tris-HCl (pH 7.5), and 150 mmol/L NaCl for 20 minutes at room temperature, followed by dialysis with 50 mmol/L Tris-Cl and 150 mmol/L NaCl overnight at 4°C. Eluted proteins were then concentrated on Microcon columns (Amicon, Neceolah, WI) according to the manufacturer's procedure. Electrophoretic mobility shift assays (EMSAs) were performed as described.35 The oligonucleotide in GATA-binding assays was GGCAACTGATAAGGATTC and was end-labeled with32P-γ-ATP (adenosine triphosphate) and polynucleotide kinase.17 The oligonucleotide for detecting octamer binding was AAGAGATTTATGCAAACGGGATGGG.36 The oligonucleotide for detecting GST-ETS binding was TCGAATA AAATCAGGAACTTG.36
PU.1 retroviral expression vector
The construction, packaging, and infection have been previously described.37 Briefly, the complete PU.1 coding region was isolated by polymerase chain reaction and inserted into the retroviral expression vector pBA8b-L1, termed pBA8b-L1.PU.1. It also contains the human placental alkaline phosphatase gene under the control of the cytomegalovirus promoter 3′ of the inserted PU.1 cDNA. Production of a stable pBA8b-L1.PU.1 amphotropic line and the generation of infectious supernatant were performed as described.37 To transduce G1ER cells, the wells of a 6-well tissue culture plate were first coated with fibronectin (Sigma), and then virus supernatant was added to allow viral particles to adhere. Cells were then added in the presence of 8 μg/mL polybrene, and the plate was centrifuged at 1000g for 3 hours at room temperature. After centrifugation, the cells were cultured in regular medium for 24 hours. The whole procedure was repeated 3 times.
Transient transfection
The protocol for transfection of CV-1 cells by the calcium phosphate precipitation method has been described.38 Lipofectamine transfections were performed according to the manufacturer's procedure (Gibco BRL, Rockville, MD). At 4 hours after transfection, cells were placed in 10% fetal bovine serum and incubated for another 40 hours, and firefly luciferase activity was measured as relative light units (RLUs). The RLUs from individual transfections were normalized by measurement of Renilla luciferase activity expressed from a cytomegalovirus-promoter–driven vector in the same samples.39 Individual transfection experiments were performed in triplicate, and the results are reported as mean firefly RLUs/Renilla (± SD).
Results
PU.1 does not affect GATA-1 mRNA, protein expression, or translocation to the nucleus
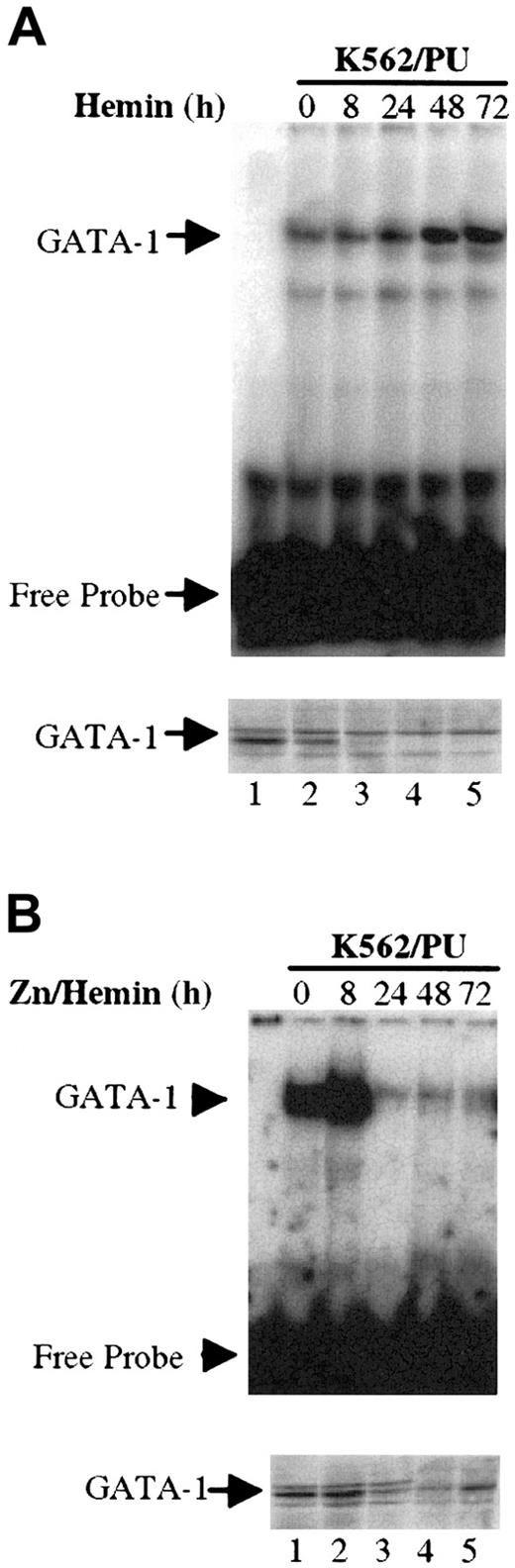
The mechanisms by which PU.1 expression blocks erythropoiesis has not been fully described.10,11,25 In the erythroid cell line MEL, PU.1 overexpression blocks dimethyl sulfoxide induction of erythroid differentiation, and the expression of β-globin decreases.9 It has been previously shown that the important regulatory elements of the β-globin gene have GATA-1 sites.22 These studies suggest that PU.1 inhibition of GATA-1 function blocks erythropoiesis. To further investigate the mechanism of PU.1 repression of GATA-1 function, we generated K562 cell lines stably transfected with the PU.1 cDNA driven by the zinc-inducible metallothionein promoter. We analyzed 2 clones (K562/mPU.1 no. 3 and K562/mPU.1 no. 7) in detail. The expression of exogenous PU.1 mRNA (data not shown) and protein (Figure1B) was induced with 100 μmol/L ZnSO4 in 8 hours. Erythroid differentiation was induced in K562 cells with 2.5 × 10−5 hemin, and the appearance of erythroid cells monitored with benzidine staining. As shown in Table1, after hemin induction, but in the absence of ZnSO4-induced PU.1 expression, we observed 35% to 46% benzidine-positive cells in K562/pC18 (a line with vector alone), K562/mPU.1 no. 3, and K562/mPU.1 no. 7 cells. When the cells were treated with hemin in the presence of ZnSO4, the percentage of benzidine-positive cells in K562/mPU.1 no. 3 and K562/mPU.1 no. 7 decreased dramatically to 6.7% and 8.9%, respectively. ZnSO4 had no significant effect on the differentiation of K562 cells transfected with the metallothionein vector alone (K562/pC18).
Effect of PU.1 on GATA-1 mRNA and protein expression in K562/PU.1 cells.
PU.1 does not inhibit GATA-1 mRNA and protein expression in K562/PU.1 cells. (A) Northern blot analysis of GATA-1 mRNA in K562/mPU.1 cells at different time points after induction of PU.1 expression with ZnSO4. The ethidium bromide–stained 18S RNA was used as a loading control as shown at the bottom of the panel. (B) Western blot analysis of PU.1 and GATA-1 protein levels at different time points after ZnSO4 induction in whole-cell lysates from K562/mPU.1 cells. β-Actin was used as a loading control. (C) Western blot analysis of PU.1 and GATA-1 in nuclear extracts from K562/mPU.1 cells after ZnSO4 induction. Sp1 was used as a loading control.
Effect of PU.1 on GATA-1 mRNA and protein expression in K562/PU.1 cells.
PU.1 does not inhibit GATA-1 mRNA and protein expression in K562/PU.1 cells. (A) Northern blot analysis of GATA-1 mRNA in K562/mPU.1 cells at different time points after induction of PU.1 expression with ZnSO4. The ethidium bromide–stained 18S RNA was used as a loading control as shown at the bottom of the panel. (B) Western blot analysis of PU.1 and GATA-1 protein levels at different time points after ZnSO4 induction in whole-cell lysates from K562/mPU.1 cells. β-Actin was used as a loading control. (C) Western blot analysis of PU.1 and GATA-1 in nuclear extracts from K562/mPU.1 cells after ZnSO4 induction. Sp1 was used as a loading control.
We next wished to determine if PU.1 decreased the expression of GATA-1 mRNA or protein in these lines. In Northern blot analysis, overexpression of PU.1 after zinc induction had no effect on the expression of GATA-1 mRNA for up to 72 hours (Figure 1A). Western analysis of whole-cell lysates at different times after ZnSO4 induction demonstrated that the PU.1 protein was induced (Figure 1B, top panel) and translocated into nuclei (Figure 1C, top panel) at 8 hours after ZnSO4 treatment. However, the level of detectable PU.1 protein remains constant in whole-cell lysates but decreases in nuclear extracts for unknown reasons. Similar results were obtained from multiple experiments. This could be due to altered reactivity of PU.1 to the antibody used for detection.40The expression of GATA-1 protein was not affected in whole-cell lysates (Figure 1B). The level of GATA-1 protein in the nuclei was assessed to discover whether PU.1 blocked GATA-1 translocation from the cytoplasm. As shown in Figure 1C, the level of GATA-1 protein in the nuclei of K562/PU.1 cells remains unchanged before and after ZnSO4induction. In summary, expression of PU.1 blocks erythroid differentiation without affecting GATA-1 mRNA, protein expression, or nuclear translocation.
The N terminus of PU.1, but not the β3/β4 region, is required for repression of GATA-1 transactivation
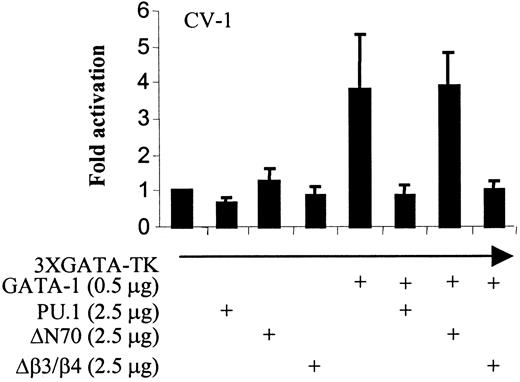
Previously, we have reported that both the N-terminal 70 amino acids and the β3/β4 domain within the C-terminal ETS domain of PU.1 interact with GATA-1, and that full-length PU.1 inhibits GATA-1 transactivation.11 To assess which domain of PU.1 contributes to the inhibition of GATA-1 function, transient transfections were performed with a plasmid consisting of multiple GATA DNA-binding sites in front of a minimal thymidine kinase promoter (3XGATA-TK) driving luciferase gene expression as a reporter. As shown in Figure 2, GATA-1 can activate the reporter gene expression about 4-fold. GATA-1 transactivation was fully repressed by cotransfection with wild-type PU.1. Deletion of the PU.1 N-terminal 70 amino acids resulted in a loss in the ability to inhibit GATA-1 function; a PU.1 polypeptide with a deletion of the β3/β4 region repressed GATA-1 function as well as wild-type PU.1. Similar results were obtained with cotransfections using PU.1 and GATA-2 expression plasmids. Therefore, the N terminus of PU.1 interacts with GATA-1 and is required for PU.1 repression of GATA-1 function. In contrast, our previous studies indicated that GATA-1 inhibits PU.1 function by directly interacting with the PU.1 β3/β4 region, thus blocking interaction with the PU.1 coactivator c-Jun.11
Effect of PU.1 expression on GATA-1 transactivation of a reporter containing GATA-1 DNA-binding sites.
PU.1 expression inhibits GATA-1 transactivation of a reporter containing GATA-1 DNA-binding sites. CV-1 cells were transiently transfected by the calcium phosphate method. The reporter construct consisted of 3 GATA DNA-binding sites in front of a minimal TK promoter driving luciferase gene expression. ΔN70 is the N-terminal 70–amino acid PU.1 deletion mutant in pcDNA3; Δβ3/β4 is the PU.1 β3/β4 region deletion mutant (amino acids 243-254) in pcDNA3. GATA-1 transactivation activity is expressed as luciferase units normalized to the cotransfected Renilla plasmid, compared with that observed with the reporter plasmid only. Luciferase activity is the mean ± SD of 4 independent transfections using 4 independent DNA preparations.
Effect of PU.1 expression on GATA-1 transactivation of a reporter containing GATA-1 DNA-binding sites.
PU.1 expression inhibits GATA-1 transactivation of a reporter containing GATA-1 DNA-binding sites. CV-1 cells were transiently transfected by the calcium phosphate method. The reporter construct consisted of 3 GATA DNA-binding sites in front of a minimal TK promoter driving luciferase gene expression. ΔN70 is the N-terminal 70–amino acid PU.1 deletion mutant in pcDNA3; Δβ3/β4 is the PU.1 β3/β4 region deletion mutant (amino acids 243-254) in pcDNA3. GATA-1 transactivation activity is expressed as luciferase units normalized to the cotransfected Renilla plasmid, compared with that observed with the reporter plasmid only. Luciferase activity is the mean ± SD of 4 independent transfections using 4 independent DNA preparations.
PU.1 blocks GATA-1 binding to DNA
Overexpression of PU.1 in the MEL erythroid cell line reduced GATA-1 DNA-binding activity in cell extracts,25 but whether this is a direct or an indirect effect was not determined. We have previously shown that PU.1 interacts with the C-finger of GATA-1,11 which serves as the DNA recognition motif for GATA-1. It is possible that PU.1 blocks GATA-1 binding to DNA owing to direct interaction with this GATA-1 C-finger. To test this hypothesis, an EMSA was performed as shown in Figure3A. Various GST-PU.1 fusion proteins were used to compete with GATA-1 for binding to an oligonucleotide containing a GATA-binding site. As assessed by SDS-PAGE, 0.1 μg and 0.5 μg of each GST-PU.1 were used for the reactions. Increasing the amount of GST-PU.1 full-length protein decreased GATA-1 DNA binding (Figure 3A, lanes 7 and 8). The PU.1 ETS domain alone did not reduce GATA-1 DNA binding (Figure 3A, lanes 10 and 11); however, it still bound to the PU.1-binding–site oligonucleotide as shown in Figure 3A, right panel (lanes 2 and 3), indicating that it is folded properly. The N-terminal 70 amino acids of PU.1 could block GATA-1 binding to DNA as effectively as the full-length PU.1 protein (Figure 3A, lanes 13 and 14). Interestingly, deletion of the N-terminal 100 amino acids and the β3/β4 region of PU.1 were not only unable to inhibit GATA-1 DNA binding, but appeared to enhance GATA-1 DNA binding and induced the formation of slower migrating complexes (Figure3A, lanes 16-17), perhaps as a result of oligomerization of GATA-1 under these conditions.43 These data suggest that PU.1 blocks GATA-1 binding to DNA by direct interaction with the GATA-1 C-finger. To exclude the possibility that PU.1 represses GATA-1 function through the GATA-1 activation domain, we performed transient transfections using a fusion protein between GATA-1 N- and C-zinc fingers (as a DNA-binding domain) and the VP16 transactivation domain (N + C–VP16) as an activator, and the 3XGATA-TK luciferase as a reporter. As shown in Figure 3B, the N + C–VP16 protein activated the GATA-1 reporter nearly 3-fold. Although this activation was modest, it was consistently observed in multiple experiments. As expected, full-length PU.1 but not the N-terminal 70–amino acid deletion of PU.1 inhibited the fusion protein transactivation function, even though the protein level of the truncated protein was higher than that of wild-type PU.1 in the transfected CV-1 cells, as shown in Figure 3B, right panel. These results demonstrate that inhibition of GATA-1 DNA binding can block transactivation.
Effect of PU.1 on GATA-1 DNA binding and the transactivation domain.
PU.1 inhibits GATA-1 DNA binding but not the transactivation domain. (A) Left panel: EMSA was performed using a32P-α-dCTP–labeled GATA-binding site17oligonucleotide as a probe. GATA-1 protein was synthesized with the use of the Promega in vitro transcription and translation kit. GST-PU.1 fusion proteins were purified with the use of glutathione agarose. PU.1: GST-PU.1 full-length fusion protein. ETS: GST-PU.1 ETS-domain fusion protein (PU.1 amino acids 171-272). ΔN70: GST-PU.1 N-terminal 70–amino acid fusion protein. Δ2: deletion mutant of both the N-terminal 70–amino acid and β3/β4 (amino acids 243 to 254) region of the PU.1-GST fusion protein. GST: GST alone. Right panel: EMSA was performed by means of a 32P-[γ]ATP–labeled PU.1 binding site from the murine PU.1 promoter as a probe.35The same amount of GST-ETS or GST protein was used for this experiment as was in the left panel. (B) Left panel: 0.2 μg of a luciferase reporter under the control of a promoter consisting of 3 GATA sites proximal to a minimal thymidine kinase (TK) promoter was cotransfected with 40 ng of N + C–VP16 with or without 0.25 μg of PU.1 or PU.1 ΔN70 with the use of the lipofectamine method in CV-1 cells. N + C–VP16 is the fusion protein consisting of the VP16 activation domain fused to the GATA-1 N + C finger region (DNA-binding domain). ΔN70: PU.1 with a deletion of the N-terminal 70 amino acids. Right panel: Whole-cell lysates from transfected CV-1 cells were analyzed by Western blots and detected with anti-PU.1 antibody. Lanes 1 and 2 are nuclear extracts from a PU.1−/− cell line transfected with PU.1 and the nontransfected parental PU.1−/− cell line41 as positive and negative controls, respectively. Lanes 3 and 4 contain whole-cell lysates from CV-1 cells transfected with wild-type and ΔN70, respectively. The asterisk indicates the position of an N-terminal degradation product of PU.1, which has been previously described.42
Effect of PU.1 on GATA-1 DNA binding and the transactivation domain.
PU.1 inhibits GATA-1 DNA binding but not the transactivation domain. (A) Left panel: EMSA was performed using a32P-α-dCTP–labeled GATA-binding site17oligonucleotide as a probe. GATA-1 protein was synthesized with the use of the Promega in vitro transcription and translation kit. GST-PU.1 fusion proteins were purified with the use of glutathione agarose. PU.1: GST-PU.1 full-length fusion protein. ETS: GST-PU.1 ETS-domain fusion protein (PU.1 amino acids 171-272). ΔN70: GST-PU.1 N-terminal 70–amino acid fusion protein. Δ2: deletion mutant of both the N-terminal 70–amino acid and β3/β4 (amino acids 243 to 254) region of the PU.1-GST fusion protein. GST: GST alone. Right panel: EMSA was performed by means of a 32P-[γ]ATP–labeled PU.1 binding site from the murine PU.1 promoter as a probe.35The same amount of GST-ETS or GST protein was used for this experiment as was in the left panel. (B) Left panel: 0.2 μg of a luciferase reporter under the control of a promoter consisting of 3 GATA sites proximal to a minimal thymidine kinase (TK) promoter was cotransfected with 40 ng of N + C–VP16 with or without 0.25 μg of PU.1 or PU.1 ΔN70 with the use of the lipofectamine method in CV-1 cells. N + C–VP16 is the fusion protein consisting of the VP16 activation domain fused to the GATA-1 N + C finger region (DNA-binding domain). ΔN70: PU.1 with a deletion of the N-terminal 70 amino acids. Right panel: Whole-cell lysates from transfected CV-1 cells were analyzed by Western blots and detected with anti-PU.1 antibody. Lanes 1 and 2 are nuclear extracts from a PU.1−/− cell line transfected with PU.1 and the nontransfected parental PU.1−/− cell line41 as positive and negative controls, respectively. Lanes 3 and 4 contain whole-cell lysates from CV-1 cells transfected with wild-type and ΔN70, respectively. The asterisk indicates the position of an N-terminal degradation product of PU.1, which has been previously described.42
We next investigated whether PU.1 blocks GATA-1 binding to DNA in K562 cells. Nuclear extracts from K562/PU.1 cells at different time points after induction with hemin alone or hemin plus ZnSO4 were used for gel shift assays. As shown in Figure4A, when K562/PU.1 cells were induced with hemin alone, the expression of GATA-1 protein did not increase, but GATA-1 DNA-binding activity increased 3.6-fold 48 hours after induction. When PU.1 expression was induced with ZnS04 at the same time as hemin, the expression of GATA-1 protein remained the same at all distinct time points, but the DNA-binding activity of GATA-1 decreased dramatically (Figure 4B).
Effect of overexpression of PU.1 on GATA-1 DNA-binding activity in K562 cells during erythroid differentiation.
Overexpression of PU.1 decreases GATA-1 DNA binding activity in K562 cells during erythroid differentiation. (A) Nuclear extracts were harvested from K562/PU cells at different time points after hemin induction (in the absence of ZnSO4 and PU.1 induction). As shown on the top panel, 1 μg of each preparation was used for EMSAs. The probe is the same as in Figure 3A. As shown in the bottom panel, 20 μg of each extract was used for Western blot analysis to detect GATA-1 protein expression. (B) Nuclear extracts were harvested from K562/PU cells at different time points after induction with both ZnSO4 and hemin. The amounts of protein used, probe, and antibody are identical to those described in Figure 4A. The bottom panel demonstrates the amount of GATA-1 protein expression.
Effect of overexpression of PU.1 on GATA-1 DNA-binding activity in K562 cells during erythroid differentiation.
Overexpression of PU.1 decreases GATA-1 DNA binding activity in K562 cells during erythroid differentiation. (A) Nuclear extracts were harvested from K562/PU cells at different time points after hemin induction (in the absence of ZnSO4 and PU.1 induction). As shown on the top panel, 1 μg of each preparation was used for EMSAs. The probe is the same as in Figure 3A. As shown in the bottom panel, 20 μg of each extract was used for Western blot analysis to detect GATA-1 protein expression. (B) Nuclear extracts were harvested from K562/PU cells at different time points after induction with both ZnSO4 and hemin. The amounts of protein used, probe, and antibody are identical to those described in Figure 4A. The bottom panel demonstrates the amount of GATA-1 protein expression.
PU.1 blocks GATA-1–dependent erythroid cell differentiation by inhibiting GATA-1 DNA binding
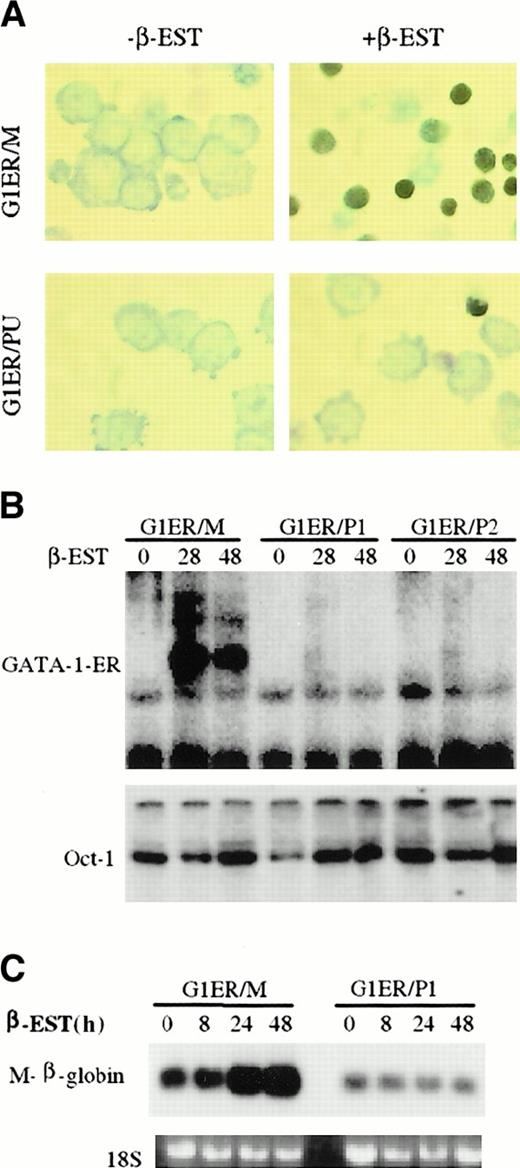
To further analyze if PU.1 inhibition of GATA-1 DNA binding blocks erythroid cell differentiation in vivo, we used G1ER cells. These cells were derived from GATA-1–deficient embryonic stem cells transfected with a GATA-1–ER fusion protein and differentiate to mature red blood cells in response to exogenous β-estradiol.2 44 G1ER cells were transduced with a viral construct harboring the PU.1 cDNA. We analyzed 2 G1ER/PU.1 lines derived from separate clones. Mock transduced G1ER cells (G1ER/M) were used as a control. As shown in Figure 5A and Table 1, when the G1ER/M cells were treated with β-estradiol, the percentage of benzidine-positive cells increased in 48 hours from below 0.5% to 52%. In contrast, only 5.6 to 6.8% benzidine-positive cells were observed in G1ER/PU lines (which express high levels of PU.1) after β-estradiol stimulation. To obtain another marker of erythroid differentiation, we measured the expression of erythroid-specific βmajor-globin mRNA expression in G1ER/M and G1ER/PU cells after β-estradiol induction (Figure 5C). The expression of βmajor-globin mRNA increased markedly in G1ER cells but remained at the same low levels in G1ER/PU cells after β-estradiol induction. Therefore, expression of PU.1 inhibited the ability of GATA-1 to induce erythroid differentiation of these cells.
Effect of overexpression of PU.1 on GATA-1 DNA binding and erythroid differentiation in an erythroid cell line dependent on GATA-1 for erythroid differentiation.
Overexpression of PU.1 blocks GATA-1 DNA binding and erythroid differentiation in an erythroid cell line dependent on GATA-1 for erythroid differentiation (A) G1ER cells were mock-infected (G1ER/M, top 2 pictures) or infected with a PU.1 retrovirus (G1ER/PU, bottom 2 pictures). Cells were stained with benzidine before (left side, EST) or after (right side, +β-EST) treatment with β-estradiol. (B) EMSA of GATA-1 DNA binding. Nuclear extracts were made from G1ER, G1ER/PU-1, and G1ER/PU-2 cells 0, 28, and 48 hours after β-estradiol induction as shown in the upper panel. The GATA-1 probe is the same as that used in Figure 3A. Oct-1 DNA binding was used as a control as shown on the lower panel. G1ER/PU-1, and G1ER/PU-2 are 2 individual clones of PU.1-virus–transduced G1ER cells. (C) Total RNA was prepared from G1ER/M and G1ER/PU cells at 0, 8, 24, and 48 hours after stimulation with β-estradiol. Then, 10 μg of each sample was loaded onto the gel and hybridized to a murine major-globin probe.31
Effect of overexpression of PU.1 on GATA-1 DNA binding and erythroid differentiation in an erythroid cell line dependent on GATA-1 for erythroid differentiation.
Overexpression of PU.1 blocks GATA-1 DNA binding and erythroid differentiation in an erythroid cell line dependent on GATA-1 for erythroid differentiation (A) G1ER cells were mock-infected (G1ER/M, top 2 pictures) or infected with a PU.1 retrovirus (G1ER/PU, bottom 2 pictures). Cells were stained with benzidine before (left side, EST) or after (right side, +β-EST) treatment with β-estradiol. (B) EMSA of GATA-1 DNA binding. Nuclear extracts were made from G1ER, G1ER/PU-1, and G1ER/PU-2 cells 0, 28, and 48 hours after β-estradiol induction as shown in the upper panel. The GATA-1 probe is the same as that used in Figure 3A. Oct-1 DNA binding was used as a control as shown on the lower panel. G1ER/PU-1, and G1ER/PU-2 are 2 individual clones of PU.1-virus–transduced G1ER cells. (C) Total RNA was prepared from G1ER/M and G1ER/PU cells at 0, 8, 24, and 48 hours after stimulation with β-estradiol. Then, 10 μg of each sample was loaded onto the gel and hybridized to a murine major-globin probe.31
To investigate whether PU.1 blocked GATA-1–ER binding to DNA in G1ER cells, nuclear proteins were isolated from G1ER/M and G1ER/PU cells at different times after β-estradiol stimulation. EMSA was performed with the use of a GATA consensus binding site oligonucleotide. As shown in Figure 5B, GATA-1–ER DNA binding was observed in G1ER/M cells 24 hours and 48 hours after β-estradiol treatment but not in either of 2 independent of G1ER/PU clones. Using an octamer binding site as a positive control, we found similar amounts of Oct-1/DNA complexes present in the different cells at all time points. These data suggest that PU.1 inhibition of GATA-1 DNA binding contributes to the observed block in erythroid differentiation.
Discussion
We and others have recently reported that PU.1 physically interacts with GATA-1.10,11 13 These physical interactions result in repression in which PU.1 and GATA-1 inhibit each other's function. Previously, we demonstrated that GATA-1 interacts with the β3/β4 region of the PU.1 ETS domain and inhibits PU.1 transactivation by blocking the PU.1 coactivator c-Jun from binding to this same region (Figure 6). However, the mechanism of how PU.1 inhibits GATA-1 function was unclear. In this paper, we have demonstrated that the mechanism of PU.1 inhibition of GATA-1 function differs from that of GATA-1 inhibition of PU.1 function and involves a different region of the PU.1 protein.
How PU.1 and GATA-1 inhibit each other's function.
PU.1 and GATA-1 inhibit each other's function through protein–protein interactions by different mechanisms. The top panel shows that GATA-1 inhibits PU.1 transactivation of PU.1 target genes (such as CD11b46) by blocking the binding of the PU.1 coactivator c-Jun to the β3/β4 region of PU.1. The bottom panel shows that the N-terminal region of PU.1 blocks GATA-1 function by binding to the GATA-1 C-finger and inhibiting GATA-1 binding to DNA.
How PU.1 and GATA-1 inhibit each other's function.
PU.1 and GATA-1 inhibit each other's function through protein–protein interactions by different mechanisms. The top panel shows that GATA-1 inhibits PU.1 transactivation of PU.1 target genes (such as CD11b46) by blocking the binding of the PU.1 coactivator c-Jun to the β3/β4 region of PU.1. The bottom panel shows that the N-terminal region of PU.1 blocks GATA-1 function by binding to the GATA-1 C-finger and inhibiting GATA-1 binding to DNA.
GATA-1 contains 2 zinc fingers. The N-terminal zinc finger mediates interaction with the GATA-1 coactivator FOG, which is essential for erythroid development.2 The C-terminal zinc finger not only serves as a DNA-recognition motif,17 but also contributes to self-association43 and protein–protein interactions with the coactivator CBP.45 This functionally important domain interacts with 2 distinct regions of PU.1, a transcription factor that is functionally divergent from GATA-1. PU.1 has been reported to decrease GATA-1 binding to DNA during the induction of apoptosis by overexpression of PU.1 in MEL cells, an erythroid cell line that expresses GATA-1, but the mechanism of how PU.1 inhibited GATA-1 DNA binding was not elucidated.25
GATA-1 inhibits PU.1 function by inhibiting binding of the PU.1 coactivator c-Jun.5 The GATA-1 N-finger, not the C-finger, interacts with the coactivator, FOG.2 Our previous results indicated that both domains of PU.1 interacted with the GATA-1 C-finger, but not the FOG-binding N-finger.11 These results, combined with the studies presented here, suggest that blocking DNA binding rather than inhibition of a coactivator mediates the repression of GATA-1 by PU.1, whereas the opposite is true for the mechanism of inhibition of PU.1 by GATA-1 (Figure 6).11
We hypothesized that the mechanism of PU.1 repression of GATA-1 function was from direct interaction with the C-finger of GATA-1, which inhibits GATA-1 DNA binding. Our hypothesis was supported by the results from our EMSA studies, in which a PU.1 polypeptide containing the first 70 amino acids inhibited GATA-1 DNA binding, and a fragment containing the C-terminal PU.1 ETS domain had no effect (Figure 3A). Interestingly, a PU.1 mutant peptide with deletions of both regions of PU.1 interacting with the GATA-1 C-finger demonstrated increased DNA binding as well as more slowly migrating complexes (Figure 3A). These results imply that the interaction of the PU.1 β3/β4 region might not inhibit the ability of the GATA-1 C-finger to bind to DNA, it might inhibit self-association of GATA-1 mediated by this domain.43 For gel shift assay, GATA-1 protein was synthesized in a reticulocyte lysate derived from erythroid cells. We cannot eliminate the possibility that this lysate contains a third protein that mediates the interaction between PU.1 and GATA-1, although PU.1 and GATA-1 physically interact in vitro and in vivo.10,11 13 While our studies indicate that each domain of PU.1 can interact with GATA-1 independently, the presence of the slower migrating forms when both GATA-1 interacting domains are deleted (Figure 3A) suggests that both regions from a single PU.1 molecule can potentially interact with GATA-1 simultaneously. Further studies using mutants of GATA-1 that bind to either the N-terminal or the C-terminal region of PU.1 will be required to answer this question.
Further evidence supporting the role of the PU.1 N terminus blocking GATA-1 DNA binding was the ability of full-length PU.1, but not a mutant that lacked the N terminus but retained the β3/β4 region, to inhibit the function of a fusion protein containing the GATA-1 DNA-binding domain and an activation domain from the VP16 protein (Figure 3B). Although both regions of PU.1 interact with GATA-1, our studies indicate that it is the N-terminal interaction, and not binding of the PU.1 β3/β4 domain, that blocks GATA-1 DNA binding both in in vitro DNA-binding assays and in stably transfected cell lines. Consistent with our findings, Rekhtman et al10 noted that a PU.1 mutant containing a deletion of amino acids 33 through 70 lost the ability to block MEL cell differentiation. Since it remains possible that GATA-1 binds to the N-terminal 33 amino acids retained in this mutant, the true in vivo relevance of our findings remain to be determined. The same N-terminal region of PU.1 binds to the retinoblastoma protein (RB) and the transcription factor TFIID,47 and therefore it is possible that the inhibition of GATA-1 DNA binding is an indirect effect mediated by one or both of these proteins. Given the ubiquitous expression of RB and TFIID, and the direct physical interaction between the N terminus of PU.1 and GATA-1,11 we believe that the inhibition of GATA-1 is likely to be directly mediated by PU.1.
K562 and G1ER cells were used as models to investigate whether PU.1 blocks GATA-1 DNA binding in erythroid cells. PU.1 blocks GATA-1 DNA binding in both cell lines and blocks erythroid differentiation as shown in Figures 4B and 5. It is interesting that PU.1 inhibits GATA-1 DNA binding in K562 cells only when the cells are induced with hemin. PU.1 does not block GATA-1 DNA binding when PU.1 expression is induced in the absence of erythroid differentiation induction in K562/PU.1 cells (data not shown). The same result was reported in MEL cells.25 These data suggest that during erythroid differentiation, GATA-1 protein might undergo modifications, such as phosphorylation or acetylation, that affect the ability of PU.1 to block DNA binding. Both phosphorylation48 and acetylation49 have been reported to increase GATA-1 DNA-binding activity. PU.1 might only inhibit phosphorylated and/or acetylated GATA-1 binding to DNA.
In summary, the N terminus of PU.1, and not the β3/β4 domain located in the PU.1 C-terminal region, is required for inhibition of GATA-1 function, even though both regions interact physically with GATA-1. The mechanism by which PU.1 represses GATA-1 function is due to an interaction with the C-finger of GATA-1, which results in inhibition of GATA-1 DNA binding, but not with the N-finger, which binds the GATA-1 coactivator FOG. This differs from the mechanism by which GATA-1 represses PU.1 function, in which GATA-1 interaction with PU.1 blocks the ability of the PU.1 coactivator c-Jun to bind to the same small region that GATA-1 binds but does not inhibit PU.1 DNA binding.11
What are the consequences of these interactions? These repressive interactions appear to be required not only for induction of differentiation in normal myelopoiesis and erythropoiesis, but also during the process of hematopoietic lineage commitment. Such interactions are consistent with a model in which hematopoietic stem cells express simultaneously low levels of “lineage-specific” transcription factors, such as GATA-1 and PU.1, and lineage commitment is characterized by activation of one transcription factor pathway and inhibition of the other.12,50 This model is consistent with studies demonstrating that overexpression of PU.1 contributes to the development of murine erythroleukemia mediated by the Friend virus,19 and blocks GATA-1–mediated erythroid differentiation. Similarly, overexpression of GATA-1 inhibits myeloid differentiation, and this requires only the C-terminal finger.7,8,13 Of relevance to human disease is the finding that expression of GATA-1 may contribute to18 or exacerbate51 the block in myeloid differentiation found in acute myeloid leukemia. Understanding the mechanisms involved in interactions between factors such as PU.1 and GATA-1 not only will contribute to our understanding of normal hematopoiesis, but also could lead to novel therapeutic strategies.
Acknowledgments
We thank Claus Nerlov for sharing ideas and results prior to publication, Margaret Baron and Tim Ley for murine globin probes, Mary Singleton for assistance with preparation of the manuscript, and other members of the Tenen laboratory for useful discussions.
Supported by National Institutes of Health (NIH) grants CA41456 (D.G.T.) and DK659381 (B.E.T.) and additional grants from NIH (S.H.O.). S.H.O. is an Investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Daniel G. Tenen, Harvard Institute of Medicine, Rm 954, 77 Ave Louis Pasteur, Boston, MA 02115; e-mail:dtenen@caregroup.harvard.edu.


![Fig. 3. Effect of PU.1 on GATA-1 DNA binding and the transactivation domain. / PU.1 inhibits GATA-1 DNA binding but not the transactivation domain. (A) Left panel: EMSA was performed using a32P-α-dCTP–labeled GATA-binding site17oligonucleotide as a probe. GATA-1 protein was synthesized with the use of the Promega in vitro transcription and translation kit. GST-PU.1 fusion proteins were purified with the use of glutathione agarose. PU.1: GST-PU.1 full-length fusion protein. ETS: GST-PU.1 ETS-domain fusion protein (PU.1 amino acids 171-272). ΔN70: GST-PU.1 N-terminal 70–amino acid fusion protein. Δ2: deletion mutant of both the N-terminal 70–amino acid and β3/β4 (amino acids 243 to 254) region of the PU.1-GST fusion protein. GST: GST alone. Right panel: EMSA was performed by means of a 32P-[γ]ATP–labeled PU.1 binding site from the murine PU.1 promoter as a probe.35The same amount of GST-ETS or GST protein was used for this experiment as was in the left panel. (B) Left panel: 0.2 μg of a luciferase reporter under the control of a promoter consisting of 3 GATA sites proximal to a minimal thymidine kinase (TK) promoter was cotransfected with 40 ng of N + C–VP16 with or without 0.25 μg of PU.1 or PU.1 ΔN70 with the use of the lipofectamine method in CV-1 cells. N + C–VP16 is the fusion protein consisting of the VP16 activation domain fused to the GATA-1 N + C finger region (DNA-binding domain). ΔN70: PU.1 with a deletion of the N-terminal 70 amino acids. Right panel: Whole-cell lysates from transfected CV-1 cells were analyzed by Western blots and detected with anti-PU.1 antibody. Lanes 1 and 2 are nuclear extracts from a PU.1−/− cell line transfected with PU.1 and the nontransfected parental PU.1−/− cell line41 as positive and negative controls, respectively. Lanes 3 and 4 contain whole-cell lysates from CV-1 cells transfected with wild-type and ΔN70, respectively. The asterisk indicates the position of an N-terminal degradation product of PU.1, which has been previously described.42](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/96/8/10.1182_blood.v96.8.2641/5/m_h82000278003.jpeg?Expires=1767962553&Signature=38qY78hvPx1IVAceS0BZxgviZgWOt-iQJNXdcn6DWeWSaJG1tOIRSmHNIBaVM4bB6Sc4UKDannpqcyXmxcCSfAaYwhsw74RKaMj9EDv5yV1FIWGlaImamCgw0Bobm8OXMJtPsSdb8v90z8~BAthB8NTJadP0sGfqMXRetaCAk1laNZkXzQyHTjOnIJejBgKrPp4vcYJHBRud8WDgtS1Uj518RkNHTu~q10FClwZR~A1F6M3GdqpmgEQe-lMtWSaa5hH-Ft~oKvLysWlvz57AzyfbFuko0ASerLrXrJ3p~Gu0wwznoBCdpuH7n~sEMezCbr5W3gCVCMtPgf2H6zhyQg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)